Abstract
Podophyllotoxin, a well-known naturally occurring aryltetralin lignan occurs in few plant species that is used as a precursor for the chemical synthesis of the anticancer drugs like etoposide, teniposide and etopophose phosphate. The availiability of this lignan is becoming increasingly limited because of the scarce occurance of its natural sources and also because synthetic approaches for its production are still commercially unacceptable. This paper reports first time the production of podophyllotoxin by an endophytic fungus Fusarium oxysporum isolated from the medicinal plant Juniperus recurva. Further confirmation and quantification of podophyllotoxin was performed by HPLC, LC-MS, and LC-MS/MS.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
In recent years there has been renewed interest in natural medicines that are obtained from plants and microbial sources. With its varied climatic zones, India has a rich diversity of medicinal herbs. The forest harbours a large number of plant species, but over-exploitation has been responsible for the rapid loss of medicinal plant wealth, such that many valuable medicinal plants are under the threat of extinction.
One such plant that is already marked in red list of threatened species by International union for conservation of Nature and Natural resources is Juniperus recurva. In India it grows in Assam, Himachal-Pradesh, Jammu and Kashmir, Sikkim and Uttar-Pradesh. Juniperus recurva (Drooping juniper) is a juniper native to the Himalaya, from northern Pakistan east to western Yunnan in southwestern china. Juniperus belongs to family cupressaceae and it grows at an altitude of approximately 3,000–4,000 m. Besides, essential oils Juniperus is also source of podophyllotoxin. Podophyllotoxin, a bioactive lignan is a valuable natural product as a precursor to three anticancer drugs etoposide, tenopside and etopophose phosphate (Schacter 1996). Podophyllotoxin has also been reported in other plant genera, such as Linum, Juniperus, Hyptis, Teuricum, Nepata, Dysosma, Jeffersonia, Thymus and Thuja, (Kupchan et al. 1965; San Feliciano et al. 1989a, b; Broomhead and Dewick 1990a, b; Yu et al. 1991; Kuhnt et al. 1994; Konuklugil 1996a, b; Muranaka et al. 1998; Bedir et al. 2001). Literature survey on podophyllotoxin revealed that the molecule occupies an important position among the plant based anti-cancer drugs, (Imbert 1998; Utsugi et al. 1996; Subrahmanyam et al. 1998; Schacter 1996; Farkya et al. 2004; Germaine et al. 2004; Kupchan et al. 1965; San Feliciano et al. 1989a, b; Broomhead and Dewick 1990a, b; Yu et al. 1991; Kuhnt et al. 1994; Konuklugil 1996a, b; Muranaka et al. 1998; Bedir et al. 2001). There are several constraints in meeting the demand of bioactive lignan because (i) its chemical synthesis is not commercially feasible due to the presence of four chiral centres along with a γ lactone and high degree of oxygenation (Dayamanti and Lawn 1998; Berkovitz et al. 2000) and (ii) difficulties in approaching towards good yield through biotechnological aspects (Berlin et al. 1988; Empt et al. 2001; Giri and Narasu 2000; Peterson and Alferman 2001).
In search for an alternative source for the production of aryl tetralin lignan, associated endophytes from this plant have been isolated and studied. Recently podophyllotoxin has been reported to be produced by endophytes from Podophyllum hexandrum (Puri et al. 2005) and Podophyllum peltatum (Eyberger et al. 2006).
In the present study, we report the isolation of endophytic fungal strain Fusarium oxysporum from Juniperus recurva growing in the Kashmir region of India, which produces podophyllotoxin. This study deals with the standardization of growth parameters of the isolated fungus and the chemo-profiling of its secondary metabolite using chromatographic and spectroscopic methods. Spectral data, obtained using HPLC, LC-MS and LC-MS/MS of the secondary metabolite was found to be identical to that of the authentic podophyllotoxin.
Material and methods
Collection of plant material
After plant selection, disease free parts of the plant i.e. stem, root, leaves were cut with the help of sterile scalpel and placed in sterile plastic bags to store the material at 4°C until isolation procedure was started.
Isolation of endophytic organism
Plant material was collected from fully matured Juniperus recurva Buch.-Ham. Ex D.Don var. recurva, growing at an altitude of 2,200 m in Gulmarg region of South Kashmir. Its endophyte was isolated using a modified method described by Schulz et al. (Arnold et al. 2000). The material was thoroughly washed using distilled water and followed by 70% ethanol for 1–2 min and 5% sodium hypochlorite for 5 min to accomplish surface sterilization. Plant material was subsequently rinsed in sterile demineralized water. Small pieces of inner tissues and needles were placed on aqueous agar (distilled water, 1.5% agar-agar) supplemented with antibiotic streptomycin (3 mg/100 ml) in petriplates (Tarsons, Kolkata, India) and incubated at 28 ± 2°C until the fungal growth was initiated. The tips of the fungal hyphae were then removed from aqueous agar and placed on mycological medium i.e. potato dextrose agar (diced potatoes, 300 g/l; dextrose, 20 g/l; agar, 20 g/l) or sabouraud agar (dextrose 40 g/l; peptone, 10 g/l; agar, 20 g/l). The pure culture obtained were transferred to a number of solid and liquid media that support fungal growth viz potato dextrose agar (diced potatoes, 300 g/l; dextrose, 20 g/l; agar, 20 g/l) malt extract agar (malt extract, 30 g/l; peptone, 5 g/l; agar, 20 g/l) or sabouraud agar (dextrose 40 g/l; peptone, 10 g/l; agar20 g/l) PYD (peptone, 2 g/l; yeast extract, 2 g/l; dextrose, 5 g/l; agar, 20 g/l). Similar procedure, but without surface sterilization, was used as negative control to check for contaminated fungi. Each isolated strain of fungi was grown in liquid sabouraud medium and screened for podophyllotoxin production.
Identification of endophyte
Microscopic slides were prepared, stained using lactophenol cotton blue (Vainio et al. 1998) and were examined under light microscope (Olympus, USA). The total genomic fungal DNA was extracted by CTAB method (Cappiccino and Sherman 1996). For DNA extraction, JRE1 was grown in 100 ml of Sabouraud Dextrose broth at 28°C with constant shaking for three days. About 100 mg of mycelial biomass was taken, washed with sterile Tris-EDTA buffer, followed by the addition of 6 ml of CTAB extraction buffer and 60 μl of β-mercaptoethanol. After vortexing, the contents were incubated at 65°C for 45 min and finally cooled to the room temperature. This was followed by extraction with equal volume of chloroform and centrifugation at 10,000g for 10 min. Equal volume of isopropanol was added to the supernatant and mixed gently. The DNA pellet was washed with ice cold 70% (v/v) ethanol and vacuum dried and dissolved in 100 μl of TE (pH 8.0). The endophytic fungus was identified by amplifying 18S, 5.8S and 28S ribosomal region of isolated DNA using primers 5′ TCCGTAGGTGAACCTGCGG 3′ and 5′ TCCTCCGCTTATTGATATGC 3′. The amplified products were purified using Microcon columns (Millipore, USA), and sequenced using ABI Prism 310 genetic analyzer (ABI, USA) as per the manufacturer’s instructions. The DNA sequence of 497 bases thus obtained was checked for its homology by BLASTN program (Altschul et al. 1997). The ribosomal gene database (http://www.rdp.cme.msu.edu) and (http://www.ncbi.nim.nih.gov) were accessed and sequence alignment was used as an underlying basis to identify the fungus. The DNA sequence has been submitted to the Gene Bank under the accession no EF591767. The fungal strain JRE1 has been deposited to Microbial Type Culture Collection (MTCC) at Institute of Microbial Technology, Chandigarh, India (accession no. yet to be assigned).
Growth of organism
The endophytic fungus (JRE1) was grown in sabouraud broth consisting of dextrose, 40 g/l; peptone, 10 g/l. Agar blocks containing fungal mycelium were used as inoculums. The endophyte was grown in 500 ml Erlenmeyer flask containing 100 ml liquid broth (pH 5.6) for a period of 7 days at 28 ± 2°C at 220 rpm on an incubatory shaker (New brunswicks, USA). The samples were collected at 24 h intervals for 7 days. Mycelia and broth were separated by filteration. The experiments were performed in triplicates and were repeated five times.
Extraction and podophyllotoxin measurements
Mycelia were thoroughly washed with sterile distilled water and homogenized in cell disintegrator. Cell homogenates were extracted four times with chloroform: methanol (4:1 v/v). Solvent was stripped off in a rotary evaporator leaving behind the organic residue. The identity of podophyllotoxin was confirmed by TLC, HPLC, LC-MS, LC-MS/MS.
Thin layer chromatographic analysis
TLC analysis was carried out using silica gel plates (Merck 0.25 mm) of 1 mm (20 × 20 cm) size, run in a (25:1 v/v) chloroform: methanol solvent system. The spots were detected under UV (254 nm) light. Standard podophyllotoxin was also run along with the samples. Podophyllotoxin exhibited Rf values of 0.2.
Experimental solvents
All the solvents used in the study were of HPLC grade. Methanol (Rankem) was purchased from Ranbaxy Chemicals Ltd. (India) and water was prepared with a Milli-Q water purification system. Standard podophyllotoxin was purchased from Sigma Aldrich-Bangalore (India).
Stock solution of (1 mg/ml) reference compound was prepared in HPLC grade methanol and stored in a refrigerator at 4°C. From the stock solution, working solutions for podophyllotoxin was prepared as desired.
Instruments
The HPLC system consisted of an Agilent series 1100 instrument equipped with a binary pump, an autosampler, an automatic electronic degasser, an automatic thermostatic column oven, a diode array detector, and a computer with Chemstation software for data analysis. HPLC was used in combination with a Quadrupole Ion Trap Mass Spectrometer (Esquire 3000) from Bruker, Bremen (Germany) equipped with an Atmospheric Pressure Ionization (API) electrospray interface. High purity nitrogen from a nitrogen generator was used as a carrier gas.
High Performance Liquid chromatographic analysis
HPLC separations were achieved using RP-18, Merck (4.6 × 250 mm) 5 μm column. The mobile phase was set as methanol: water, 65:35 and separation was carried out at a flow rate of 0.6 ml/min over a period of 60 min and then set at 35:65 up to 70 min. The compounds were detected using a UV detector at λ max of 290 nm.
LC-MS analysis
Good results were obtained in the positive mode ES-MS. The compound exhibited identical positive ion mass spectra at m/z 437 [M + Na] + along with some of the diagnostic fragments attributable to the parent compound.
Result and discussion
Isolation of endophyte
Juniperus recurva was chosen as a source plant for isolation of endophyte. The plant parts were cut into pieces; surface sterilized and inner tissues were placed on aqueous agar. The emerging fungal hyphae on day 7 were transferred to Sabouraud agar medium plates. As a result, seven pure strains of endophytic fungi were obtained. After screening these strains for the production of podophyllotoxin only one was found positive. This was finally selected and coded as JRE1.
Identification of endophyte
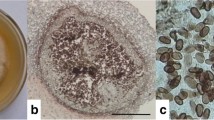
The endophytic fungus JRE1 was investigated on the basis of morphological characteristics and fungal ribotyping. The endophyte showed white cottony growth when mycelia were young and later appeared coloured. Microscopic view of endophytic hyphae was septate and hyaline. The alignment of rDNA sequence obtained from JRE1 showed 98% homology with the corresponding gene sequences of F. oxysporum. The taxonomical position has been illustrated in Fig. 1.
Growth kinetics, isolation and identification of podophyllotoxin
Shake flask experiment with the fungal culture was conducted in order to standardize culture conditions for optimum growth and production of podophyllotoxin. The fungal growth period was observed up to 9 days and the highest mycelial biomass was observed on day 7 (Fig. 2a). Thin layer chromatography was performed and the spot identified (Rf 0.2) was collected and extracted with methanol.
HPLC analysis followed by LC-MS and MS-MS fragmentation pattern resulted in authentication of podophyllotoxin. HPLC analysis of fractions collected on day 4 showed retention time that corresponds to that of standard podophyllotoxin (Fig. 3). Characteristically, authentic podophyllotoxin yield an m/z (M + Na) peak at 437 (M + Na), which correspond to fungal podophyllotoxin (Fig. 4).
Maximum production of podophyllotoxin was observed on day 4 (μg/g dry weight of mycelia). The highest yield fungal cell-associated podophyllotoxin content was found to be 28 μg/g of dry mass (Fig. 2b) on the day 4. No podophyllotoxin was observed in uninoculated culture broth and processed culture broth at 0 h. This eliminated the possibility that podophyllotoxin had been carried from the original plant material source of the fungus via the mycelium (inoculum plugs).
The production of podophyllotoxin by Fusarium oxysporum supports the theory that during the long co evolution of endophytes and their host plants, endophytes adapted themselves to their special microenvironments by genetic variation, including uptake of some plant DNA into their own genomes (Germaine et al. 2004). This could have led to the ability of certain endophytes to biosynthesise some phytochemicals originally associated with the host plant (Stierle et al. 1993). It is possible to isolate hundreds of endophytic species from a single plant, and among them, at least one generally shows host-specificity (Tan and Zou 2001). Previous workers have reported the production of the antileukemic and antitumor drug taxol from the endophytes of Taxus spp. like Taxomyces andreanae and Pestalotiopsis microspora (Stierle et al. 1993; Strobel et al. 1996a, b).
The results indicate that Fusarium oxysporum can be a promising candidate for large scale production of podophyllotoxin. Further optimization of fermentation conditions and molecular studies of JRE1 is under way.
References
Arnold AE, Maynard Z, Gilbert GS, Coley PD, Kursar TA (2000) Are tropical fungal endophytes hyperdiverse? Ecol Lett 3:267–274
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST, A new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Bedir E, Khan I, Moraes RM (2001) Bioprospecting for podophyllotoxin. In: Janik J, Whipkey A (eds) Trends in new crops and new uses. ASHS, Aleandria, pp 545–549
Berkovitz DB, Choi S, Maeng JH (2000) Enzyme assisted asymmetric total synthesis of (−)-podophyllotoxin and (−) picropodophyllotoxin. J Org Chem 65:847–860
Berlin J, Bedorf N, Mollenschott C, Wray V, Sasse F, Hofle G (1988) On the podophyllotoxins of root cultures of Linum flavum. Plant Med 54:204–206
Broomhead AJ, Dewick PM (1990a) Tumour—inhibitory aryltetralin lignans in Podophyllum versipelle, Diphylleia cymosa and Diphylleia grayi. Phytochemistry 29:3831–3838
Broomhead AJ, Dewick PM (1990b) Aryltetralin lignans in Linum flavum and Linum capitatum. Phytochemistry 29:3839–3844
Cappiccino JG, Sherman N (1996) Microbiology a laboratory manual, VIth ed. The Benjamin/Cummings Publishing Company, Redwood City, CA, USA
Damayanti Y, Lown JW (1998) Podophyllotoxins: current status and recent developments. Curr Med Chem 5:205–252
Eyberger AL, Dondapati R, Porter JR (2006) Endophyte fungal isolates from Podophyllum peltatum produce podophyllotoxin. J Nat Prod 69(8):1121–1124
Empt U, Alfermann AW, Pras N, Peterson M (2001) The use of plant cell cultures for the production of podophyllotoxin and related lignans. J Appl Bot 74:145–150
Farkya S, Bisaria VS, Shrivastava AK (2004) Biotechnological aspects of the production of the anticancer drug podophyllotoxin. Appl Microbiol Biotechnol 65:504–519
Germaine K, Keogh E, Garcia-Cabellos G, Borremans B, Lelie D, Barac T, Oeyen L, Vangronsveld J, Moore FP, Moore ERB, Campbell CD, Ryan D, Dowling DN (2004) Colonisation of poplar trees by gfp expressing bacterial endophytes. FEMS Microbiol Ecol 48:109
Giri A, Narasu ML (2000) Production of podophyllotoxin from Podophyllum hexandrum: a potential natural product for clinically useful anticancer drugs. Cytotechnology 34:17–26
Imbert TF (1998) Discovery of podophyllotoxins. Biochimie 80:207–222
Konuklugil B (1996a) Aryltetralin lignans from genus Linum. Fitoterpia 67:379–381
Konuklugil B (1996b) Investigation of podophyllotoxin in some plants in Lamiaceae using HPLC. J Fac Pharm Ankara 25:23–27
Kuhnt M, Rimpler H, Henrich M (1994) Lignans and other compounds from the mixed Indian medicinal plant Hyptis verticillata. Phytochemistry 36:485–489
Kupchan SM, Hemingway JC, Knox JR (1965) Tumour inhibitors VII podophyllotoxin, the active principle of Juniperus verginiana. J Pharma Sci 54:659–660
Muranaka T, Miyata M, Kazutaka I, Tachibana S (1998) Production of podophyllotoxin in Juniperus chinensis callus cultures treated with oligosaccharides and a biogenetic precursor. Phytochemistry 49:491–496
Peterson M, Alferman AW (2001) The production of cytotoxic lignans by plant cell cultures. Appl Microbiol Biotechnol 55:135–142
Puri SC, Nazir A, Chawla A, Arora R, Riyaz-ul-Hasan S, Amna T, Ahmed B, Verma V, Singh S, Sagar R, Sharma A, Kumar R, Sharma RK, Qazi GN (2005) The endophytic fungus Trametes hirsuta as a novel alternative source of podophyllotoxin and related aryl tetralin lignans. J Biotechnol 122:494–510
San Feliciano A, Del Corral JMM, Gordaliza M, Castro MA (1989a) Acetylated lignans from Juniperus sabinai. Phytochemistry 28:659–660
San Feliciano A, Medarde M, Lopez JL, Puebla P, Del Corral JMM, Barrero AF (1989b) Lignans from Juniperus thurifera. Phytochemistry 28:2863–2866
Schacter L (1996) Etoposide phosphate; what, why, where, and how? Semin Oncol 6(Suppl 13):1–7
Stierle A, Strobel GA, Stierle D (1993) Taxol and taxanes production by Taxomyces andreanae, an entophytic fungus of pacific yew. Science 260:214
Strobel G, Yang X, Sears J, Kramer R, Sidhu RS, Hess WM (1996a) Taxol from Pestalotiopsis microspora, an endophytic fungus of Taxus wallichiana. Microbiology 142:435–440
Strobel G, Hess WM, Ford EJ, Sidhu RS, Yang X (1996b) Taxol from fungal endophytes and the issue of biodiversity. J Ind Microbiol 17:417–423
Subrahmanyam D, Renuka B, Rao CB, Sagar PS, Deevi DS, Babu JM, Vyas K (1998) Novel d-ring analogues of podophyllotoxin as potent anticancer agents. Bioorg Med Chem Lett 8:1391
Tan RX, Zou WX (2001) Endophytes: as rich source of fuctional metabolite. Nat Prod Rep 18:448
Utsugi T, Shibata J, Sugimoto Y, Aoyagi K, Wierzba K, Kobunani T, Terada T, Oh-hara T, Tsuruo T, Yaada Y (1996) Antitumour activity of a novel podophyllotoxin derivative (Top-53) against lung cancer and lung metastic cancer. Cancer Res 56:2809–2814
Vainio EJ, Korhonen K, Hantula J (1998) Genetic variation in Phlebiopsis giganteam as detected with random amplified microsatellite (RAMS) markers. Mycol Res 2:187–192
Yu P, Wang L, Chen Z (1991) A new podophyllotoxin type lignan from Dysosma versipllis var. tomentosa. J Nat Prod 54:1422–1424
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kour, A., Shawl, A.S., Rehman, S. et al. Isolation and identification of an endophytic strain of Fusarium oxysporum producing podophyllotoxin from Juniperus recurva . World J Microbiol Biotechnol 24, 1115–1121 (2008). https://doi.org/10.1007/s11274-007-9582-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-007-9582-5