Abstract
Studies of abundance, diversity and distribution of antibiotic-resistant bacteria and their resistance determinants are necessary for effective prevention and control of antibiotic resistance and its dissemination, critically important for public health and environment management. In order to gain an understanding of the persistence of resistance in the absence of a specific antibiotic selective pressure, microbiological surveys were carried out to investigate chloramphenicol-resistant bacteria and the chloramphenicol acetyltransferase resistance genes in Jiaozhou Bay after chloramphenicol was banned since 1999 in China. About 0.15–6.70% cultivable bacteria were chloramphenicol resistant, and the highest abundances occurred mainly in the areas near river mouths or sewage processing plants. For the dominant resistant isolates, 14 genera and 25 species were identified, mostly being indigenous estuarine or marine bacteria. Antibiotic-resistant potential human or marine animal pathogens, such as Escherichia coli, Klebsiella pneumoniae, Proteus mirabilis and Shewanella algae, were also identified. For the molecular resistance determinants, the cat I and cat III genes could be detected in some of the resistant strains, and they might have the same origins as those from clinical strains as determined via gene sequence analysis. Further investigation about the biological, environmental and anthropogenic mechanisms and their interactions that may contribute to the persistence of antibiotic-resistance in coastal marine waters in the absence of specific antibiotic selective pressure is necessary for tackling this complicated environmental issue.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Extensive use and misuse of antibiotics in medication, veterinary, agriculture and aquaculture have caused antibiotic-resistant bacteria to be widespread (Kummerer 2004). Environmental and zoonotic antibiotic-resistant bacterial pathogens may directly cause human illness and the spread of food-borne diseases (Barza 2002; Sorum and L’Abee-Lund 2002). Resistance genetic material transfer from environmental bacteria to commensal microflora may also cause bacterial pathogens to carry antibiotic resistance, complicating disease prevention and treatment (Levy and Marshall 2004). Elevated number of resistant bacteria, especially clinical strains, could be found near civic or hospital sewage discharging locations (Choi et al. 2003). High incidences of resistant bacteria in response to antibiotic usage have also been reported in coastal maricultural areas (Herwig et al. 1997). Thus, aquatic environments may serve as reservoirs of antibiotic resistance (Biyela et al. 2004), and coastal resistant bacteria represent a serious biotic contamination and a means for the spread and evolution of resistance genes and their vectors (Young 1993).
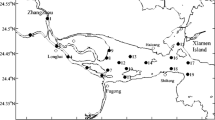
Jiaozhou Bay, located on the western coast of the Yellow Sea (N35°38′—36°18′, E120°04′—120°23′), is a shallow semi-enclosed water with a total area of 367 km2 and an average water depth of 7 m on the eastern part of the Shandong Peninsula of China. Environmental quality of Jiaozhou Bay has deteriorated dramatically in the past three decades due to rapid development in agriculture, industry, urbanization and mariculture in the surrounding areas (Shen 2001; Ren et al. 2006). More than ten small rivers used to form the major sources of discharged industrial and sewage wastewater. Wastewater discharge from maricultural practices makes the situation even worse. Improperly processed hospital effluent and civic wastewater could be a source of resistant bacteria. Excessive usage of antibiotics in mariculture might also stimulate the propagation of resistant bacteria in Jiaozhou Bay.
Chloramphenicol was once extensively used until it was banned in 1999 in China. The molecular determinants of chloramphenicol resistance were well studied (Schwarz et al. 2004), and the cat I, II, III and IV genes that encode the chloramphenicol acetyltransferases are the most common resistance mechanisms found in aquatic bacteria (Yoo et al. 2003). However, most antibiotic resistance studies focused on pathogenic bacteria (Schwarz et al. 2004). Very few studies paid attention to the environmental aspect (Mudryk 2005; Henriques et al. 2006). In order to gain an understanding of the current status of chloramphenicol resistance in a typical coastal environment of China, and to define the corresponding resistance determinants, molecular techniques were employed to determine the phylogenies of the typical culturable resistant bacteria and their resistance genes in Jiaozhou Bay.
Materials and methods
Sample collection and processing
Surface seawater from ten stations (Fig. 1) of Jiaozhou Bay was collected aseptically on the 12th and 13th of September and the 11th and 12th of October of 2004. A multi-function water quality monitoring system (model U-2001, Horiba, Japan) was used for in situ environmental factor measurements (Table 1), except for water depth and diaphaneity. Water samples were kept on ice before being transferred to the laboratory, where samples for total microbial counts were fixed with formaldehyde (2% final concentration) and stored at 4°C in the dark, and samples for bacterial cultivations were processed immediately.
Microbial number counts
For total microbial and cultivable bacteria counts, epifluorescence microscopy and 2216 marine agar plating techniques were used as described previously (Dang et al. 2006a). For chloramphenicol-resistant bacteria counts, a membrane filter incubation technique was used (Rompre et al. 2002). Water sample from each station was diluted into a ten-fold series in sterilized artificial seawater and 3 ml portions were filtered onto sterilized 0.22-μm pore-size polycarbonate membrane filters. Triplicates filters were placed onto tryptic soy agar (TSA, Difco formula) plates supplemented with 3% (w/v) NaCl and 10 μg/ml chloramphenicol (CP10). CP10-resistant colonies on the highest or the second highest dilution (if there were too few colonies on the highest) positively growing plates were further screened with 30 μg/ml chloramphenicol (CP30) based on the method described for antibiotic susceptibility tests (Yoo et al. 2003). Only CP30-resistant isolates were counted as chloramphenicol-resistant bacteria, which were further assayed for higher resistance to 100 μg/ml chloramphenicol (CP100).
Phylogenetic analysis of typical chloramphenicol-resistant isolates
CP100-resistant bacteria were picked based on unique colony morphology and color features. Because all of these isolates were originally screened from the highest or the second highest dilution positively growing CP10 plates, they represented the dominant chloramphenicol-resistant bacteria in Jiaozhou Bay during our sampling period. Eventually, 60 isolates were selected for 16S rDNA sequencing and cat gene screening.
A simple boiling method was used for rapid bacterial genomic DNA extraction (De Medici et al. 2003). Primers 27F and 785R were used for 16S rDNA amplifications, and the standard PCR reactions and program were followed as described previously (Dang et al. 2006a). Primer 27F was also used for sequencing with purified PCR products as templates using an ABI 3770 automatic sequencer (Applied BioSystems, USA). The DNA fragments sequenced were usually about 700 bp long, covering at least the V1–V3 hypervariable regions of bacterial 16S rDNA. Bioinformatic determination of sequence affiliations followed standard methods (Dang and Lovell 2000; Dang et al. 2006a). Phylogenetic trees were constructed by using the DNADIST and NEIGHBOR programs within the PHYLIP package (version 3.6) (Felsenstein 1989).
Multiplex PCR detection of the cat genes
The above selected 60 CP100-resistant isolates were screened for the cat I, II, III and IV genes via a multiplex PCR method developed by Yoo et al. (2003). The original primers (Table 2), experimental procedure and PCR amplification condition were used, only with a minor modification for total DNA (including genomic and plasmid DNA) extraction (De Medici et al. 2003). Representative PCR products were sequenced to confirm their identities as the cat genes.
Nucleotide sequence accession numbers
The 16S rDNA sequences determined in the current study have been deposited in the NCBI GenBank database under accession numbers DQ319006–DQ319065, and the representative cat gene sequences determined under accession numbers DQ319066 and DQ319067.
Results
Microbial abundance
During the sampling period, aquatic microbial density ranged from 2.7 × 106 to 3.8 × 107 cells/ml, cultivable bacterial abundance ranged from 1.9 × 102 to 5.2 × 105 CFUs/ml, and the density of chloramphenicol-resistant bacteria ranged from 1 to 9 × 102 CFUs/ml, which accounted for 0.15–6.70% of the total cultivable bacteria (Table 3). The highest abundance of chloramphenicol-resistant bacteria occurred at station A5, D6 and B2 in September, and A5, Y1 and D6 in October. Most of these stations are located close to a river mouth and/or sewage processing plant. Station A5 is near the river mouth of Licun and the Licun River sewage processing plant (Fig. 1), which discharged about 50,000 tons processed sewage into the bay daily. Station Y1 is close to the river mouths of Loushan and Moshui in the Hongdao maricultural area. Both stations are in the most polluted area of the Jiaozhou Bay (Li et al. 2005). Station B2 is located near the Dagu river mouth. Only station D6 is located outside of the Jiaozhou Bay.
Phylogenetic affiliations of the dominant chloramphenicol-resistant bacteria
Totally 348 CP30-resistant bacteria (143 from September and 205 from October) were isolated from surface seawater samples of Jiaozhou Bay, and 100 isolates (43 from September and 57 from October) showed resistance to at least 100 μg/ml chloramphenicol. Based on colony morphology and color features, a total of 60 unique CP100-resistant bacterial isolates (27 from September and 33 from October) were selected for further molecular analyses, which represent the most predominant chloramphenicol-resistant bacteria in Jiaozhou Bay during our sampling period.
All the 60 dominant chloramphenicol-resistant bacteria had more than 97% 16S rDNA sequence similarity to their best-match sequences retrieved from the GenBank database, and 85% of these isolates had 99% or higher 16S rDNA sequence similarity to their best-match sequences (data not shown). The phylogenetic tree constructed further verified their phylogenetic affiliations (Fig. 2). The chloramphenicol-resistant bacteria were quite diverse, at least 14 genera and 25 species could be identified. Most of the isolates belonged to the γ-Proteobacteria, and the remaining isolates belonged to the α-Proteobacteria, Actinobacteria or Firmicutes. Forty eight of the 60 sequenced chloramphenicol-resistant isolates were Gram-negative, and bacteria affiliated to Pseudomonadaceae (40%) and Pseudoalteromonadaceae (18.3%) formed the majority of the resistant microflora. Pseudomonas strains occurred in 3 and 9 of the 10 sampling stations in September and October respectively (Table 4). Pseudoalteromonas strains occurred in four stations in both sampling months. Gram-positive bacteria affiliated to Micrococcaceae and Bacillaceae only occurred in September, except for the Kocuria strain. Besides common estuarine and marine species, chloramphenicol-resistant bacteria affiliated to Enterobacteriaceae, such as Escherichia coli, Klebsiella pneumoniae and Proteus mirabilis, were also found to occur in the surface seawaters of Jiaozhou Bay during our sampling period, mainly in stations B2, C4 and Y1.
Phylogenetic tree of the 60 dominant chloramphenicol-resistant bacteria isolated from Jiaozhou Bay constructed based on partial 16S rDNA sequences in a 620-bp frame length using neighbor-joining method with the Kimura 2-parameter model for nucleotide change. The tree branch distances represent nucleotide substitution rate, and the scale bar represents the expected number of changes per homologous nucleotide position. Bootstrap values greater than 70% of 100 resamplings are shown near nodes as ‘**’, and those greater than 90% are shown as ‘*’. Bacterial strains are named with combinations of sampling time (SC for September and OC for October) and sampling station (such as A3, A5 and so on). The cat genes detected are labeled in “[ ]” besides the corresponding strains
Molecular screening of the cat genes
Only 20% of the 60 dominant chloramphenicol-resistant bacteria isolated from Jiaozhou Bay were found to harbor the cat genes screened. Of the 12 cat-positive isolates, 10 carried cat I, and the remaining 2 carried cat III (Fig. 2). The molecular mechanisms encoding for chloramphenicol resistance of the remaining 48 strains could not be identified by the current multiplex PCR method, other mechanisms might exist.
Six of the 12 Gram-positive CP100-resistant bacterial isolates carried a cat gene, actually cat I (Fig. 2). On the contrary, only 6 of the 48 Gram-negative CP100-resistant bacteria carried a cat gene, including 3 of the 4 Enterobacteriaceae bacteria identified. The cat I gene occurred in Aeromonas, Arthrobacter, Bacillus, Escherichia, Micrococcus, Proteus and Pseudomonas. The cat III gene occurred only in Klebsiella and Pseudoalteromonas, and only in October. Spatially, the cat genes mainly occurred in the sampling stations of A5, B2, C1, C3, C4 and D1 (Table 4), the main water body of the central Jiaozhou Bay.
The identities of the DNA fragments detected via the multiplex PCR method that might be the target cat genes were further verified by DNA sequencing of two representative PCR products, one for the possible cat I gene and the other for the possible cat III gene (Table 5). Both sequences had 99% or higher similarity to the best-match known cat genes retrieved from the GenBank database, and they also had 100% similarity to the best-match known chloramphenicol acetyltransferase sequences after they were translated into conceptual amino acid sequences. The multiplex PCR method (Yoo et al. 2003) provides a rapid and reliable approach for typical cat gene detection and differentiation in the marine environment studied.
Discussion
A basic understanding of the abundance, diversity and distribution of antibiotic-resistant bacteria and their resistance determinants is necessary for effective prevention and control of antibiotic resistance and its dissemination, critically important for public health and environment management. Without specific selective pressure from chloramphenicol per se, high abundance (at least 100 CFUs/ml of seawater) of resistant bacteria still could be found at some of the sampling stations in Jiaozhou Bay, such as A5, B2, D6 and Y1 (Table 3). River flow and/or wastewater discharge from the land might be the major source of antibiotic resistance in Jiaozhou Bay, as most of these stations are located near river mouths and/or sewage processing plants. Station D6, which is located outside of the Jiaozhou Bay, also had quite high abundance of resistant bacteria. About 100,000 tons of processed sewage was discharged daily into the seawater from the Tuandao sewage processing plant, located at the mouth of the Jiaozhou Bay (Fig. 1). Tide and/or current might be the major mechanism for carrying resistance bacteria to the D6 station from the nearby contaminated areas. Thus, antibiotic resistance contamination in the coastal environment studied is complicated with a variety of factors. Comprehensive study and modeling might help understand the processes and mechanisms involved.
Two sources of chloramphenicol-resistant bacteria could be identified in Jiaozhou Bay, mainly the indigenous estuarine or marine bacteria and some of the terrestrial bacteria that might be related to anthropogenic activities. Terrestrial bacteria entering into seawater with antibiotic-resistant plasmids were proposed to be partially responsible for the prevalence of resistance genes in marine environments (Chandrasekaran et al. 1998). In Jiaozhou Bay, two sources of cat genes could be identified, one from the indigenous estuarine or marine bacteria and another from the terrestrial bacteria potentially related to anthropogenic activities. The cat-positive Enterobacteriaceae bacteria were mainly detected in stations B2 and C4, close to the Dagu river mouth and the Haipo River sewage processing plant, which discharged about 80,000 tons of processed sewage into the bay daily. Although the seawater in the interior of Jiaozhou Bay is highly dynamic and water mixing happens frequently due to currents, tides and other hydrological factors, confined distribution of certain bacteria, especially the terrestrial resistant strains, still could be identified, indicating a strong terrestrial or anthropogenic impact on the distribution of certain antibiotic resistant bacteria. This finding is congruent with our previous study result about the spatial distribution of bacterial diversity in Jiaozhou Bay investigated via the microbial community 16S rDNA T-RFLP analysis method (Ren et al. 2006). The result of our current investigation indicates that sewage contamination might be a serious environmental problem of Jiaozhou Bay, at least in our sampling period. All the sewage processing plants in Qingdao had only the secondary treatment capacity. Wastewater collected from the nearby hospitals and communities might lack appropriate biological treatment, further processing of its bacteria load is needed before it can be discharged safely into the bay water.
The cat genes isolated from marine environments might have the same origins as those from clinical strains. Escherichia coli strains that harbored the cat I type of chloramphenicol resistance determinant have been identified from various sources around the world, including human and animal pathogens (Maynard et al. 2004; Bartoloni et al. 2006; Travis et al. 2006). A Proteus mirabilis strain was identified to harbor a variant of the cat I gene (Charles et al. 1985). Several Nigerian Klebsiella pneumoniae strains were identified to carry cat III-positive plasmids isolated from patients with community-acquired urinary tract infections (Soge et al. 2006). The isolation of similar Enterobacteriaceae bacteria carrying similar cat genes from Jiaozhou Bay indicates the wide distribution of these chloramphenicol-resistant bacteria in the world. Certain Shewanella algae strains were also found to be pathogenic (Holt et al. 2005). However, the molecular mechanism could not be identified for the chloramphenicol-resistant strains isolated from Jiaozhou Bay. To our knowledge, the current investigation is the first identification of the cat I gene in Aeromonas punctata, Arthrobacter mysorens, Bacillus spp., Micrococcus sp. and Pseudomonas pachastrellae, and the first identification of the cat III gene in Pseudoalteromonas sp.
While both Gram-positive and Gram-negative bacteria carried the cat I gene, only some Gram-negative bacteria carried the cat III gene in Jiaozhou Bay, at least in our sampling period. The cat-positive Gram-positive bacteria only occurred in September of our sampling period. What environmental and/or biological factors might influence the change of these resistance populations? Dramatic drops of temperature, pH and dissolved O2 were observed in October (Table 1). How these changes were related to antibiotic-resistant bacteria composition in Jiaozhou Bay need to be further studied. Changes of the source of terrestrial resistance bacteria or resistance determinants via river runoff and sewage discharge might also contribute to the temporal and spatial shift of antibiotic-resistant bacteria populations in Jiaozhou Bay.
Chloramphenicol-resistant molecular determinants varied with geological locations and/or environments along the China coast of the Pacific Ocean. In our previous investigations, it was found that the detectable resistance genes were cat II and cat IV in the studied mariculture pond waters from the Dalian coast of China (Dang et al. 2006a, b), where the cat-positive bacteria were affiliated to marine Vibrio and Pseudoalteromonas. In the currently studied coastal environment, cat I and cat III were detected and more diverse resistant bacteria species were identified. The Dalian mariculture ponds received frequent application of various antibiotics, such as oxytetracycline, penicillin, streptomycin, enrofloxacin and furanzolidone, for bacterial disease prevention and treatment of the cultured marine animals. Persistent and high combinational antibiotic selective pressure might eliminate some of the bacterial diversity. On the contrary, it would be normal to expect a higher diversity of antibiotic-resistant bacteria in natural marine environments, though the currently studied Jiaozhou Bay was highly disturbed by anthropogenic activities and terrestrial resistant bacteria contaminations.
It has been found that chloramphenicol-resistant bacteria might persist in the environments even after the drug use has been stopped (Schwarz et al. 2004). This is probably due to the phenomenon that chloramphenicol resistant genes derived from limited sources could be transferred among aquatic microbes without a high specific selective pressure (Yoo et al. 2003). Cross-resistance or coresistance caused by cross-selection or coselection might be the mechanism involved (Alonso et al. 2001; Courvalin and Trieu-Cuot 2001; Schwarz et al. 2004). Besides chloramphenicol acetyltransferases encoded by the cat genes, chloramphenicol resistance could also be caused by multidrug transporters (Poole 2005), which might provide the major cross-resistance of chloramphenicol in the environment studied, as the cat genes were detected in only 20% of the resistant isolates. For the mechanism of coresistance, the use of one antibiotic could increase levels of resistance not only to that specific drug but also to many others, even those using very different modes of antibacterial action (Kummerer 2004). It has been found that coselection by other antibiotics, such as oxytetracycline, could be the molecular mechanism responsible for the maintenance of the cat genes in some of the maricultural environments in China (Dang et al. 2006a, b). Besides antibiotics, other reagents might also contribute to the cross-selection and/or coselection mechanisms of chloramphenicol resistance. Jiaozhou Bay, especially its eastern coast, was highly polluted by heavy metals and other contaminants (Chen et al. 2005; Li et al. 2006). Exposure to heavy metals and other toxicants may confer resistance to antibiotics in the environments (Schwarz et al. 2004; Baker-Austin et al. 2006). Plasmids harboring resistance to both heavy metals and antibiotics have been reported in Enterobacteriaceae bacteria, such as avian pathogen Escherichia coli (Johnson et al. 2005) and human pathogen Salmonella enterica serovar Typhi CT18 (Parkhill et al. 2001), and in marine Pseudomonas (Rajini Rani and Mahadevan 1992). Heavy metal contamination might represent a long-standing, widespread and recalcitrant selection pressure, potentially contributing to the maintenance and spread of antibiotic resistance in Jiaozhou Bay.
Deterioration of coastal environmental quality is becoming a global issue due to various pollutions and contaminations. Oxygen-starved areas in bays and coastal waters have been expanding since the 1960s, and the number of known dead zones around the world has doubled since 1990 (UNEP 2004). The occurrence and persistence of diverse antibiotic-resistant bacteria and their resistance genes makes the situation even worse. Epidemics of pathogens in coastal marine environments may spread in large scale and at extremely rapid speed, due to their potential for long-term survival outside the host and the lack of barriers to dispersal (McCallum et al. 2003). Long-term comprehensive surveys and molecular mechanism and process studies will be needed for most of the coastal environments having similar situations to Jiaozhou Bay, in order to decode the biological, environmental and anthropogenic factors and their interactions that may contribute to the antibiotic resistance issue, for winning the battle for the health of humans and environments.
References
Alonso A, Sanchez P, Martinez JL (2001) Environmental selection of antibiotic resistance genes. Environ Microbiol 3:1–9
Baker-Austin C, Wright MS, Stepanauskas R, McArthur JV (2006) Co-selection of antibiotic and metal resistance. Trends Microbiol 14:176–182
Bartoloni A, Pallecchi L, Benedetti M, Fernandez C, Vallejos Y, Guzman E, Villagran AL, Mantella A, Lucchetti C, Bartalesi F, Strohmeyer M, Bechini A, Gamboa H, Rodriguez H, Falkenberg T, Kronvall G, Gotuzzo E, Paradisi F, Rossolini GM (2006) Multidrug-resistant commensal Escherichia coli in children, Peru and Bolivia. Emerg Infect Dis 12:907–913
Barza M (2002) Potential mechanisms of increased disease in humans from antimicrobial resistance in food animals. Clin Infect Dis 34:S123–S125
Biyela PT, Lin J, Bezuidenhout CC (2004) The role of aquatic ecosystems as reservoirs of antibiotic resistant bacteria and antibiotic resistance genes. Water Sci Technol 50:45–50
Chandrasekaran S, Venkatesh B, Lalithakumari D (1998) Transfer and expression of a multiple antibiotic resistance plasmid in marine bacteria. Curr Microbiol 37:347–351
Charles IG, Keyte JW, Shaw WV (1985) Nucleotide sequence analysis of the cat gene of Proteus mirabilis: comparison with the type I (Tn9) cat gene. J Bacteriol 164:123–129
Chen ZX, Dong HP, Li SQ, Yuan HM, Wang BJ (2005) Distribution pattern of heavy metals in the surface sediments of the Jiaozhou Bay. Mar Sci Bull 7:41–56
Choi S, Chu W, Brown J, Becker SJ, Harwood VJ, Jiang SC (2003) Application of enterococci antibiotic resistance patterns for contamination source identification at Huntington Beach, California. Mar Pollut Bull 46:748–755
Courvalin P, Trieu-Cuot P (2001) Minimizing potential resistance: the molecular view. Clin Infect Dis 33:S138–S146
Dang HY, Lovell CR (2000) Bacterial primary colonization and early succession on surfaces in marine waters as determined by amplified rRNA gene restriction analysis and sequence analysis of 16S rRNA genes. Appl Environ Microbiol 66:467–475
Dang HY, Song L, Chen M, Chang Y (2006a) Concurrence of cat and tet genes in multiple antibiotic-resistant bacteria isolated from a sea cucumber and sea urchin mariculture farm in China. Microb Ecol 52:634–643
Dang HY, Zhang XX, Song LS, Chang YQ, Yang GP (2006b) Molecular characterizations of oxytetracycline resistant bacteria and their resistance genes in mariculture waters of China. Mar Pollut Bull 52:1494–1503
De Medici D, Croci L, Delibato E, Di Pasquale S, Filetici E, Toti L (2003) Evaluation of DNA extraction methods for use in combination with SYBR green I real-time PCR to detect Salmonella enterica serotype enteritidis in poultry. Appl Environ Microbiol 69:3456–3461
Felsenstein J (1989) PHYLIP – phylogeny inference package (version 3.2). Cladistics 5:164–166
Henriques I, Moura A, Alves A, Saavedra MJ, Correia A (2006) Analysing diversity among β-lactamase encoding genes in aquatic environments. FEMS Microbiol Ecol 56:418–429
Herwig RP, Gray JP, Weston DP (1997) Antibacterial resistant bacteria in surficial sediments near salmon net-cage farms in Puget Sound, Washington. Aquaculture 149:263–283
Holt HM, Gahrn-Hansen B, Bruun B (2005) Shewanella algae and Shewanella putrefaciens: clinical and microbiological characteristics. Clin Microbiol Infect 11:347–352
Johnson TJ, Siek KE, Johnson SJ, Nolan LK (2005) DNA sequence and comparative genomics of pAPEC-O2-R, an avian pathogenic Escherichia coli transmissible R plasmid. Antimicrob Agents Chemother 49:4681–4688
Kummerer K (2004) Resistance in the environment. J Antimicrob Chemother 54:311–320
Levy SB, Marshall B (2004) Antibacterial resistance worldwide: causes, challenges and responses. Nat Med 10:S122–S129
Li GY, Ye SY, Gao ZJ, Zhang ZX (2005) Nutrient distributional characteristics and organic pollution analysis in bottom water in Jiaozhou Bay. Global Geol 24:194–199 (in Chinese with English abstract)
Li Y, Yu Z, Song X, Mu Q (2006) Trace metal concentrations in suspended particles, sediments and clams (Ruditapes philippinarum) from Jiaozhou Bay of China. Environ Monit Assess 121:491–501
Maynard C, Bekal S, Sanschagrin F, Levesque RC, Brousseau R, Masson L, Lariviere S, Harel J (2004) Heterogeneity among virulence and antimicrobial resistance gene profiles of extraintestinal Escherichia coli isolates of animal and human origin. J Clin Microbiol 42:5444–5452
McCallum H, Harvell D, Dobson A (2003) Rates of spread of marine pathogens. Ecol Lett 6:1062–1067
Mudryk ZJ (2005) Occurrence and distribution antibiotic resistance of heterotrophic bacteria isolated from a marine beach. Mar Pollut Bull 50:80–86
Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, Churcher C, Mungall KL, Bentley SD, Holden MT, Sebaihia M, Baker S, Basham D, Brooks K, Chillingworth T, Connerton P, Cronin A, Davis P, Davies RM, Dowd L, White N, Farrar J, Feltwell T, Hamlin N, Haque A, Hien TT, Holroyd S, Jagels K, Krogh A, Larsen TS, Leather S, Moule S, O’Gaora P, Parry C, Quail M, Rutherford K, Simmonds M, Skelton J, Stevens K, Whitehead S, Barrell BG (2001) Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848–852
Poole K (2005) Efflux-mediated antimicrobial resistance. J Antimicrob Chemother 56:20–51
Rajini Rani DB, Mahadevan A (1992) Plasmid mediated metal and antibiotic resistance in marine Pseudomonas. BioMetals 5:73–80
Ren J, Dang HY, Song LS, Sun S, An LG (2006) Bacterial and cyanobacterial diversities determined by T-RFLP analyses in Jiaozhou Bay. Acta Oceanol Sin 25:113–123
Rompre A, Servais P, Baudart J, de-Roubin MR, Laurent P (2002) Detection and enumeration of coliforms in drinking water: current methods and emerging approaches. J Microbiol Methods 49:31–54
Schwarz S, Kehrenberg C, Doublet B, Cloeckaert A (2004) Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol Rev 28:519–542
Shen ZL (2001) Historical changes in nutrient structure and its influences on phytoplankton composition in Jiaozhou Bay. Estuar Coast Shelf Sci 52:211–224
Soge OO, Adeniyi BA, Roberts MC (2006) New antibiotic resistance genes associated with CTX-M plasmids from uropathogenic Nigerian Klebsiella pneumoniae. J Antimicrob Chemother 58:1048–1053
Sorum H, L’Abee-Lund TM (2002) Antibiotic resistance in food-related bacteria – a result of interfering with the global web of bacterial genetics. Int J Food Microbiol 78:43–56
Travis RM, Gyles CL, Reid-Smith R, Poppe C, McEwen SA, Friendship R, Janecko N, Boerlin P (2006) Chloramphenicol and kanamycin resistance among porcine Escherichia coli in Ontario. J Antimicrob Chemother 58:173–177
UNEP (2004) GEO Year Book 2003. Global Environment Outlook (GEO)/United Nations Environment Programme (UNEP). ISBN: 9280724150. United Nations, New York, NY, USA
Yoo MH, Huh MD, Kim EH, Lee HH, Jeong HD (2003) Characterization of chloramphenicol acetyltransferase gene by multiplex polymerase chain reaction in multidrug-resistant strains isolated from aquatic environments. Aquaculture 217:11–21
Young HK (1993) Antimicrobial resistance spread in aquatic environments. J Antimicrob Chemother 31:627–635
Acknowledgements
This work was financially supported by the Pilot Projects of Knowledge Innovation Project of Chinese Academy of Sciences grants KZCX2-YW-211-03, KZCX3-SW-214, KZCX3-SW-223 and KZCX3-SW-233, the National Natural Science Foundation of China grants 40476058 and 40576069, and the LSC2-3-1 grant from IOCAS.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Dang, H., Ren, J., Song, L. et al. Dominant chloramphenicol-resistant bacteria and resistance genes in coastal marine waters of Jiaozhou Bay, China. World J Microbiol Biotechnol 24, 209–217 (2008). https://doi.org/10.1007/s11274-007-9458-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-007-9458-8