Abstract
Detailed vegetation mapping of wetlands, both natural and restored, can offer valuable information about vegetation diversity and community structure and provides the means for examining vegetation change over time. We mapped vegetation at six tidal marshes (two natural, four restored) in the San Francisco Estuary, CA, USA, between 2003 and 2004 using detailed vegetation field surveys and high spatial-resolution color-infrared aerial photography. Vegetation classes were determined by performing hierarchical agglomerative clustering on the field data collected from each tidal marsh. Supervised classification of the CIR photography resulted in vegetation class mapping accuracies ranging from 70 to 92%; 10 out of 12 classification accuracies were above 80%, demonstrating the potential to map emergent wetland vegetation. The number of vegetation classes decreased with salinity, and increased with size and age. In general, landscape diversity, as measured by the Shannon’s diversity index, also decreased with salinity, with an exception for the most saline site, a newly restored marsh. Vegetation change between years is evident, but the differences across sites in composition and pattern were larger than change within sites over two growing seasons.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nearly half of the world’s wetlands have been diked, drained, filled, or otherwise lost (Zedler and Kercher 2005), with an 80% loss in developed countries (Pennings and Bertness 2001). Furthermore, much of the remaining wetland area has been degraded by human activities (Zedler and Kercher 2005). Recently, there have been significant endeavors to restore wetland habitat throughout the world (Zedler and Kercher 2005), particularly within major estuaries such as the San Francisco Estuary, where there is the potential to restore large expanses of tidal marsh (Williams and Faber 2001). Tidal marshes provide multiple ecosystem services, habitat for endangered species, flood-control benefits, and, under the right conditions, can sequester carbon at high rates (Chmura et al. 2003), providing a potential additional economic incentive for restoration with the emergence of global carbon markets.
Fine-scaled mapping of tidal marsh vegetation is important to restoration efforts because it enables vegetation inventory and change detection, which informs overall habitat quality for numerous species and other important processes like sedimentation. Wetland vegetation classification using remote-sensing imagery analysis can be challenging, however, because of the high level of spectral confusion with other land cover classes, among different types of wetlands (Andresen et al. 2002; Ozesmi and Bauer 2002), and among different vegetation species (Ozesmi and Bauer 2002; Ramsey and Laine 1997; Schmidt and Skidmore 2003). Floristically-detailed mapping of tidal marsh vegetation is especially challenging in brackish marshes, which exhibit high levels of patchiness in plant species diversity and distribution.
The value of remote sensing for wetland monitoring has long been recognized (Hinkle and Mitsch 2005; Phinn et al. 1996), and recent advancements have made the methods and tools more applicable and cost-effective. Automated (computer-assisted) image analysis approaches allow for objective, consistent, and repeatable results, making them more scientifically defensible, especially across large, heterogeneous areas. The use of automated image classification reduces inconsistencies and error introduced through visual photo-interpretation of imagery. For this reason, automated image classification across multiple sites and time periods is more consistent and cost-effective than visual delineation and classification (Thomson et al. 2003).
Automated pixel-based classification methods are typically unsupervised, supervised, or a hybrid of both approaches. Unsupervised classification performs spectral clustering without a priori input from the analyst. Supervised classification is informed by ground reference field data as the analyst “trains” the classification algorithm. Both types have been used successfully in wetlands but there is no consensus regarding the best method (Belluco et al. 2006; Thomson et al. 1998, 2003). Our study had detailed and abundant field data collected to inform mapping and vegetation analyses, so we elected to use a supervised classification approach with high spatial resolution color infrared digital imagery to map dominant vegetation types.
Our overall goal was to characterize the range of variation in vegetation composition and diversity among sites and years and to demonstrate the accessibility of remote sensing to restoration ecologists for monitoring and assessment of marsh vegetation. Our specific objective was to map tidal marsh vegetation in marshes along a salinity gradient using high resolution (1-m pixel) color infrared (CIR) imagery, and to examine changes in vegetation composition and pattern over two growing seasons. First, we separated each site into vegetation, non-vegetation, and bare areas based on a simple normalized difference vegetation index (NDVI) threshold. Second, we used detailed, extensive, and targeted field-based vegetation data to determine the land cover classification scheme for each site so that it corresponded to the floristic data at the site, and cross-walked with alliances found in the California Manual of Vegetation (Sawyer et al. 2009). We then used the field data to train a supervised classification algorithm to map the landcover classes at each site across 2 years, and assessed the accuracy of the resulting maps. Finally, we calculated landscape metrics at both natural and restoring tidal marshes over two growing periods.
Methods
Study sites
This research was performed as part of the Integrated Regional Wetland Monitoring (IRWM) Pilot Project (http://www.irwm.org/), a multi-investigator interdisciplinary research project with the goal of evaluating how wetland restoration efforts throughout the San Francisco Estuary, California, USA are affecting ecosystem processes at different spatial and temporal scales; and to prepare for subsequent longer-term monitoring. The IRWM project vegetation component focused on data collection between 2003 and 2004.
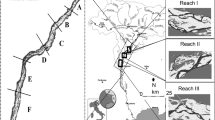
Our study sites were located in the Sacramento-San Joaquin Delta and San Pablo Bay, within the greater San Francisco Estuary (Fig. 1; hereafter called the Estuary). The Estuary is comprised of both natural and restored wetlands, as well as potential restoration sites such as diked bayland areas, former salt ponds, and seasonal and perennial wetlands. Mean salinities across our study sites ranged from approximately 15.40‰ at the western-most study site to 0.17‰ at the easternmost site (Table 1). The Estuary is one of the most modified estuaries in the United States, with history of land and water development and reclamation, over-fishing, and waste disposal since 1850 (Nichols et al. 1986).
The six sites that we mapped were (from west to east): Petaluma River Marsh (PRM) on the Petaluma River; Pond 2A (P2A), Coon Island (CI), and Bull Island (BuI) on the Napa River; and Browns Island (BrI) and Sherman Lake (SL) in the western Sacramento-San Joaquin River Delta (Fig. 1). Browns Island and Sherman Lake are located at the confluence of the Sacramento and San Joaquin Rivers, a combined watershed of 257,000 km2, and receive considerable freshwater input. These two sites are oligohaline (i.e. characterized by low salinity (0.5–5 ppt)). under most climate conditions but during drought years can be subject to higher salinity levels. Two of the six sites, Coon Island and Browns Island, are mature marshes (a combination of ancient and centennial marshland); the remaining four sites were restored within the last 90 years (Table 1). Study sites were chosen because they were representative of the dominant salinity gradient in the Estuary.
It is widely accepted that in coastal wetlands plant species richness decreases with salinity due to salinity and inundation-driven osmotic stresses; brackish and freshwater tidal marshes are characterized by diverse species mixtures due to reduced osmotic stresses (Engels and Jensen 2009; Sharpe and Baldwin 2009). These latter marshes usually are composed of several co-dominant plant species with numerous sub-dominant species and configured in a heterogeneous mosaic of vegetation community types, often with poorly defined boundaries between patch types. In the Estuary, salt marshes have between 2 and 22 plant species, brackish marshes contain 27–65 species, and freshwater marshes have upwards of 117 species (Parker, unpublished data).
Among our study sites, dominant species composition was variable. Petaluma River Marsh is the most saline marsh and consisted of four dominant species: California cordgrass (Spartina foliosa Trin.), common perennial pickleweed (Sarcocornia pacifica (Standl.) A. J. Scott), annual pickleweed (Salicornia depressa Standl.), and alkali bulrush (Bolboschoenus maritimus Palla). The three Napa River sites are brackish marshes, and consisted primarily of the same salt marsh species with the addition of more salt-sensitive marsh species: three-square common bulrush (Schoenoplectus americanus (Persoon) Volkart ex Schinz & R. Keller), common tule (S. acutus var. occidentalis (Muhlenberg ex Bigelow) Á. Löve & D. Löve), California tule/bulrush (S. californicus (C. A. Mey.) Soják), and three cattail species (Typha angustifolia L., T. domengensis Persoon, and T. latifolia L.). Perennial pepperweed (Lepidium latifolium Linn.) also exists at Coon, Bull, and Browns Island sites. In the freshwater sites, dominant vegetation consisted of S. americanus, S. acutus, S. californicus, and all Typha species, with the addition of alkali common reed (Phragmites australis (Cav.) Trin. ex Steud.). Gumplant (Grindelia stricta var. stricta (A. Gray) M. A. Lane) occurred at all the study sites along channel margins, levees, and upland areas.
Image acquisition and pre-processing
We used CIR aerial photography that was acquired for all of the IRWM project sites in October of 2003 and August of 2004. Imagery was captured with a Zeiss RMK Top 15 film camera, equipped with forward motion compensation and a Pleogon A3/4 lens with 153 mm focal length. The imagery was captured at the lowest possible tide in 2003 to aid vegetation and geomorphic mapping; in 2004, imagery was taken at mid-tide to aid channel delineation. Mid-tide level had the advantage of helping delineate channels because they had some water in them, but the level was not high enough to flood the wetland plane and subcanopy, and thus the vegetation classification should be comparable between years. This image had three bands: near infrared, red, and green. All sites were flown at a scale of 1:9,600 and the hardcopy CIR photography was scanned at a resolution of 1,200 dots per inch (dpi), resulting in a pixel size of 0.2-m for all sites. The only exception was that in 2003 only, Petaluma River Marsh was flown for use with another project and had a slightly different flight scale (1:7,200). Imagery is available for download at http://www.irwm.org/. The overall goal was to achieve the same scale and pixel resolution for all sites, regardless of site size, in order to achieve uniform data for subsequent analyses. We used digital photography for this project for two reasons: first, we could control the time of image acquisition; and second there is less problems with atmospheric influence with aerial photography.
We utilized at least four Trimble GPS-located (sub-meter accuracy) geospatial ground control points to rectify each photo. These points generally were located in different corners of each site, in order to maximize the accuracy of the rectification process (Trimble Inc. 2005). The images were orthorectified and digital elevation models (DEMs) were used in the rectification process to account for the local terrain’s effect on image distortion, using publicly available USGS 10-m DEMs. The accuracy standards employed for ortho-rectification were such that approximately 90% of all control points on the photos were within two meters of their corresponding ground coordinates.
In preparation for image analysis and metric calculation, we re-sampled all the imagery from 0.2-m pixel size to 1-m pixel size. The images were resampled to a coarser pixel size for three reasons. First, the imagery contained high pixel variability that is common with hyperspatial data, and we wanted to reduce the amount of spurious pixels in the classification output. Second, 1-m resolution was considered appropriate for the development of avian habitat models (Dechka et al. 2002; Stralberg et al. 2010). Third, we wanted to enable better GIS computing using the classification output, including the calculation of landscape pattern metrics.
Field data collection
We collected ground reference data to guide the classification scheme, train the classification of the imagery, and to assess the accuracy of the final maps. Ground reference data points were stratified by vegetation classes. In 2003, the field effort was carried out in two phases. During the first phase, we collected ground data for training the image classification using a supervised approach, in which a classification algorithm assigns a class to each pixel across using correlations between ground data and image spectral properties (Justice and Townshend 1981). We collected data at points that were randomly-generated in a Geographic Information System (GIS) prior to field visits, and points were chosen during the field visit at the discretion of the field samplers to train the image classification more effectively, especially for rare or underrepresented communities. The randomly-generated points were stratified across general vegetation groups that we created prior to our field visit using a simple unsupervised classification. We employed this step to account for the limited prior knowledge of the vegetation that occurred in the sites. Handheld Garmin GPS units with an average recorded location error of 5 m were used to navigate to each point. During the second 2003 field phase, we collected data at randomly-generated points to assess the accuracy of the 2003 classified vegetation map. In 2004, we conducted only one field phase that consisted of accuracy assessment data collection at randomly-generated points that were stratified along 2003 vegetation classes and area-weighted.
At all points visited during both years, the field crew recorded absolute percent cover of all species within a three-meter diameter circular plot. We chose this plot size in order to accurately represent species coverage based on observations of the average size and distribution of tidal wetland plants in this region and because these data were collected for purposes other than vegetation mapping. Percent cover was estimated using a seven-category modified Daubenmire ranking system (Daubenmire 1959). Species composition and cover data have been used frequently as indicators of ecosystem processes and are a useful component of ecosystem classifications (Mueller-Dombois and Ellenberg 1974). Between 50 and 300 plots were visited at each site in 2003 and 2004 (Table 2). Due to delicate conditions at some of the sites, including soft sediments and bird nesting territories, the field crew had the choice to observe the plot from a few meters away. We used this practice on less than 10% of the data points.
Image classification
Our classification involved several steps that are outlined in Fig. 2. We performed all image classification work with Leica Geosystem’s Erdas Imagine software (Leica Geosystems Inc. 2006).
Vegetation classification scheme
The most important first step in image classification is to choose or create a relevant classification scheme that is ecologically meaningful and as unbiased as possible (Jensen 2000). Many remote sensing mapping projects use a pre-defined classification scheme (e.g. USGS Landcover scheme (Anderson et al. 1976)); but such course landcover schemes do not capture the detailed and complex floristic patterns of each site. Instead we matched our field data to the alliance level listed in the Manual of California Vegetation (Sawyer and Keeler-Wolf 2004; Sawyer et al. 2009). The concept of the alliance is a physiognomically uniform plant group with one or more dominant/diagnostic species. We accomplished this by using the field data from all the randomly-generated points from both years, and performed an agglomerative hierarchical clustering measure in PC-ORD (McCune and Mefford 1999) for each site. These kinds of cluster analyses partition heterogeneous data into more manageable groups—in our cases, classes that we could use in our classification scheme. This algorithm builds a hierarchy of similar clusters from individual elements by progressively merging clusters. The PC-ORD implementation also builds a distance matrix that shows how different the clusters are from each other using a user defined distance criteria. Ward’s linkage method and Euclidean distance were used as a group linkage method and a distance measure, respectively. The resulting dendrograms were scaled using Wishart’s objective function, which measures loss of information at each step of cluster formation (McCune and Mefford 1999). We used a subjective criterion for pruning the dendrograms so that the resulting classes chosen matched an alliance in the Manual of California Vegetation. One species, perennial pepperweed, was not one of the primary groups from the hierarchical clustering but was added to the classification scheme because it was considered a target monitoring species due to its invasiveness in Estuary marshes (Grossinger et al. 1998). Although we were primarily concerned with wetland plants, we also mapped floating aquatic vegetation and upland species. We did this to be comprehensive in our mapping, and to make sure that we could use all the field data collected.
Separation of vegetated and non-vegetated areas
Vegetated and non-vegetated areas were separated first using Normalized Difference Vegetation Index (NDVI), which was calculated for each raw 3-band image using the red and NIR bands. Many studies have used vegetation indices to aid in wetland vegetation mapping (Eastwood et al. 1997; Thomson et al. 2004; Zhang et al. 1997). NDVI is considered an effective way to indicate presence of vegetation (Tuxen et al. 2008), as it is usually highly correlated with green leaf biomass and green leaf area index and is often considered to be a proxy for primary production (Hardisky et al. 1983, 1984; Jensen 2000, Jensen et al. 1998, 2002). The vegetated class included all the wetland areas, as well as the vegetated upland areas. The non-vegetated areas were masked out of the dataset to render the raw imagery for areas with vegetation only (Fig. 2).
Step 3: Supervised classification of individual species
All vegetated pixels were then classified into detailed vegetation classes using a supervised classification approach. We first reclassed the plot data according to the dominant vegetation type in each plot. This was necessary in order to simplify and reduce the number of potential vegetation classifications, since many field plots had up to 12 species recorded in them. We considered each plot to belong to a certain vegetation class if that class occupied more than 50% of its area. These dominance-based plot data were used as training sets for the maximum likelihood classifier. Training pixels were created using the “seed pixel” approach, where a pixel chosen at the center of a relatively homogenous feature and whose spectral properties are compared to its neighboring pixels. Given enough similarity between neighboring pixels, a training region, or “signature,” expands in the direction of spectrally similar pixels. The appropriate vegetation class was then assigned to each signature. We examined the spectral separability for all classes using a transformed divergence measure, a common index of signature separability (ERDAS 1999). The separability analysis was necessary in order to verify that the signatures for different classes were not too spectrally similar so as to produce errors in the classification output. Transformed divergence ranges from 0 to 2,000, and is easy to interpret because any value under 1,700 indicates poor separation between signatures (ERDAS 1999). If any confusion existed in the separability analysis, signatures were added or removed to allow for better representation of the classes in the spectral signatures. When adequate signatures were finalized (i.e. the separation analysis was satisfactory), each vegetation group image was classified using the Maximum Likelihood Classifier (MLC). Finally, all vegetation groups were combined together with the non-vegetated areas, to create the vegetation map for each site for each of the 2 years.
Mosaicking images
All but two sites had more than one image tile (i.e. more than one image to make up the entire site), and for several sites, the different tiles had varying airplane tilt, causing brightness gradients from one side of the tile to the other (Devereux et al. 1990) that were impossible to remove completely using standard radiometric corrections such as illumination correction. Since images were not color-matched during mosaicking, some seams were extremely noticeable and would have caused abrupt spectral changes, causing false vegetation classification. Therefore, for all sites with more than one image tile, each tile was classified separately, and then mosaicked before accuracy assessment using the mosaic function in Erdas Imagine (Leica Geosystems Inc. 2006).
Spatial filtering of spurious pixel effect
The vegetation map was filtered to smooth out the speckled effect inherent and ubiquitous with all pixel-based classifications (Blaschke and Strobl 2001). This is apparent especially in fine-scaled data and can include shadows of leaves and plants, patches with highly mixed species, and differences in reflectance values on the same species caused by different reflectance angles off of leaves (Jensen 2000). The smoothing technique used was an elimination filter, a function that allows for the specification of a minimum “clump,” or patch of pixels of a certain number or area, and clumps smaller than the minimum are filled in from their neighboring large clumps in an iterative fashion until all small patches are accounted for. The minimum clump size used was four pixels, resulting in a minimum mapping unit (MMU) of approximately four square-meters, an adequate size to capture the patchy and heterogeneous nature of Pacific coast wetlands (Phinn et al. 1996).
Accuracy assessment
We performed an accuracy assessment for each map by comparing field data to classified data; all field data used in accuracy assessment had not been used for classification training. For each ground point, the field-based dominant vegetation type was compared to the mapped dominant vegetation type in order to assess accuracy. Since each 3-m-diameter plot often contained a mixture of mapped species, we used the thematic mapped class with the majority of pixels that fell inside plot as the mapped class for that point. Standard remote sensing accuracy measures, including the overall accuracy and errors of omission and commission for each class at each site for each year were calculated using the error matrix method (Congalton and Green 1999; Congalton 2009).
Metrics calculation
We calculated site-level diversity metrics as a way to characterize differences amongst sites and between years. We used FRAGSTATS 3.3 (McGarigal et al. 2002) to calculate the percent of landscape for each vegetation class, as well as site-wide (landscape) metrics: Shannon’s Diversity Index and Shannon’s Evenness Index. Shannon’s Diversity measures landscape (vegetation class) diversity; it equals zero when the landscape contains only one vegetation class, and increases as the number of different vegetation classes increases and/or the proportional distribution of area among vegetation classes becomes more equitable. Shannon’s Evenness measures landscape evenness; it equals zero when the landscape contains only one vegetation class and approaches one as the distribution of area among the different vegetation classes becomes increasingly uneven (McGarigal et al. 2002).
To understand the similarity of vegetation cover across sites and between years, we calculated a Bray-Curtis dissimilarity metric (Faith et al. 1987) for each site/year combination, using the percent cover of each vegetation type for each site and each year. The Bray Curtis dissimilarity is a statistic used to quantify the compositional dissimilarity between two different sites. It is equivalent to the total number of species that are unique to any one of the two sites divided by the total number of species over the two sites. The metrics were calculated using the ‘vegan’ package, version 1.3 (J. Oksanen) for R (R_Development_Core_Team 2005).
We did not perform a traditional change detection in this project for two main reasons. First, conventional change detection products, such as a land cover transition matrix, are usually performed on larger scale classifications; our fine scale would result in unwieldy transition matrix. In addition, we are mostly interested in overall changes in land cover class and particularly in overall pattern changes.
Results
Hierarchical clustering
We used hierarchical clustering of all field data to develop our classification scheme by site. The hierarchical dendrogram output for PRM is shown in Fig. 3, and is a typical output from PC-Ord’s hierarchical agglomerative clustering routine. The vertical dashed line indicates the pruning point on the tree, which resulted in five classes in this case. These five classes were used to train a supervised classification algorithm to map the marsh vegetation. For each additional site, we repeated this analysis, finding the dominant vegetation classes to serve as landcover classes in our remote sensing process. The complete analysis yielded many additional vegetation classes (Table 4).
Cluster analysis dendrogram used for the creation of the classification scheme for Petaluma River Marsh. This is one of six dendrograms created with hierarchical agglomerative clustering using PC-ORD to create classification schemes for the vegetation maps for 2003 and 2004. The vertical dashed line indicates the area where tree was pruned to show which groups or classes should be used for the classification scheme
Classification results
Map accuracies ranged from 69.8 to 91.5% (Table 3). The two largest sites, BrI and SL, were the most problematic to accurately classify. The remaining sites were all classified with over 80% accuracy in both years. Of the individual vegetation classes, the most consistently accurately classified was the perennial pickleweed, although there was some confusion with annual pickleweed at some sites; the most problematic classes were the bulrush, cattail and reed classes, which had overlap.
Our classification shows some vegetation change between 2003 and 2004 in each of the six sites. PRM and BuI, for example, shows clear growth in pickleweed along channel margins. On the larger sites (BrI and SL), this change is largely on the marsh plain, and is likely accounted for by classification confusion between some of the more spectrally similar classes mentioned above (Figs. 4 and 5).
Vegetation maps for Petaluma River Marsh (PRM), Bull Island (BuI), and Coon Island (CI) for 2003 (left image) and 2004 (right image). Locations of each site are found in Fig. 1. Petulama River Marsh is the most saline site; Bull Island and Coon are brackish
Vegetation maps for Browns Island (BrI), Pond 2A (P2A), and Sherman Lake (SL) for 2003 (left image) and 2004 (right image). Locations of each site are found in Fig. 1. Brown Island and Sherman are the largest and freshest sites; Pond 2A is brackish
We examined each site’s vegetation composition and the trends in vegetation composition across site age, size and salinity, and time. The sites were increasingly diverse (in terms of number of vegetation classes) as site salinity decreased, site size increased, and with age; salinity accounts for the strongest trend (Table 4). Many of the common brackish marsh vegetation types were mapped at almost all of the sites (including cattail, common perennial pickleweed, and bulrush) but no vegetation class was present at all six sites. Some vegetation types were mapped at only one or two sites, such as annual pickleweed at the most saline site (PRM) and alkali common reed, saltgrass, and floating aquatic vegetation at the two freshest sites (BrI and SL).
The common perennial pickleweed class increased the most in cover between years at four of the six sites (BuI, CI, PRM and P2A), with a corresponding decrease in mudflat at two of these sites, PRM and P2A—both young, restored sites, although the difference in tide stage between the respective images needs to be considered. At sites CI and BuI, the increase in common perennial pickleweed likely occurred at the expense of alkali bulrush; at PRM this expansion likely was offset by losses in alkali bulrush and mudflat, reflecting the colonization of pickleweed onto bare ground following restoration (Tuxen et al. 2008). At the freshest site, SL, there was a change in the cover of some of the fresh water vegetation classes: a decrease in bulrush and three-square and an increase in alkali common reed.
Diversity metrics
In both years, landscape diversity, as measured by Shannon’s Diversity, was slightly greater at fresher, larger sites, but this was not a strong trend. PRM was an exception; the smallest, most saline site had as diverse a landscape as the two large, fresh sites. Browns Island, a large, old, mature marsh, had the highest vegetation landscape diversity in both years. These results were the opposite for landscape evenness, which generally declined with age, salinity and size.
Based on Bray-Curtis dissimilarity metrics, differences among sites were much greater than differences between years within the same site (Table 5), but there was no clear relationship between site dynamics and salinity, size or age. Restoration sites yielded both the highest (PRM) and lowest (P2A) change (dissimilarity metric) between years. The most similar sites, BuI, CI, and P2A, are all brackish marshes that vary in size and age. The most saline site, PRM, was most different from all other sites.
Discussion
Mapping wetlands with high spatial resolution imagery
Using high spatial resolution multispectral imagery, we were successful in producing detailed vegetation maps of both restored and natural wetlands that had varying levels of salinity and vegetation heterogeneity and composition. Field calibration and validation of vegetation maps was necessary to increase mapping detail and accuracy. Pixel-based methods applied in detailed vegetation type mapping are effective at measuring large-scale pattern change, including site-wide richness, diversity, and evenness. These methods can face challenges when mapping small-scale vegetation changes, particularly when the time interval between measurements is short. This might suggest that vegetation mapping with this level of detail should take place less frequently than annually, and for a longer duration than two time periods. This limitation is largely due to spectral similarity between tidal marsh species assemblages in the spectral bands available to us. Although high spatial resolution imagery is necessary for detailed mapping, a per-pixel classification approach is limiting because of high local-scale spectral variability of the pixels, and the inability of most pixel-based algorithms to take spatial and contextual information into account in the classification. Where only a few species comprise a relatively homogeneous patch mosaic, CIR imagery is effective and relatively straightforward. However, for sites with small-scale diversity, using high spatial resolution CIR imagery requires different types of classification algorithms such as object based image analysis, or hyperspectral data with a high spatial resolution (Blaschke 2010). These alternate approaches may increase success of discriminating within-patch species.
Map accuracy
The overall vegetation map accuracy increased from 2003 to 2004 at four out of six sites. Additional ground reference points and acquiring imagery with better spectral separation between vegetation types could explain this increase (2004 imagery was acquired at a later date for these two sites than in 2003—October rather than August/September). Laba et al. (2005) found, in contrast, that wetland vegetation community types could be distinguished best with imagery acquired in August for measuring peak biomass. Our accuracy rates for October 2003 imagery are less than 80% are likely are due to the underrepresentation of some classes in the field data due to their small area or habitat conditions that made field ground-truthing more difficult.
Though the classification approach used effectively mapped marsh vegetation types, each tile was processed individually, which produced considerable work due to the idiosyncrasies of each individual image (e.g., brightness gradients). We recommend acquiring imagery for a site that incorporates as few tiles as possible (preferably one tile), while still being able to achieve the desired pixel size for mapping. Modern high resolution scanners can help achieve this goal by enlarging the footprint for each photo tile. This practice will reduce processing time and image problems due to mosaicking seams. If a larger spatial extent needed processing, which requires numerous photograph tiles to capture the entire area, supervised or unsupervised methods may not be possible, and visual interpretation, albeit time-consuming, may produce the most accurate results (Zharikov et al. 2005).
Certain vegetation classes were identified with low accuracy, indicating high commission and omission errors. Shrub species, including both gumplant and coyote brush, usually existed as one or two plants, creating very small patches that were either too small for the one-meter pixel size to detect, or were eliminated using the filtering technique for smoothing out the speckled effect. Both of these species often existed on levees and in upland areas, so they were incorporated into an “upland” class.
Diversity metrics
Vegetation pattern in tidally influenced marshes responds to the interaction between hydrodynamics and elevation, and can present clear zonation in vegetation type, local-scale variations in microtopography (and hence inundation), which can create more complex vegetation patterns (Hickey and Bruce 2010, Pratolongo et al. 2009). But pattern is more difficult to quantify than diversity, thus there are fewer studies examining pattern and structural differences across salinity gradients, or across time. Engles and Jensen (2009) looked at wetlands across salinity gradients along the Elbe (Germany), and the Connecticut (USA) Rivers, and found clear differences in pattern as expressed by Shannon’s Diversity and Evenness metrics.
We measured overall vegetation similarity between sites and years with the Bray-Curtis index (Table 5). In Table 5, we are comparing each site’s vegetation with all the other sites’ vegetation in the same year (e.g. PRM 2003 with P2A 2003), and each site’s vegetation in 2003 with its vegetation in 2004 (e.g. PRM 2003 with PRM 2004). The higher Bray-Curtis score indicates large differences, either between sites, or across years. In our study, overall vegetation similarity, showed small differences between years for any given site, compared to the large differences observed between sites in a given year. Salinity seemed to play a role in this: across sites, the most similar sites were those that are geographically close and have similar salinities, illustrating the effect of position along the estuarine salinity gradient.
Changes in wetland vegetation pattern over time, especially in restored wetland sites, are also not straightforward. In many cases, species diversity and dominance patterns do not match those of natural sites, even decades after restoration (Garbutt and Wolters 2008). In the long term, disturbance (both chronic and extreme) will change wetland vegetation pattern (Byrd and Kelly 2006; Byrd et al. 2004; Frieswyk and Zedler 2007; Zedler 2009), in many cases shifting toward fewer species and increased dominance. In our study, site-level diversity metrics demonstrated several important differences across sites and between years that are detailed in the next sections. Though these metrics may highlight actual change in vegetation type, it is also possible that any changes in a two-year study, such as this one, are depicting annual variability and do not indicate long-term trends. These metrics are effective at measuring differences from year to year and, used over the long term as a monitoring technique, can allow for land cover change quantification and ultimately the measurement of restoration success.
A recently restored marsh (PRM) ranked as high or higher than mature marshes in vegetation class diversity and number of patches in both years (Shannon’s diversity of 1.66 and 1.47 respectively), in contrast to the relatively low plant species diversity at this high-salinity site (Callaway et al. 2007). In other words, the recently restored site had a more complex vegetation patch structure than a more mature marsh. At least at this site, we suggest that processes of vegetation colonization produce a more complex vegetation structure in the early period after restoration and that over longer time periods this structure simplifies, due to hydrologic and salinity patterns, or other factors such as competition.
Bull Island, a restored site, had variable diversity and evenness between years, while other more recently restored sites (PRM and P2A) were less variable (Table 5). This finding could reflect an actual true increase in vegetation type diversity at BuI, or the fact that the earlier image acquisition in 2004 (August 2004 vs. October 2003) allowed for better separation of the vegetation types. These results emphasize the importance of performing these measures consistently over many years to measure long-term changes through the marsh’s restoration, in order to notice long-term changes that could be lost due to interannual variability.
The three oldest sites, two mature marshes (BrI, CI) and one 83-year old restoration site (SL), exhibited very small changes in vegetation class diversity, evenness, and patch density. This finding was expected, as these mature sites should not show as much change (i.e., colonization and succession) as the recently restored sites. For example, at PRM, the large increase of the perennial pickleweed compared to the smaller increase in the early-colonizing annual pickleweed reflects marsh maturation as more sediment accumulates and elevation increases at the site.
Conclusions
Based on the findings of this study, we have several recommendations for applying this kind of vegetation mapping method. First, images should be collected at the same time of year when conducting multi-year evaluations to reduce variability introduced by growing season spectral variability. Third, except at locations undergoing very rapid vegetation changes (newly restored sites), intervals between mapping should be greater than 1 year in order to capture actual vegetation changes rather than method variability. Two consecutive years of image acquisition and analysis might not be sufficient to show substantial vegetation change with both natural and restoring sites. Fourth, avoiding the need for multiple image tiles through higher altitude image acquisition and higher resolution scanning improves mapping results and reduces mapping effort. Sixth, it is beneficial to have ample field data for use in both training image processing software and in validating classification results. With these guidelines, there is potential value to mapping restored wetlands sites using high resolution imagery over the long term.
References
Anderson JR, Hardy EH, Roach JT, Whitmer RE (1976) A land use and land cover classification system for use with remote sensing data. Washington, DC: U.S. Government Printing Office. Report: Geological Survey Professional Paper 964
Andresen T, Mott C, Zimmermann S, Schneider T, Melzer A (2002) Object-oriented information extraction for the monitoring of sensitive aquatic environments. IEEE 3083–3085
AS ERD (1999) Erdas field guide. Erdas Inc, Atlanta
Belluco E, Camuffo M, Ferrari S, Modenese L, Silvestri S, Marani A, Marani M (2006) Mapping salt-marsh vegetation by multispectral and hyperspectral remote sensing. Remote Sens Environ 105:54–67
Blaschke T (2010) Object based image analysis for remote sensing. ISPRS J Photogramm Remote Sens 65:2–16
Blaschke T, Strobl J (2001) What’s wrong with pixels? Some recent developments interfacing remote sensing and GIS. GeoBIT/GIS 6:12–17
Byrd K, Kelly M (2006) Salt marsh vegetation response to edaphic and topographic changes from upland sedimentation in a Pacific estuary. Wetlands 26:813–829
Byrd KB, Kelly NM, Dyke EV (2004) Decadal changes in a Pacific estuary: a multi-source remote sensing approach for historical ecology. GIScience Remote Sens 41:347–370
Callaway JC, Parker TV, Vasey MC, Schile LM (2007) Emerging issues for the restoration of tidal marsh ecosystems in the context of predicted climate change. Madroño 54:234–248
Chmura GL, Anisfeld SC, Cahoon DR, Lynch JC (2003) Global carbon sequestration in tidal, saline wetland soils. Global Biogeochem Cycles 17:1111
Congalton RG (2009) Accuracy assessment of spatial data sets. In: Madden M (ed) Manual of geographic information systems. The American Society of Photogrammetry and Remote Sensing, Bethesda, pp 225–234
Congalton RC, Green K (1999) Assessing the accuracy of remotely sensed data: principles and practices. CRC Press, Inc, Boca Raton
Daubenmire RF (1959) Canopy coverage method of vegetation analysis. Northw Sci 33:43–64
Dechka J, Franklin S, Watmough M, Bennett R, Ingstrup D (2002) Classification of wetland habitat and vegetation communities using multitemporal IKONOS imagery in sourthern Saskatchewan. Can J Remote Sens 28:679–685
Devereux BJ, Fuller RM, Carter L, Parsell RJ (1990) Geometric correction of airborne scanner imagery by matching Delaunay triangles. Int J Remote Sens 11:2237–2251
Eastwood JA, Yates MG, Thomson AG, Fuller RM (1997) The reliability of vegetation indices for monitoring saltmarsh vegetation cover. Int J Remote Sens 18:3901–3907
Engels J, Jensen K (2009) Patterns of wetland plant diversity along estuarine stress gradients of the Elbe (Germany) and Connecticut (USA) Rivers. Plant Ecol Divers 2:301–311
Faith DP, Minchin PR, Belbin L (1987) Compositional dissimilarity as a robust measure of ecological distance. Vegetatio 69:57–68
Frieswyk C, Zedler J (2007) Vegetation change in Great Lakes coastal wetlands: deviation from the historical cycle. J Great Lakes Res 33:366–380
Garbutt A, Wolters M (2008) The natural regeneration of salt marsh on formerly reclaimed land. Appl Veg Sci 11:335–344
Grossinger R, Alexander J, Cohen A, Collins JN (1998) Introduced tidal marsh plants in the San Francisco Estuary: regional distribution and priorities for control. San Francisco Estuary Institute, Richmond, CA
Hardisky MA, Smart RM, Klemas V (1983) Seasonal spectral characteristics and aboveground biomass of the tidal marsh plant, Spartina alterniflora. Photogramm Eng Remote Sens 49:85–92
Hardisky MA, Daiber FC, Roman CT, Klemas V (1984) Remote sensing of biomass and annual net aerial primary productivity of a salt marsh. Remote Sens Environ 16:91–106
Hickey D, Bruce E (2010) Examining tidal inundation and salt marsh vegetation distribution patterns using spatial analysis. Botany Bay, Australia
Hinkle RL, Mitsch WJ (2005) Salt marsh vegetation recovery at salt hay farm wetland restoration sites on Delaware Bay. Ecol Eng 25:240–251
Jensen JR (2000) Remote sensing of the environment: an earth resource perspective, 2nd edn. Prentice Hall, Upper Saddle River
Jensen JR, Coombs C, Porter D, Jones B, Schill S, White D (1998) Extraction of smooth cordgrass (Spartina alterniflora) biomass and leaf area index parameters from high resolution imagery. Geocarto Int 13:25–34
Jensen JR, Olson G, Schill SR, Porter DE, Morris J (2002) Remote sensing of biomass, leaf-area-index, and chlorophyll a and b content in the ACE Basin National Estuarine Research Reserve using sub-meter digital camera imagery. Geocarto Int 17:25–34
Justice CO, Townshend JRG (1981) Integrating ground data with remote sensing. In: Townshend JRG (ed) Terrain analysis and remote sensing. George Allen & Unwin, London, p 232
Laba M, Tsai F, Ogurcak D, Smith S, Richmond ME (2005) Field determination of optimal dates for the discrimination of invasive wetland plant species using derivative spectral analysis. Photogramm Eng Remote Sens 71:603–611
Leica Geosystems Inc (2006) (17 January 2007 URL http://gis.leica-geosystems.com/)
McCune B, Mefford MJ (1999) PC-ORD for windows. MjM Software Design, Gleneden Beach
McGarigal K, Cushman SA, Neel MC, Ene E (2002) FRAGSTATS: Spatial Pattern Analysis Program for Categorical Maps. Pages Computer software program produced by the authors at the University of Massachusetts, Amherst. Available at the following web site: www.umass.edu/landeco/research/fragstats/fragstats.html
Mueller-Dombois D, Ellenberg H (1974) Aims and methods in vegetation ecology. Wiley, New York
Nichols FH, Cloern JE, Luoma SN, Peterson DH (1986) The modification of an estuary. Science 231:567–573
Ozesmi SL, Bauer ME (2002) Satellite remote sensing of wetlands. Wetl Ecol Manage 10:381–402
Pennings SC, Bertness MD (2001) Salt marsh communities. In: Bertness MD, Gaines SD, Hay ME (eds) Marine community ecology. Sinauer, Sunderland, pp 289–316
Phinn SR, Stow DA, Zedler JB (1996) Monitoring wetland habitat restoration in southern California using airborne multispectral video data. Restor Ecol 4:412–422
Pratolongo PD, Kirby JR, Plater A, Brinson MM (2009) Temperate coastal wetlands: morphology, sediment processes, and plant communities. In: Perillo GME, Wolanski E, Cahoon DR, Brinson MM (eds) Coastal wetlands: an integrated ecosystem approach. Elsevier, Amsterdam, pp 89–118
R_Development_Core_Team (2005) R: a language and environment for statistical computing. http://www.r-project.org/
Ramsey EW III, Laine S (1997) Comparison of Landsat Thematic Mapper and high resolution photography to identify change in complex coastal wetlands. J Coastal Res 13:281–292
Sawyer JO, Keeler-Wolf T (2004) A manual of California vegetation. California Native Plant Society, Sacramento
Sawyer JO, Keeler Wolf T, Evens JM (2009) A manual of California vegetation, 2nd edn. California Native Plant Society, Sacramento
Schmidt KS, Skidmore AK (2003) Spectral discrimination of vegetation types in a coastal wetland. Remote Sens Environ 85:92–108
Sharpe P, Baldwin A (2009) Patterns of wetland plant species richness across estuarine gradients of Chesapeake Bay. Wetlands 29:225–235
Stralberg D, Herzog MP, Nur N, Tuxen KA, Kelly M (2010) Predicting avian abundance within and across tidal marshes using fine-scale vegetation and geomorphic metrics. Wetlands 30:475–487
Thomson AG, Fuller RM, Sparks TH, Yates MG, Eastwood JA (1998) Ground and airborne radiometry over intertidal surfaces: waveband selection for cover classification. Int J Remote Sens 19:1189–1205
Thomson AG, Fuller RM, Yates MG, Brown SL, Cox R, Wadsworth RA (2003) The use of airborne remote sensing for extensive mapping of intertidal sediments and saltmarshes in eastern England. Int J Remote Sens 24:2717–2737
Thomson AG, Huiskes A, Cox R, Wadsworth RA, Boorman LA (2004) Short-term vegetation succession and erosion identified by airborne remote sensing of Westerschelde salt marshes, The Netherlends. Int J Remote Sens 25:4151–4176
Trimble Inc (2005) (17 January 2007 URL http://www.trimble.com/)
Tuxen K, Schile L, Kelly M, Siegel S (2008) Vegetation colonization in a restoring tidal marsh: a remote sensing approach. Restor Ecol 16:313–323
Williams PB, Faber PM (2001) Salt marsh restoration experience in the San Francisco Bay Estuary. J Coastal Res 27:203–211
Zedler J (2009) How frequent storms affect wetland vegetation: a preview of climate-change impacts. Front Ecol Environ 8:540–547
Zedler JB, Kercher S (2005) Wetland resources: status, trends, ecosystem services, and restorability. Ann Rev Environ Resour 30:39–74
Zhang M, Ustin SL, Rejmankova E, Sanderson EW (1997) Monitoring Pacific coast saltmarshes using remote sensing. Ecol Appl 7:1039–1053
Zharikov Y, Skilleter GA, Loneragan NR, Taranto T, Cameron BE (2005) Mapping and characterising subtropical estuarine landscapes using aerial photography and GIS for potential application in wildlife conservation and management. Biol Conserv 125:87–100
Acknowledgments
This work was supported by the CALFED California Bay-Delta Program. The CALFED Science Program supported most of this work under Grant Number 4600002970. The CALFED Ecosystem Restoration Program supported manuscript preparation under Grant Number P0685516. Jake Schweitzer was instrumental in processing and georectifying aerial imagery from HJW GeoSpatial, Inc.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tuxen, K., Schile, L., Stralberg, D. et al. Mapping changes in tidal wetland vegetation composition and pattern across a salinity gradient using high spatial resolution imagery. Wetlands Ecol Manage 19, 141–157 (2011). https://doi.org/10.1007/s11273-010-9207-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11273-010-9207-x