Abstract
Chlorpyrifos (CP) is a widely used organophosphate (OP) insecticide and a potent environmental neurotoxin. This research focuses on the potential of bacteria, both native to agricultural soil and part of a designed consortium composed of laboratory strains, to completely degrade CP and its toxic metabolites. Metabolite production and degradation kinetics analysis through gas chromatography-mass spectrophotometry (GCMS) analysis was conducted on native soil samples and compared to soil spiked with different combinations of bacterial consortia over 7 days to determine the effectiveness of CP degradation in both non-augmented and augmented soil. GCMS analysis of augmented soil samples inoculated with putative microbial CP degradation activity identified four CP metabolites, including 3,5,6-trichloropyridinol (TCP), phosphorothioic acid, fumaric acid, and ethanol. Non-augmented ranch and crop field soil also displayed a greater native degradation capacity than garden soil, possibly due to greater pesticide exposure. CP-inoculated soil spiked with a 3-strain consortium exhibited the highest degradation rate, with 78.55% of CP degraded within 48 h. Overall, degradation kinetics for augmented and non-augmented soil samples showed that CP had an average half-life of 1.03 and 5.45 days, respectively. The outcome of this study suggests that while native agricultural populations are capable of CP degradation, supplementing contaminated soil with a bacterial consortium consisting of Pseudomonas putida CBF10-2, Ochrobactrum anthropi FRAF13, and Rhizobium radiobacter GHKF11 could be a highly efficient and safe biological approach to facilitating rapid and efficient CP degradation in agriculture.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Organophosphate (OP) compounds are highly toxic acetylcholinesterase inhibitors and among the most widely used pesticides in agriculture. The adverse effects of their wide usage worldwide directly contribute to approximately 300,000 deaths, with more than three million people adversely affected annually (Eyer, 2003; Iyer et al., 2015a; Karalliedde, 1999; Singh & Walker, 2006; Sogorb et al., 2004). The persistence and retention of these pesticides and their metabolites in the environment is a significant concern for the safety of both human and environmental health.
Chlorpyrifos (CP), O, O-Diethyl O-3, 5, 6-trichloropyridin-2-yl phosphorothioate, is an OP insecticide used as the primary ingredient in several commercial pesticide formulations that kill insect pests across a wide variety of food crops. CP has been used in the USA as a pesticide in this manner since 1965. Non-agricultural uses include controlling insect pests on golf courses, turf, greenhouses, non-structural wood treatments, as mosquito adulticide, and in roach and ant bait stations. CP is registered for use in nearly 100 countries and is applied to approximately 8.5 million crop acres (DOW Chemical Company, 2020). CP remains one of the most used insecticides in the USA (Vogel, 2016). However, greater restrictions have been placed on the insecticide in the USA, limiting its range of application predominantly to agriculture. Most notably, residential use of CP was banned in the USA in 2001 (US EPA, 2002). Furthermore, due to its impact on child development, in August 2018, the US 9th Circuit Court of Appeals ordered the EPA to ban CP’s sale in the USA within 60 days. However, no governmental action at the national level has been taken to date, leaving states to make a decision on an independent basis (Washington Examiner, 2018).
As a highly toxic neurotoxin, CP can adversely impact many non-target organisms. Agricultural soil is often contaminated due to the widespread use of CP as well as through mishandling of the compound and accidental spillage (Briceño et al., 2012). In such instances, soil microbiota is likely to be affected. This can lead to the loss of soil fertility and, by association, plant growth resulting in losses to crop productivity (Fang et al., 2009). CP residue and its metabolites cause widespread contamination of aquatic ecosystems, often leaching into surface water and nearby groundwater sources. Many reports indicate that a wide range of water and terrestrial ecosystems are persistently co-contaminated through this process, impacting different biota and disrupting biogeochemical cycling (Horne et al., 2002; Tse et al., 2004).
CP’s persistence in soil depends on the formulation, rate of application, soil type, climate, native microbial community, and availability of desired genes among microbial populations to degrade CP (Gao et al., 2012; Kamrin, 1997; Roberts & Hutson, 1999; Singh & Walker, 2006). As such, the half-life of CP in the soil can vary between 7 and 120 days (Singh & Walker, 2006). The predicted pathway of the aerobic transformation of CP can be seen in Fig. 1 and leads to the formation of two toxic metabolites, TCP and DETP (Singh & Walker, 2006). TCP is persistent and mobile with a half-life ranging from 65 to 360 days in the soil, which can also depend on various factors such as soil type, pH, moisture, microbial activity, climate, and other conditions. The accumulation of TCP in soil or liquid medium often limits CP degradation and inhibits microorganisms’ proliferation. Some microbial species can further metabolize TCP and DETP as a carbon and phosphorus source as needed (Singh et al., 2004) (see Fig. 1).
Proposed pathway for microbial chlorpyrifos degradation. As shown in Singh & Walker, 2006
We hypothesized that natural biodegradation of CP, where it exists, would be a relatively slow process as few microorganisms in each population are likely to possess the requisite CP degradation genes. However, this process may be expedited by the addition of a specialized bacterial consortium. In this manuscript, we first identify metabolites related to microbial degradation of CP in different agricultural soil types to determine the rate at which chlorpyrifos is metabolized in these samples by native bacterial populations. Lastly, we discuss the impact of a specialized soil consortium developed from three OP-degrading strains, Pseudomonas putida CBF 10–2, Ochrobactrum anthropi FRAF13, and Rhizobium radiobacter GHKF11 from the Iyer Laboratory on its removal through enhanced bioremediation (Iyer et al., 2018).
2 Materials and Methods
2.1 Chemicals, Equipment, and Microorganisms
CP was purchased from Sigma-Aldrich (USA). Hexane was used as a solvent to dissolve 100 mg pure white CP powder to make 1 mL final volume of CP solution stored in an amber glass bottle at room temperature. Cobalt chloride, zinc sulfate, magnesium sulfate, calcium nitrate, iron sulfate, Luria–Bertani media, acetonitrile, and hexane were all purchased from Sigma-Aldrich (USA). A gas chromatography-mass spectrophotometer (GCMS) unit was purchased from Shimadzu Corporation (Japan). The UV–Vis spectrophotometer and costar 96 flat transparent plates used for optical density readings were purchased from Tecan and the centrifuge unit from Eppendorf.
Consortia microorganisms, P. putida CBF 10–2, O. anthropi FRAF13, and R. radiobacter GHKF11 (Iyer et al., 2018), were grown from glycerol stock taken from a library of putative OP-degrading bacteria housed in the Iyer Laboratory (Iyer et al., 2015b).
2.2 Growth Media and Culture Condition
Bacterial cultures were grown in Luria–Bertani (LB) medium, and mineral salt medium (MSM) used to conduct all CP biodegradation tests (pH 7.0 and temp 30 °C) contained (g/L) K2HPO4, 1.5; KH2PO4, 0.5; (NH4)2SO4, 0.5; MgSO4.7H2O, 0.2; and 5 μL/L of 1 M CoCl2.6H2O solution. One milliliter of K12 trace element solution composed of (g/L) NaCl, 5.0; ZnSO4.7H2O, 4.0; MnCl2.4H2O, 4.0; FeCl3.6H2O, 4.7; CuSO4.5H2O, 4.0; H3BO3, 0.575; NaMoO4.2H2O, 0.5; and 12.5 mL of 6 N H2SO4 was also added to the MSM.
2.3 Sample Collection
Three soil samples were collected from three separate locations: a ranch near Fairchild, TX, (29.4225° N, 95.8007° W), a residential garden in Jersey Village, TX, (29.8979° N, 95.5703° W), and a cotton field near Pecan Grove, TX, (29.6239° N, 95.7191° W). The soil was collected only from the upper layer (7–12 cm) and placed in a 50 mL amber tube to minimize degradation from light exposure. Soil samples were stored for the duration of the project at room temperature in amber tubes.
2.4 Growth of Consortia Strains and Biodegradation of Chlorpyrifos
Selected strains were tested for their capacity to utilize CP as a carbon source when inoculated into MSM medium. The bacterial strains P. putida CBF10-2, O. anthropi FRAF13, and R. radiobacter GHKF11 were each inoculated into LB (10 mL) and then incubated at 37 °C for 24 h. Next, 200 μL of each sample was transferred into Costar 96 flat transparent wells, and the optical density was read at a wavelength of 600 nm in a Tecan UV–Vis spectrophotometer. Once all samples had shown an optical density reading of 1.0, they were spun down at 4000 rpm for 8 min. The supernatant was discarded, and the pellet resuspended into 5 mL MSM media supplemented with CP (400 mg/L). The MSM culture tubes were incubated on a rotary shaker at 200 rpm for 6 days at 30 °C. Optical density readings at 600 nm were taken every 48 h.
Biodegradation capacity of different soil populations against CP was analyzed with respective soils in MSM medium over 7 days. This experiment was carried out to compare sterilized, non-sterilized, and augmented (spiked with putative CP degradation bacteria) soil samples when amended with CP. CP-inoculated sample soils were designated as non-augmented cultures. CP-inoculated sample soils spiked with P. putida CBF10-2, O. anthropi FRAF13, and R. radiobacter GHKF11 bacterial strains were defined as augmented cultures. All tests were conducted in triplicates. Degradation analysis was performed in 50-mL culture tubes containing 5 g of sample soil and 20 mL of autoclaved MSM inoculated with 200 mg/L concentrations of CP. The samples were then incubated in the dark at 30 °C on an orbital shaker at 150 rpm for 7 days. For degradation analysis, an aliquot of 1 mL culture was withdrawn at regular intervals for 7 days, and degradation was evaluated through GCMS analysis. Parathion solubilized in hexane was used as a series of internal standards ranging from 0.1 to 100 mg/mL, while CP-inoculated sterilized soil and MSM media were used as controls.
2.5 Sample Extraction for GCMS Analysis
Samples obtained from culture tubes were extracted with hexane prior to GCMS analysis. Samples were thoroughly shaken for 1 min to obtain homogeneity, and then 1 mL aliquots of each sample were placed into centrifuge tubes. An equal amount of hexane (1 mL) was added for extraction of the organic layer, and centrifugation was conducted at 4000 rpm for 10 min. The organic layer was extracted and transferred to a new clean storage tube. For GCMS analysis, the organic layer was diluted 1:10 with additional hexane and transferred into a 1.5 mL GCMS sample vial.
2.6 Real-Time GCMS Analysis
Gas chromatography-mass spectrometry (GCMS) analyses were performed with a Shimadzu GCMS-TQ-8040, equipped with auto-sampler, on-column, auto-injector, split/splitless capillary injection system, and an Rtx-5MS capillary column (30.0 m length by 0.25 mm diameter and 0.25 μm film thickness). Helium was used as the carrier gas with a constant flow rate of 1.0 mL/min and at a pressure of 89.4 kPa. The flow control mode was under linear velocity 40.7 cm/s, the total flow was 28.1 mL/min, and column flow was set to 1.20 mL/min. Purge flow was set to 3.0 mL/min, and the split ratio was 20.0. The chromatography program was as follows: initial column oven temperature 100 °C, a temperature increase of 10 °C/min, and final heating to 240 °C. The injector and detection temperatures were 250 °C and 300 °C, respectively. Sampling was done at 1-min intervals, with a sampling start time set at 2 min and an end time 25 min later for a total run time of 28 min. Under these conditions, chlorpyrifos eluted as a peak at a retention time of 16.26 min. The instrument detection limit was approximately 100 pg for chlorpyrifos. CP and other metabolites were identified using NIST MS library software and by comparing retention time (RT) and the MS fragmentation profiles generated from standardized pure samples of each target compound.
2.7 Kinetics Analysis
Kinetics was determined by plotting log CP residues against time. The following algorithms were used, as seen in Eqs. (1) and (2).
where Ct is the concentration at time t, C0 is the initial concentration, e is the base, k is the rate constant of decline 1/days, and t is time.
where ln is natural log.
In C was plotted against time t, and a straight-line regression equation was obtained.
The slope of the regression equation gives value for rate constant “k.”
The concentration of CP was calculated using the following equations:
3 Results
3.1 Growth Patterns of Bacterial Strains in Chlorpyrifos-Supplemented MSM
The growth pattern of P. putida CBF10-2, O. anthropi FRAF13, and R. radiobacter GHKF11 (individually and combined) in MSM supplemented with CP and without CP (control) is shown in Fig. 2. The growth rate of each strain in CP was significantly higher than the control after 48 h. Following 48 h, both the test samples show falling cell densities that continue to decline until the end of the experiment. Overall, P. putida CBF10-2 and R. radiobacter GHKF11 show comparable increases in cell density. O. anthropi shows only a small increase compared to its control, but cell density does not decline as steeply after 48 h. A comparable increase in cell density is measured in the bacterial consortium in CP-supplemented MSM, shown in Fig. 3. However, cell growth is extended for 24 h, as only after 96 h does the consortium begin to decline.
Growth of bacterial strains in chlorpyrifos-supplemented MSM.
 P. putida CBF10-2,
P. putida CBF10-2,
 control for P. putida CBF10-2,
control for P. putida CBF10-2,
 O. anthropi FRAF13,
O. anthropi FRAF13,
 control for O. anthropi FRAF13,
control for O. anthropi FRAF13,
 R. radiobacter GHKF11,
R. radiobacter GHKF11,
 control for R. radiobacter GHKF11. Error bars represent a confidence limit of 95%. The optical density of isolates was measured at 600 nm
control for R. radiobacter GHKF11. Error bars represent a confidence limit of 95%. The optical density of isolates was measured at 600 nm
3.2 Results of GCMS Analysis: Chlorpyrifos Metabolites and Pathway Prediction
Chromatographic analysis reveals CP’s molecular weight of 350.59 g/mol and a retention time of 16.247 min. The primary metabolite TCP or 2-hydroxy-3,5,6-trichloropyridine was found with a molecular weight of 197 g/mol and retention time of 18.273, as seen in Fig. 4. TCP, which is considered the most toxic and primary metabolite of CP, was more abundant in augmented soil samples but only within the first 24 h after inoculation. After 24 h, the presence of TCP disappears from the test medium. Multiple secondary metabolites at low concentrations were observed after 24 h, including phosphorothioic acid and ethanol, which are derived from the breakdown of DETP. Fumaric acid was also observed and is a probable breakdown product of TCP.
3.3 Biodegradation of Chlorpyrifos in Soil Samples
The ability of the selected bacterial strains to degrade CP in soil was studied using parathion as an internal standard, sterilized soil as a control, and non-augmented natural soil and augmented natural soil inoculated with the three-strain bacterial consortium. The results obtained from GCMS analysis showed that the CP degradation dynamics differ significantly between test conditions. CP degradation in soil suggested a time-dependent disappearance that followed first-order kinetics. Multiple combinations of bacterial consortia were tested to determine differences in CP biodegradation. The results are shown in Figs. 5, 6, and 7. Composition of bacterial consortia was as follows: A + B + C: P. putida, + R. radiobacter + O. anthropi, A + B: P. putida, + R. radiobacter, B + C: R. radiobacter + O. anthropi, and C + A: O. anthropi + P. putida. Ranch soil samples resulted in complete degradation in all test samples by the end of the experiment on day 7. The rate of degradation varied based on the composition of the consortium used. In general, the addition of P. putida and an additional strain to the consortium increased the degradation rate. Garden soil samples show comparatively less biodegradation over time. However, applied CP was eventually completely degraded in all augmented samples except for a two-strain bacterial consortium composed of Pseudomonas putida and Ochrobactrum anthropi. Farm soil samples reveal that the applied CP was completely degraded in all samples except in two-strain bacterial consortia composed of P. putida and R. radiobacter or R. radiobacter and O. anthropi.
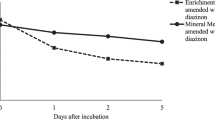
Augmented soil with a complete three-strain consortium exhibited higher CP degradation as compared to non-augmented soil samples, as seen in Fig. 8. In uninoculated sterilized soil, it was found that approximately 78.02% of the initial CP persisted till the end of the 7-day incubation period. Augmented soil samples were able to degrade 100% of CP within 7 days, while non-augmented native soil showed significant if comparatively less degradation over the same period (see Fig. 8).
3.4 Kinetics of Chlorpyrifos Biodegradation
Ranch soil demonstrated the most rapid degradation among non-augmented soil samples throughout this experiment. Notably, while all augmented soil samples improved the overall degradation rate, ranch soil showed the least improvement when spiked with bacterial consortia. In contrast, garden soil appeared to benefit the most from the addition of the bacterial consortium.
As seen in Table 1, both controls and non-augmented soil show significantly less degradation than augmented soil samples over the 7-day test duration. The average half-life for controls and non-augmented soil samples were 25.02 and 5.45 days, respectively. Among augmented soil, the average half-life ranged from 4.52 to 1.03. The differences between augmented, non-augmented, and sterile soil can be seen in Fig. 9.
4 Discussion
The present study describes the functional characterization of soil bacteria to degrade CP and its metabolites in contaminated soil. Degradation of CP involves both chemical hydrolysis and microbial activity. Maintaining an optimal culture environment (pH 7–8, 30 °C) is critical as the temperature and pH of the soil and water significantly affect the efficiency of the microorganisms to degrade chlorpyrifos (Racke et al., 1996; Singh et al., 2003, 2006). Metabolites produced during CP degradation throughout this study confirm that the first step of its metabolic pathway was hydrolysis of the O-P ester linkage to make TCP and likely DETP. The presence of TCP was readily detected in both non-augmented and augmented soil samples. However, throughout this study, DETP was not identified, possibly due to its polarity and lack of retention with the utilized chromatographic conditions. Biodegradation of TCP is a crucial part of the remediation of CP-contaminated sites. If left accumulated, TCP will affect the soil’s underlying microbial populations because of its inherent antimicrobial properties. Notably, TCP was not persistently found after 48 h, particularly in augmented soil, due to its subsequent degradation. As such, it was unlikely to have significantly repressed CP biodegradation.
Average CP degradation was observed to be higher in non-augmented soils than the sterilized control soil by day 2. This finding confirms that the indigenous microbes in each soil sample did contribute to the overall degradation of CP. Among the different soil-tested types, both ranch and crop native field soil samples exhibited higher biodegradation properties than the garden soil samples. This result is not unexpected given that CP is banned from residential use but not from agricultural use. Therefore, garden soil is not expected to be as exposed to CP, which would have a significant impact on the overall distribution of OP-degrading and non-OP-degrading bacteria in such an environment. By comparison, both ranch soil and crop field soil are likely to be more abundant sources of OP-degrading bacteria.
Degradation of CP was significantly higher in augmented soil than non-augmented soil indicating the high bioremediation potential of the strains composing the bacterial consortia. The observed enhancement in CP utilization can be attributed to the fact that the inoculation of soil with our consortium led to increased catabolic potential without negatively impacting existing population dynamics. While the biodegradation properties of two-strain bacterial consortia showed variance in their biodegradation ability depending on the source and nature of the soil, in all instances, the three-strain consortium exhibited improved biodegradation properties over all other combinations. Specifically, a consortium composed of P. putida CBF10-2, O. anthropi FRAF13, and R. radiobacter GHKF11 degraded an average of 78.55% of CP within 2 days and had obtained complete degradation by the end of the test duration. Degradation kinetics were determined using a first-order rate equation Ct = C0 × e − kt because the disappearance of CP was noted to be time-dependent. The rate constant (K) for CP degradation by the three-strain bacterial consortium in soil ranged from 0.45 to 1.20 days with a half-life (T1/2) of 0.58–1.54 days, depending on the soil source. By comparison, one notable CP-degrading strain of Serratia marcescens was capable of degrading CP at a rate constant ranging from 0.017 to 0.052 d−1 with T1/2 of 13.6–37 days in several types of soils (Cycon et al., 2013).
In summary, this study provided novel insights into the bacterial capacity for bioremediation of CP under simulated environmental settings, which represents a positive step in designing an effective field strategy and protocol for remediation of CP-contaminated agricultural soil. The appearance of 4 metabolites, 2 hydroxy-3,5,6-trichloropyridine (TCP), phosphorothioic acid, fumaric acid, and ethanol found through GCMS analysis suggests that native bacteria soil populations readily work in concert to mineralize CP, with populations putatively exposed to more pesticides demonstrating increased biodegradation characteristics. In addition, the three-strain consortium composed of P. putida CBF10-2, O. anthropi FRAF13, and R. radiobacter GHKF11 could be a highly desirable candidate for use in CP bioremediation strategies across a variety of agricultural settings given their overall efficiency and low pathogenicity.
Data Availability
All 3 bacterial strains associated with this study are housed in the Iyer microbial library at the University of Houston.
Code Availability
Not applicable.
Abbreviations
- CP:
-
Chlorpyrifos
- DETP:
-
Diethylthiophosphate
- EPA:
-
Environmental Protection Agency
- GC-MS:
-
Gas chromatography-mass spectrometry
- IS:
-
Internal standard
- MRM:
-
Multiple reaction monitoring
- MSM:
-
Modified mineral medium
- OP:
-
Organophosphate
- PTE:
-
Phosphotriesterase
- TCP:
-
2-Hydroxy-3,5,6-trichloropyridine
References
Briceño, G., Fuentes, M. S., Palma, G., Jorquera, M. A., Amoroso, M. J., & Dieza, M. C. (2012). Chlorpyrifos biodegradation and 3,5,6-trichloro-2-pyridinol production by actinobacteria isolated from soil. International Biodeterioration and Biodegradation, 73, 1–7. https://doi.org/10.1016/j.ibiod.2012.06.002.
Cycon, M., Zmijowska, A., Wojcik, M., & Piotrowska-Seget, Z. (2013). Biodegradation and bioremediation potential of diazinon-degrading Serratia marcescens to remove other organophosphorus pesticides from soils. Journal of Environmental Management, 117, 7–16. https://doi.org/10.1016/j.jenvman.2012.12.031.
Dow Chemical Company. Chlorpyrifos and Responsible Use. Retrieved 2020–07–24.
Eyer, P. (2003). The role of oximes in the management of organophosphorus pesticide poisoning. Toxicological Reviews, 22(3), 165–190. https://doi.org/10.2165/00139709-200322030-00004.
Fang, H., Yu, Y., Chu, X., Wang, X., Yang, X., & Yu, J. (2009). Degradation of chlorpyrifos in laboratory soil and its impact on soil microbial functional diversity. Journal of Environmental Sciences, 21, 380–386. https://doi.org/10.1016/S1001-0742(08)62280-9.
Gao, J., Wang, Y., Gao, B., Wu, L., & Chen, H. (2012). Environmental fate and transport of pesticides. Pesticides—evaluation of environmental pollution (pp. 29–41). CRC Press Taylor and Francis Group.
Horne, I., Sutherland, T. D., Harcourt, L. R., Russell, R. J., & Oakeshott, J. G. (2002). Identification of an opd (organophosphate degradation) gene in an Agrobacterium isolate. Applied and Environment Microbiology, 68, 3371–3376. https://doi.org/10.1128/AEM.68.7.3371-3376.2002.
Iyer, R., Iken, B., Damania, A., & Krieger, J. (2018). Whole-genome analysis of six organophosphate-degrading rhizobacteria reveals putative agrochemical degradation enzymes with broad substrate specificity. Environmental Science and Pollution Research, 25(14), 13660–13675. https://doi.org/10.1007/s11356-018-1435-2.
Iyer, R., Iken, B., & Leon, A. (2015a). Developments in alternative treatments for organophosphate poisoning. Toxicology Letters, 233(2), 200–206. https://doi.org/10.1016/j.toxlet.2015.01.007.
Iyer, R., Smith, K., Kudrle, B., & Leon, A. (2015b). Detection and location of OP-degrading activity: A model to integrate education and research. New Biotechnology, 32(4), 403–411. https://doi.org/10.1016/j.nbt.2015.03.010.
Kamrin, M. A. (1997). Pesticide profiles toxicity, environmental impact, and fate (pp. 147–152). Lewis Publishers.
Karalliedde, L. (1999). Organophosphorus poisoning and anaesthesia. Anaesthesia, 54(11), 1073–1088. https://doi.org/10.1046/j.1365-2044.1999.01061.x.
Racke, K. D., Steele, K. P., Yoder, R. N., Dick, W. A., & Avidov, E. (1996). Factors affecting the hydrolytic degradation of chlorpyrifos in soil. Journal of Agriculture and Food Chemistry, 44, 1582–1592. https://doi.org/10.1021/jf9506141.
Roberts, T. R., & Hutson, D. H. (1999). Metabolic pathways of agrochemicals - Part 2: Insecticides and fungicides (pp. 235–242). The Royal Society of Chemistry.
Singh, B. K., & Walker, A. (2006). Microbial degradation of organophosphorus compounds. FEMS Microbiology Reviews, 30(3), 428–471. https://doi.org/10.1111/j.1574-6976.2006.00018.x.
Singh, B. K., Walker, A., Morgan, J. A. W., & Wright, D. J. (2003). Effects of soil pH on the biodegradation of chlorpyrifos and isolation of a chlorpyrifos-degrading bacterium. Applied and Environment Microbiology, 69(9), 5198–5206. https://doi.org/10.1128/AEM.69.9.5198-5206.2003.
Singh, B. K., Walker, A., Morgan, J. A. W., & Wright, D. J. (2004). Biodegradation of chlorpyrifos by Enterobacter strain B-14 and its use in biodegradation of contaminated soils. Applied and Environment Microbiology, 70, 4855–4863. https://doi.org/10.1128/AEM.70.8.4855-4863.2004.
Singh, B. K., Walker, A., & Wright, D. J. (2006). Bioremedial potential of fenamiphos and chlorpyrifos degrading isolates: Influence of different environmental conditions. Soil Biology & Biochemistry, 38, 2682–2693. https://doi.org/10.1016/j.soilbio.2006.04.019.
Sogorb, M. A., Vilanova, E., & Carrera, V. (2004). Future application of phosphotriesterases in the prophylaxis and treatment of organophosphorus insecticide and nerve agent poisoning. Toxicology Letters, 151, 219–233. https://doi.org/10.1016/j.toxlet.2004.01.022.
Tse, H., Comba, M., & Alaee, M. (2004). Methods for the determination of organophosphate insecticides in water, sediments and biota. Chemosphere, 54, 41–47. https://doi.org/10.1016/S0045-6535(03)00659-3.
US EPA. (2002). Interim reregistration eligibility decision for chlorpyrifos. Archived from the original on November 19, 2012. Retrieved 2020–02–28.
Vogel, D. (2016). Chlorpyrifos Status Update. EPA. Retrieved 2020–10–15.
Washington Examiner. (2018). Federal appeals court orders EPA to ban pesticide. Retrieved 2018–08–09.
Acknowledgements
The authors wish to thank Brian Iken, a lecturer at the University of Houston, for his help in project management while research was conducted in the Iyer Laboratory and the editing of the final manuscript.
Author information
Authors and Affiliations
Contributions
All research work was conducted in RI’s research laboratory at the University of Houston. RI supervised and directed all research work conducted in the laboratory. NI performed all relevant experimentation and drafted the current manuscript. Both authors contributed to the proofreading of the final manuscript draft.
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Islam, N., Iyer, R. Functional Analysis of Chlorpyrifos Biodegradation in Agricultural Soils Augmented with a Three-Strain Bacterial Consortium. Water Air Soil Pollut 232, 425 (2021). https://doi.org/10.1007/s11270-021-05349-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11270-021-05349-z









 (-) control,
(-) control,
 Non-augmented samples,
Non-augmented samples,
 A + B + C,
A + B + C,
 A + B,
A + B,
 B + C,
B + C,
 A + C. Error bars represent a confidence limit of 95%
A + C. Error bars represent a confidence limit of 95%
 (-) control,
(-) control,
 non-augmented samples,
non-augmented samples,
 A + B + C,
A + B + C,
 A + B,
A + B,
 B + C,
B + C,
 A + C. Error bars represent a confidence limit of 95%
A + C. Error bars represent a confidence limit of 95%
 (-) control,
(-) control,
 non-augmented samples,
non-augmented samples,
 A + B + C,
A + B + C,
 A + B,
A + B,
 B + C,
B + C,
 A + C. Error bars represent a confidence limit of 95%
A + C. Error bars represent a confidence limit of 95%
 (-) control,
(-) control,
 non-augmented ranch soil,
non-augmented ranch soil,
 Non-augmented garden soil,
Non-augmented garden soil,
 Non-augmented farm soil,
Non-augmented farm soil,
 augmented ranch soil,
augmented ranch soil,
 Augmented garden soil,
Augmented garden soil,
 Augmented farm soil. Error bars represent a confidence limit of 95%
Augmented farm soil. Error bars represent a confidence limit of 95%