Abstract
Biopurification systems (BPS) are employed for the treatment of pesticide-containing wastewaters. In this work, a biomixture (active core of BPS) complemented by the addition of the fungus Trametes versicolor was evaluated for the elimination of a mixture of pesticides under different treatment conditions. The biomixture achieved high removal of all the pesticides assayed after 16 d: atrazine (68.4%, t1/2: 9.6 d), carbendazim (96.7%, t1/2: 3.6 d), carbofuran (98.7%, t1/2: 3.1 d) and metalaxyl (96.7%, t1/2: 3.8 d). Variations in the treatment conditions including addition of the antibiotic oxytetracycline and co-bioaugmentation with a bacterial consortium did not significantly affect the removal performance of the biomixture. Bacterial and fungal community profiles determined by DGGE analyses revealed changes that responded to biomixture aging, and not to antibiotic or pesticide addition. The proposed biomixture exhibits very efficient elimination during simultaneous pesticide application; moreover, the matrix is highly stable during stressful conditions such as the co-application of antibiotics of agricultural use.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Diverse approaches (biological and physicochemical) have been designed for the mitigation of effects derived from contamination by pesticides; nevertheless, most of them are not cost-effective, which restricts their field use in farms (Rodríguez-Rodríguez et al. 2013). For this reason, biologically active systems, named pesticide biopurification systems (BPS) were developed. BPS aim to treat pesticide-containing wastewater derived from the management of concentrated formulations and the diluted application solutions, to avoid point-source pollution from events such as spills, leaks or the cleansing of application equipment (Castillo et al. 2008).
BPS promote microbial degradation and pesticide adsorption to decrease the adverse effects of agrochemicals in the ecosystems (Castillo et al. 2008). Accelerated removal of pesticides occurs in the biomixture, which is the biologically active matrix of BPS.
The biomixture is composed by three materials, each in charge of specific functions: soil, a humic material, and a lignocellulosic substrate. Soil provides both adsorption capacity and degrading-microbiota; therefore, soil pre-exposed to the target pesticide is usually preferred (Sniegowski et al. 2011). The humic material contributes to the adsorption too, but also favors the humidity control and pesticide removal (Castillo et al. 2008). Finally, lignocellulosic wastes enhance the colonization by ligninolytic fungi, whose enzymatic activity participates in the transformation of several organic contaminants.
Operational BPS are usually exposed to various pesticides with different modes of action at one given time. Studies have shown that the elimination of several pesticides might be slower when applied simultaneously than when applied individually to biomixture systems (Fogg et al. 2003). Furthermore, antimicrobial agents (fungicides and antibiotics) are often simultaneously discarded with pesticides in BPS systems and can potentially disrupt their pesticide removal capabilities by affecting their indigenous degrading microbiota.
In order to face these challenges and improve the removal of complex mixtures of compounds in BPS, bioaugmentation has arisen as a promising option. Bioaugmentation involves the addition of high concentrations of specialized microbial populations to a natural environment with the purpose of increasing the rate or extent to which a pollutant is removed or metabolized (Leahy and Colwell 1990). It is advantageous when compared with other treatment alternatives because of a relatively low cost and the fact that it is environmentally friendly (Carter and Jewell 1993; Edwards and Cox 1997). However, it has often been observed that biotic and abiotic stresses may cause the number of added organisms to decrease sharply not long after being added (Alexander 1999).
In this context, ligninolytic fungi have emerged as effective bioaugmentation agents. Ligninolytic fungi are capable of removing a wide range of organic contaminants thanks to their unspecific enzymatic activity, which includes lignin-modifying enzymes and the cythochrome P450 complex (Doddapaneni and Yadav 2004; Pointing 2001). Their ability to degrade diverse pesticides such as lindane, atrazine and diuron has been demonstrated in the past (Bending et al. 2002; Hickey et al. 1994; Kennedy et al. 1990; Quintero et al. 2007; Singh and Kuhad 1999). Given the multiple substrates that may be used in BPS composition and the great variety of compounds to be treated, relatively few reports in the literature have dealt with degradation of pesticides by ligninolytic fungi in BPS (Bending et al. 2002; Madrigal-Zúñiga et al. 2016; Rigas et al. 2007; von Wirén-Lehr et al. 2001).
Likewise, the use of bacterial strains or consortia for improving pesticide-degradation efficiency in BPS have only been marginally studied showing mixed results (Briceño et al., 2013; Verhagen et al., 2013). Recently, a biomixture bioaugmented with a carbofuran-degrading consortium was assessed for the removal of a mixture of pesticides (atrazine, carbendazim, carbofuran and metalaxyl) while the antibiotic oxytetracycline was also applied (Castro-Gutiérrez et al., 2018). Even though very good rates and extent of removal were obtained for the pesticides, these were not affected by the addition of the bacterial degrading consortium.
Aiming to improve pesticide removal rates, we explored the use of a biomixture with an identical composition as the one just described, but in which the lignocellulosic component had been pre-colonized by the ligninolytic fungus Trametes versicolor. The objectives were to assess the pesticide removal capabilities and microbial community changes of this amended biomixture under co-bioaugmentation with a pesticide-degrading bacterial consortium, while also assessing the effect of the addition of the antibiotic oxytetracycline.
2 Materials and Methods
2.1 Chemicals
Commercial formulations and standards of carbofuran, atrazine, carbendazim, metalaxyl and oxytetracycline were employed in the removal assays and analytical determinations, respectively; details can be found in Supplementary Material. Surrogate (carbofuran-d3) and internal standard (linuron-d6) were used in the analysis. Solvents and extraction chemicals are listed elsewhere (Ruiz-Hidalgo et al. 2014).
2.2 T. versicolor and Biomixture Preparation
A biomixture composed of coconut fiber precolonized with T. versicolor, garden compost and agricultural soil (45:13:42% v/v respectively) was prepared and used for all systems throughout the study. A blended mycelial suspension of T. versicolor (ATCC 42530) was prepared according to Font-Segura et al. (1993), using Sabouraud broth. The precolonization of the coconut fiber with T. versicolor was performed before biomixture preparation. Containers (1.0 L) with humidified coconut fiber were sterilized (121 °C, 15 min) before inoculation of the fungal suspension (ratio: 1 mL suspension in 10 g coconut fiber). After fungal colonization for 18 d at 25(±1) °C (with manual homogenization every 6 d), biomixtures were prepared by adding soil (with a history of carbofuran application) and garden compost, at a ratio of precolonized-coconut fiber/compost/soil of 45:13:42% (v/v), followed by manual homogenization. Biomixture composition had been previously optimized for the elimination and detoxification of carbofuran (Chin-Pampillo et al. 2015a, Chin-Pampillo et al. 2015b).
2.3 Experimental Set-up and Sampling
Biomixture systems were set up to follow pesticide removal and changes in the microbial community throughout time. The effects of the addition of a mixture of pesticides (atrazine, carbofuran, carbendazim and metalaxyl), bioaugmentation with a previously obtained carbofuran-degrading bacterial consortium (Castro-Gutiérrez et al. 2016) and the addition the antibiotic oxytetracycline were studied. Six different treatments were set up as shown in Table 1.
The treatments were assayed in systems consisting of 5 g of the T. versicolor-amended biomixture (pH 6.4) in 50 mL Falcon tubes. For each treatment, triplicate systems per time point were set up for pesticide quantification; additional replicates were used for DNA extraction and DGGE analyses. All systems were homogenized and incubated (25 °C) for a period of 64 d. Humidity was adjusted every 4 d with sterile Mili-Q water.
The bacterial consortium had been obtained from a pesticide-degrading biomixture (coconut fiber, garden compost, soil pre-exposed to carbofuran; 50:25:25, v/v), and the inocula were prepared as previously described (Castro-Gutiérrez et al. 2016; Castro-Gutiérrez et al. 2018) and, where appropriate, added to specific biomixture systems at a concentration of 1.14 × 106 CFU g−1 as determined by plate counts.
2.4 Analytical Procedures
2.4.1 Extraction and Quantification of Pesticides
Pesticide extraction and quantification were carried out exactly as described elsewhere (Castro-Gutiérrez et al. 2018) using UPLC-MS/MS for analysis. Residual concentrations were employed to determine the extent of pesticide removal (%). Data was adjusted to a first order model (using SigmaPlot 12.5) to estimate removal rate constants (k) and pesticide half-lives (t1/2). ANOVA tests were used to compare removal rate constants (by comparison of regression lines with the STATGRAPHICS Centurion software XVII, Statpoint Technologies, Inc.).
2.5 Analysis of Microbial Communities
Biomixture samples of 0.40 g were employed for the extraction of total genomic DNA, using the NucleoSpin Soil DNA Isolation Kit (Macherey-Nagel GmbH & Co., Germany). For DNA extraction of the strain of T. versicolor, the protocol described by Płaza et al. (2004) was strictly followed. Total genomic DNA from the bacterial consortium was obtained as previously described (Castro-Gutiérrez et al. 2018). Purity and concentration of DNA extracted was analyzed in a 2000c NanoDrop spectrophotometer (Thermo Fischer Scientific, Wilmington, DE). Extracted DNA was stored at −20 °C until use. PCR amplification and DGGE analyses were performed according to Castro-Gutiérrez et al. (2017).
3 Results and Discussion
3.1 Removal of Pesticides in the Different Biomixture Treatments
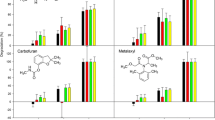
The biomixture was exposed to a combination of pesticides with different modes of action: carbendazim and metalaxyl (fungicides), atrazine (herbicide), and carbofuran (insecticide). Moreover, selected systems were also exposed to oxytetracycline and bioaugmented with a carbamate-degrading bacterial consortium.
Figure 1 illustrates the pesticide removal profiles for the different compounds and treatments; their respective half-lives and removal rate constants are shown in Table 2. For each one of the pesticides, removal half-lives showed no statistically significant differences (p > 0.05) between all the treatments; i.e., bacterial bioaugmentation, oxytetracycline co-application or the combination of both caused no significant changes on pesticide removal rates for this biomixture. Even so, carbendazim, carbofuran and metalaxyl were almost completely degraded for all systems at the 16 d time point, while atrazine removal was lower (67.2–72.2%).
In general, low removal half-lives were obtained when compared to other studies in soil and biomixture systems. For carbendazim, values are similar to those found for biomixtures and soil after previous exposure to the compound (Castillo-González et al. 2017; Huete-Soto et al. 2017b; Tortella et al. 2013b; Yu et al. 2009), which highlights its applicability for carbendazim removal. The removal of carbofuran was efficient and in line with half-life values obtained for biomixtures prepared with carbofuran-primed soil (Chin-Pampillo et al. 2015a). Half-lives for metalaxyl in non-primed biomixtures range from 26 to 37 d, while studies for soils have found values between 36 and 39 d (De Wilde et al. 2010; Lewis et al. 2016; Vischetti et al. 2008), all of them higher if compared to 3.8 to 4.0 d in this study. For atrazine, half-lives are similar to those found in other studies using biomixtures prepared with previously exposed soils (Chu and Eivazi 2015; Tortella et al. 2013a; Tortella et al. 2013c), while unexposed soils alone generally exhibit longer half-lives, even reaching 75 d (Lewis et al. 2016; Zablotowicz et al. 2006).
Castro-Gutiérrez et al. (2018) had previously evaluated a biomixture with an overall identical composition to the one used in this study, using the same batches of materials, but without fungal pre-colonization in the coconut fiber (coconut fiber, garden compost and agricultural soil 45:13:42% v/v respectively) and determined pesticide half-lives. Overall similar values to this study were obtained for atrazine (t1/2: 8.9–9.9 d), carbendazim (t1/2: 2.9–3.1 d), carbofuran (t1/2: 2.5–2.8 d) and metalaxyl (t1/2: 2.7–2.8 d).
T. versicolor has been shown to be capable of degrading atrazine and carbofuran in different conditions in the past. The fungus degraded atrazine on clay soil and sawdust (85–90%) and in a sterile biomixture (Bending et al. 2002; Bastos and Magan 2009). For carbofuran, Mir-Tutusaus et al. (2014) found extensive degradation by mycelial pellets of T. versicolor in defined liquid medium. Madrigal-Zúñiga et al. (2016) found addition of T. versicolor to be more effective when assayed for carbofuran removal in a peat-based biomixture as opposed to a compost-based one. The authors cited the higher C/N ratio and lower pH values derived from using peat instead of compost as part of the reasons for this finding; these factors have been found to promote the colonization and production of degrading enzymes by ligninolytic fungi (Eggert et al., 1996; Tavares et al., 2006). Furthermore, crude laccase extracts produced by T. versicolor have been found to express maximum activity at pH 4.5 (Stoilova et al. 2010). As compost was used as humic substance in this work, the biomixture had a relatively high pH of 6.4, which may explain why the addition of the fungus did not seem to improve atrazine and carbofuran removal vs. the biomixture without it.
As to the fungicides (metalaxyl and carbendazim), results from other studies regarding the efficiency of their removal using T. versicolor have been relatively poor. The fungus degraded metalaxyl in a sterile biomixture at low levels (Bending et al. 2002). Negligible carbendazim removal by T. versicolor has been reported in defined medium (Murillo-Zamora et al. 2017). Furthermore, carbendazim and other benzimidazole fungicides have been used in formulations for wood preservation against white rot fungi (WRF) such as Trametes sp. (González-Laredo et al. 2015).
No significant differences in the removal half-lives in all the assayed treatments for each of the four pesticides indicate that the addition of oxytetracycline is not interfering with pesticide-removal capabilities of this particular biomixture.
In a recent report (Huete-Soto et al. 2017a) no effect of oxytetracycline on atrazine half-lives was found in a biomixture of the same proportional composition as the one used in this study but with a different mixture of pesticides. Similarly, Jiménez-Gamboa et al. (2018) found that removal or accelerated degradation of carbofuran were not negatively affected by the addition of oxytetracycline in a similar biomixture. Regarding the fungicides, Castillo-González et al. (2017) found that the combination of oxytetracycline and gentamycin from commercial formulations did not affect the removal of carbendazim and metalaxyl in a biomixture of similar composition as the one employed in this work. Contrastingly, oxytetracycline has been shown to delay the removal of carbendazim and metalaxyl also in a similar matrix (Huete-Soto et al. 2017b), nevertheless with a different mixture of pesticides. Altogether, data suggest that the impact of oxytetracycline on the removal of these fungicides in this biomixture may be affected by the combination of pesticides applied at a given time.
In this case, the lack of effect of the antibiotic on pesticide removal efficiency may be determined by a complex combination of factors that includes the particular autochthonous microbial communities, physicochemical characteristics of the pesticides and matrix, and the addition of specific combinations of agrochemicals.
Regarding bacterial bioaugmentation, the lack of difference in pesticide removal rate constants and half-lives between the treatments with and without the carbofuran-degrading consortium (P vs PBB and PO vs PBBO) indicates that the carbofuran-degrading capabilities of the biomixture were not enhanced. Literature cites small inoculum, failure to adapt to new environmental conditions, competition with native microorganisms and predation as some of the main reasons for a bioaugmentation failure (Bouchez et al. 2000). The degrading consortium was originally isolated from a biomixture with a similar composition to the one used in this study and successful bioaugmentation of other biomixtures has been carried out with similar inoculum levels (Goux et al. 2003; Struthers et al. 1998). Therefore, the lack of enhancement in carbofuran removal by bacterial bioaugmentation might have been due to competition with native microflora.
3.2 Alteration of Microbial Communities during Different Treatments in the Biomixture
DGGE analyses were employed to determine changes for both fungal and bacterial communities (Fig. S1, Supplementary Material). For all profiles, changes in the community were more evident at the 16, 32 and 64 d time points. As to fungal DGGE profiles, the main band corresponding to T. versicolor was clearly seen during all the treatments, however in some of them two prominent bands appeared associated with T. versicolor, even though the samples were generated from the same template DNA in the same single round of PCR amplification. Neilson et al. (2013) showed that a single amplicon may present multiple bands in DGGE corresponding to different structural conformations, which may explain the observation in this study. Regardless of the number, the intensity of the band(s) usually decreased slightly at time 32 d, while the intensity of multiple other bands in the fingerprint increased; nonetheless, it still remained as the most prominent band in the profile. At time 64 d the magnitude of other bands consistently surpassed that of T. versicolor. This indicates that even though the fungus remains in the biomixture throughout the whole assay, its relative importance in the fungal community decreases through time. Persistence of T. versicolor has not been corroborated for other BPS applications; however, its success in soil colonization varies widely (Borràs et al. 2010; McErlean et al. 2006, Novotný et al., 1999). Even though the ability of T. versicolor to persist in a compost-based biomixture with diverse resident microbiota was demonstrated, it did not effectively translate in enhanced pesticide removal.
UPGMA cluster and nMDS analyses for paired treatments of bacterial amplicons are shown in Fig. 2. Figure 2A represents the treatments lacking pesticides: NP and NPBB (with bacterial consortium). DGGE profiles for consortium-bioaugmented treatments did not show the persistence of the inoculum throughout time, probably because the magnitude of the inoculum was low relative to the overall bacterial population in the biomixture. Cluster analysis revealed high similarity between the consortium-bioaugmented samples and their respective non-bioaugmented controls for each time point (81–88%) which indicates that bacterial bioaugmentation is not generating substantial changes in the main bacterial populations of the matrix. However, moderate changes, probably related to aging (Castillo et al. 2008; Castro-Gutiérrez et al. 2017) and/or environmental factors (relative drought, rewetting) (Sniegowski et al. 2011), are visible through time in the nMDS analysis.
Cluster and non-metric multidimensional scaling (nMDS) analysis of DGGE profiles of bacterial amplicons of a WRF-amended biomixture under different pesticide addition, oxytetracycline addition and bacterial consortium bioaugmentation schemes. NP: no pesticides; P: pesticides; PO: pesticides + oxytetracycline; NPBB: no pesticides + bacterial bioaugmentation; PBB: pesticides + bacterial bioaugmentation; PBBO: pesticides + bacterial bioaugmentation + oxytetracycline. Scale on UPGMA graphs indicates similarity. Numbers on nMDS plots indicate sampling day
DGGE profiles for two pesticide-amended treatments, one with oxytetracycline (PO) and one without it (P), are illustrated in Fig. 2B. Similarity values between 83% and 95% between time-paired samples show that addition of the antibiotic formulation did not generate considerable shifts in the overall profile of the main bacterial communities. However, as in the previous pair of biomixture treatments, aging and environmental factors are seemingly driving bacterial community drift. Given the elevated similarity values between time-paired samples for treatments PBB and PBBO (83–93%), similar conclusions can be drawn from the cluster and nMDS analysis of paired samples, depicted in Fig. 2C.
In an attempt to reveal differences between biomixture configurations, an additional DGGE analysis was performed with samples from times 16, 32 and 64 for all conditions simultaneously (Fig. 3). For the bacterial profiles (Fig. 3A) clusters with similarities of 77%, 84% and 82% can be observed for all the samples from times 16 d, 32 d, and 64 d respectively. Little divergence for all samples for the same time points indicates that none of the assayed factors in the initial conditions, namely pesticide mixture addition, bacterial bioaugmentation or oxytetracycline addition, or their combinations are generating significant changes in the overall main bacterial community structure of the biomixture.
Cluster and non-metric multidimensional scaling (nMDS) analysis of DGGE profiles of bacterial (A) and fungal (B) amplicons of a WRF-amended biomixture for all pesticide addition, oxytetracycline addition and bacterial consortium bioaugmentation schemes, restricted to sampling days 16, 32 and 64 for bacteria, or days 32 and 64 for fungi. NP: no pesticides; P: pesticides; PO: pesticides + oxytetracycline; NPBB: no pesticides + bacterial bioaugmentation; PBB: pesticides + bacterial bioaugmentation; PBBO: pesticides + bacterial bioaugmentation + oxytetracycline. Scale on UPGMA graphs indicates similarity. Numbers on nMDS plots indicate sampling day
Cluster and nMDS analyses of fungal fingerprints for the different treatments are shown in Fig. 4. A common observation throughout the sample patterns is that at time 0 d, the main bands associated with T. versicolor clearly dominated the fingerprint, while gradually, as commented before, their relative importance decreased; usually at 32 d a relatively complex banding pattern was present for all the treatments. Figure 4A illustrates fungal community structure changes throughout the assay for the treatments without pesticide application (NP and NPBB). Similarity levels from 74% to 96% were obtained for time-paired samples, indicating that bacterial bioaugmentation did not exert significant shifts on the main fungal community of these samples. Nonetheless, as with previous samples, aging effect was clearly visible.
Cluster and non-metric multidimensional scaling (nMDS) analysis of DGGE profiles of fungal amplicons of a WRF-amended biomixture under different pesticide addition, oxytetracycline addition and bacterial consortium bioaugmentation schemes. NP: no pesticides; P: pesticides; PO: pesticides + oxytetracycline; NPBB: no pesticides + bacterial bioaugmentation; PBB: pesticides + bacterial bioaugmentation; PBBO: pesticides + bacterial bioaugmentation + oxytetracycline. Scale on UPGMA graphs indicates similarity. Numbers on nMDS plots indicate sampling day
Analyses for paired treatments P and PO, and PBB and PBBO, are displayed on Fig. 4B and C respectively, to illustrate the effect of oxytetracycline addition on the fungal community of the samples not bioaugmented and bioaugmented with the bacterial consortium. Similarity levels from 69% to 87% (P and PO) and from 68% to 91% (PBB and PBBO) were obtained for each series of time-paired samples. While low to moderate divergence between samples was observed halfway through the assay, nMDS analyses failed to show consistent distinction between systems containing or lacking oxytetracycline; hence the differences could not be attributed to the addition of the antibiotic formulation. In order to reveal additional differences between the treatments, samples in which higher fungal ribotype diversity was observed (32 and 64 d) were compared for all conditions in an additional DGGE run (Fig. 3B). As was the case with bacterial samples, grouping of samples from the different treatments in time-related clusters indicates that biomixture aging is the main factor guiding fungal population modifications.
No clear pattern for pesticide addition, oxytetracycline addition or bacterial bioaugmentation can be recognized to be substantially shaping bacterial or fungal community structure throughout the assay. Instead most changes appear to be associated with biomixture aging. Previous studies have determined that after the addition of pesticides to BPS the microbial community may undergo mild changes (Castro-Gutiérrez et al. 2017; Chen et al. 2015; Tortella et al. 2013a; Tortella et al. 2013b) or in other cases may suffer stronger transient effects on microbial community structure (Coppola et al. 2011, Marinozzi et al. 2013).
As mentioned before, DGGE profiles show that T. versicolor remains in the biomixture throughout the whole assay. Even though degradation of atrazine, carbofuran and metalaxyl by the fungus has been demonstrated in some conditions (Bastos and Magan 2009; Bending et al. 2002) or disproven for metalaxyl and carbendazim in others (Murillo-Zamora et al. 2017), its presence is not causing significant changes in pesticide removal rate constants.
4 Conclusions
Altogether, each of the four pesticides contained in the mixture was effectively removed by the biomixture. This was true regardless of oxytetracycline addition; however bacterial bioaugmentation did not improve removal rates in any of these cases. Bacterial and fungal community structure was modified mainly as a consequence of biomixture aging, with pesticide addition and antibiotic addition having no palpable effect. Additionally, the presence of the fungal inoculant was corroborated throughout the entire assay for all treatments. The results indicate that the biomixture formulation is robust and has a high efficiency of pesticide removal.
References
Alexander, M. (1999). Biodegradation and bioremediation. San Diego: Academic Press.
Bastos, A. C., & Magan, N. (2009). Trametes versicolor: Potential for atrazine bioremediation in calcareous clay soil, under low water availability conditions. International Biodeterioration and Biodegradation, 63, 389–394.
Bending, G. D., FrilouxM., & Walker, A. (2002). Degradation of contrasting pesticides by white rot fungi and its relationship with ligninolytic potential. FEMS Microbiology Letters, 212, 59–63.
Borràs, E., Caminal, G., Sarrà, M., & Novotný, Č. (2010). Effect of soil bacteria on the ability of polycyclic aromatic hydrocarbons (PAHs) removal by Trametes versicolor and Irpex lacteus from contaminated soil. Soil Biology and Biochemistry, 42, 2087–2093.
Bouchez, T., Patureau, D., Dabert, P., Juretschko, S., Dore, J., Delgenes, P., Moletta, R., & Wagner, M. (2000). Ecological study of a bioaugmentation failure. Environmental Microbiology, 2, 179–190.
Briceño, G., Rubilar, O., & Tortella, G. (2013). Bioaumentación de una biomezcla con actinobacterias degradadoras de residuos de plaguicidas organofosforados. Pucón: Workshop Internacional y Taller Nacional Valorización.
Carter, S. R., & Jewell, W. J. (1993). Biotransformation of tetrachloroethylene by anaerobic attached films at low temperatures. Water Research, 27, 607–615.
Castillo, M. D. P., Torstensson, L., & Stenström, J. (2008). Biobeds for environmental protection from pesticide use, a review. Journal of Agricultural and Food Chemistry, 56, 6206–6219.
Castillo-González, H., Pérez-Villanueva, M., Masís-Mora, M., Castro-Gutiérrez, V., & Rodríguez-Rodríguez, C. E. (2017). Antibiotics do not affect the degradation of fungicides and enhance the mineralization of chlorpyrifos in biomixtures. Ecotoxicology and Environmental Safety, 139, 481–487.
Castro-Gutiérrez, V., Masís-Mora, M., Caminal, G., Vicent, T., Carazo-Rojas, E., Mora-López, M., & Rodríguez-Rodríguez, C. E. (2016). A microbial consortium from a biomixture swiftly degrades high concentrations of carbofuran in fluidized-bed reactors. Process Biochemistry, 51, 1585–1593.
Castro-Gutiérrez, V., Masís-Mora, M., Diez, M. C., Tortella, G. R., & Rodríguez-Rodríguez, C. E. (2017). Aging of biomixtures: Effects on carbofuran removal and microbial community structure. Chemosphere, 168, 418–425.
Castro-Gutiérrez, V., Masís-Mora, M., Carazo-Rojas, E., Mora-López, M., & Rodríguez-Rodríguez, C. E. (2018). Impact of oxytetracycline and bacterial bioaugmentation on the efficiency and microbial community structure of a pesticide-degrading biomixture. Environmental Science and Pollution Research, 25, 11787–11799.
Chen, Q., Yang, B., Wang, H., He, F., Gao, Y., & Scheel, R. A. (2015). Soil microbial community toxic response to atrazine and its residues under atrazine and lead contamination. Environmental Science and Pollution Research, 22, 996–1007.
Chin-Pampillo, J. S., Ruiz-Hidalgo, K., Masís-Mora, M., Carazo-Rojas, E., & Rodríguez-Rodríguez, C. E. (2015a). Adaptation of biomixtures for carbofuran degradation in on-farm biopurification systems in tropical regions. Environmental Science and Pollution Research, 22, 9839–9848.
Chin-Pampillo, J. S., Ruiz-Hidalgo, K., Masís-Mora, M., Carazo-Rojas, E., & Rodríguez-Rodríguez, C. E. (2015b). Design of an optimized biomixture for the degradation of carbofuran based on pesticide removal and toxicity reduction of the matrix. Environmental Science and Pollution Research, 22, 19184–19193.
Chu, B., & Eivazi, F. (2015). Enhancing biodegradation of herbicides using biobed systems. Journal of Environmental Indicators 9, 32–33.
Coppola, L., Castillo, P., & Vischetti, C. (2011). Degradation of isoproturon and bentazone in peat and compost-based biomixtures. Pest Management Science, 67, 107–113.
De Wilde, T., Spanoghe, P., Sniegowksi, K., Ryckeboer, J., Jaeken, P., & Springael, D. (2010). Transport and degradation of metalaxyl and isoproturon in biopurification columns inoculated with pesticide-primed material. Chemosphere, 78, 56–60.
Doddapaneni, H., & Yadav, J. S. (2004). Differential regulation and xenobiotic induction of tandem P450 monooxygenase genes pc-1 (CYP63A1) and pc-2 (CYP63A2) in the white-rot fungus Phanerochaete chrysosporium. Applied Microbiology and Biotechnology, 65, 559–565.
Edwards, E. A., & Cox, E. E. (1997). Field and laboratory evidence of sequential aerobic chlorinated solvent biodegradation. In In situ and on site bioreclamation (pp. 261–265). Columbus: Batelle Press.
Eggert, C., Temp, U., & Eriksson, K. E. (1996). The ligninolytic system of the white rot fungus Pycnoporus cinnabarinus: Purification and characterization of the laccase. Applied and Environmental Microbiology, 62, 1151–1158.
Fogg, P., Boxall, A. B., Walker, A., & Jukes, A. A. (2003). Pesticide degradation in a ‘biobed’composting substrate. Pest Management Science, 59, 527–537.
Font-Segura, X., Gabarrell-Durany, X., Lozano, R., & Vicent-Huguet, T. (1993). Detoxification pretreatment of black liquor derived from non-wood feedstock with white-rot fungi. Environmental Technology, 14, 681–687.
González-Laredo, R. F. G., Castro, M. R., Guzmán, N. E. R., Infante, J. A. G., Moreno-Jiménez, M. R., & Karchesy, J. J. (2015). Wood preservation using natural products. Madera Bosques, 21, 63–76.
Goux, S., Shapir, N., El Fantroussi, S., Lelong, S., Agathos, S. N., & Pussemier, L. (2003). Long-term maintenance of rapid atrazine degradation in soils inoculated with atrazine degraders. Water, Air, and Soil Pollution, 3, 131–142.
Hickey, W. J., Fuster, D. J., & Lamar, R. T. (1994). Transformation of atrazine in soil by Phanerochaete chrysosporium. Soil Biology and Biochemistry, 26, 1665–1671.
Huete-Soto, A., Castillo-González, H., Masís-Mora, M., Chin-Pampillo, J. S., & Rodríguez-Rodríguez, C. E. (2017a). Effects of oxytetracycline on the performance and activity of biomixtures: Removal of herbicides and mineralization of chlorpyrifos. Journal of Hazardous Materials, 321, 1–8.
Huete-Soto, A., Masís-Mora, M., Lizano-Fallas, V., Chin-Pampillo, J. S., Carazo-Rojas, E., & Rodríguez-Rodríguez, C. E. (2017b). Simultaneous removal of structurally different pesticides in a biomixture: Detoxification and effect of oxytetracycline. Chemosphere, 169, 558–567.
Jiménez-Gamboa, D., Castro-Gutiérrez, V., Fernández-Fernández, E., Briceño-Guevara, S., Masís-Mora, M., Chin-Pampillo, J. S., Mora-López, M., Carazo-Rojas, E., & Rodríguez-Rodríguez, C. E. (2018). Expanding the application scope of on-farm biopurification systems: Effect and removal of oxytetracycline in a biomixture. Journal of Hazardous Materials, 342, 553–560.
Kennedy, D. W., Aust, S. D., & Bumpus, J. A. (1990). Comparative biodegradation of alkyl halide insecticides by the white rot fungus, Phanerochaete chrysosporium (BKM-F-1767). Applied and Environmental Microbiology, 56, 2347–2353.
Leahy, J. G., & Colwell, R. R. (1990). Microbial degradation of hydrocarbons in the environment. Microbiology and Molecular Biology Reviews, 54, 305–315.
Lewis, K. A., Tzilivakis, J., Warner, D. J., & Green, A. (2016). An international database for pesticide risk assessments and management. Human and Ecological Risk Assessment, 22, 1050–1064.
Madrigal-Zúñiga, K., Ruiz-Hidalgo, K., Chin-Pampillo, J. S., Masís-Mora, M., Castro-Gutiérrez, V., & Rodríguez-Rodríguez, C. E. (2016). Fungal bioaugmentation of two rice husk-based biomixtures for the removal of carbofuran in on-farm biopurification systems. Biology and Fertility of Soils, 52, 243–250.
Marinozzi, M., Coppola, L., Monaci, E., Karpouzas, D. G., Papadopoulou, E., Menkissoglu-Spiroudi, U., & Vischetti, C. (2013). The dissipation of three fungicides in a biobed organic substrate and their impact on the structure and activity of the microbial community. Environmental Science and Pollution Research, 20, 2546–2555.
McErlean, C., Marchant, R., & Banat, I. M. (2006). An evaluation of soil colonisation potential of selected fungi and their production of ligninolytic enzymes for use in soil bioremediation applications. Antonie Van Leeuwenhoek, 90, 147–158.
Mir-Tutusaus, J. A., Masís-Mora, M., Corcellas, C., Eljarrat, E., Barceló, D., Sarrà, M., Caminal, G., Vicent, T., & Rodríguez-Rodríguez, C. E. (2014). Degradation of selected agrochemicals by the white rot fungus Trametes versicolor. Science of The Total Environment, 500, 235–242.
Murillo-Zamora, S., Castro-Gutiérrez, V., Masís-Mora, M., Lizano-Fallas, V., & Rodríguez-Rodríguez, C. E. (2017). Elimination of fungicides in biopurification systems: Effect of fungal bioaugmentation on removal performance and microbial community structure. Chemosphere, 186, 625–634.
Neilson, J. W., Jordan, F. L., & Maier, R. M. (2013). Analysis of artifacts suggests DGGE should not be used for quantitative diversity analysis. Journal of Microbiological Methods, 92, 256–263.
Novotný, Č., Erbanová, P., Šašek, V., Kubátová, A., Cajthaml, T., Lang, E., Krahl, J., & Zadražil, F. (1999). Extracellular oxidative enzyme production and PAH removal in soil by exploratory mycelium of white rot fungi. Biodegradation, 10, 159–168.
Płaza, G. A., Upchurch, R., Brigmon, R. L., Whitman, W. B., & Ulfig, K. (2004). Rapid DNA extraction for screening soil filamentous fungi using PCR amplification. Polish Journal of Environmental Studies, 13, 315–318.
Pointing, S. (2001). Feasibility of bioremediation by white-rot fungi. Applied Microbiology and Biotechnology, 57, 20–33.
Quintero, J. C., Lu-Chau, T. A., Moreira, M. T., Feijoo, G., & Lema, J. M. (2007). Bioremediation of HCH present in soil by the white-rot fungus Bjerkandera adusta in a slurry batch bioreactor. International Biodeterioration & Biodegradation, 60, 319–326.
Rigas, F., Papadopoulou, K., Dritsa, V., & Doulia, D. (2007). Bioremediation of a soil contaminated by lindane utilizing the fungus Ganoderma australe via response surface methodology. Journal of Hazardous Materials, 140, 325–332.
Rodríguez-Rodríguez, C. E., Castro-Gutiérrez, V., Chin-Pampillo, J. S., & Ruiz-Hidalgo, K. (2013). On-farm biopurification systems: Role of white rot fungi in depuration of pesticide-containing wastewaters. FEMS Microbiology Letters, 345, 1–12.
Ruiz-Hidalgo, K., Chin-Pampillo, J. S., Masís-Mora, M., Carazo, E., & Rodríguez-Rodríguez, C. E. (2014). Degradation of carbofuran by Trametes versicolor in rice husk as a potential lignocellulosic substrate for biomixtures: From mineralization to toxicity reduction. Process Biochemistry, 49, 2266–2271.
Singh, B. K., & Kuhad, R. C. (1999). Biodegradation of lindane (γ-hexachlorocyclohexane) by the white-rot fungus Trametes hirsutus. Letters in Applied Microbiology, 28, 238–241.
Sniegowski, K., Bers, K., Van Goetem, K., Ryckeboer, J., Jaeken, P., Spanoghe, P., & Springael, D. (2011). Improvement of pesticide mineralization in on-farm biopurification systems by bioaugmentation with pesticide-primed soil. FEMS Microbiology Ecology, 76, 64–73.
Stoilova, I., Krastanov, A., & Stanchev, V. (2010). Properties of crude laccase from Trametes versicolor produced by solid-substrate fermentation. Advances in Bioscience and Biotechnology, 1, 208–215.
Struthers, J. K., Jayachandran, K., & Moorman, T. B. (1998). Biodegradation of atrazine by Agrobacterium radiobacter J14a and use of this strain in bioremediation of contaminated soil. Applied and Environmental Microbiology, 64, 3368–3375.
Tavares, A. P. M., Coelho, M. A. Z., Agapito, M. S. M., Coutinho, J. A. P., & Xavier, A. M. R. B. (2006). Optimization and modeling of laccase production by Trametes versicolor in a bioreactor using statistical experimental design. Applied Biochemistry and Biotechnology, 134, 233–248.
Tortella, G. R., Mella-Herrera, R. A., Sousa, D. Z., Rubilar, O., Acuña, J. J., Briceño, G., & Diez, M. C. (2013a). Atrazine dissipation and its impact on the microbial communities and community level physiological profiles in a microcosm simulating the biomixture of on-farm biopurification system. Journal of Hazardous Materials, 260, 459–467.
Tortella, G. R., Mella-Herrera, R. A., Sousa, D. Z., Rubilar, O., Briceño, G., Parra, L., & Diez, M. C. (2013b). Carbendazim dissipation in the biomixture of on-farm biopurification systems and its effect on microbial communities. Chemosphere, 93, 1084–1093.
Tortella, G. R., Rubilar, O., Stenström, J., Cea, M., Briceño, G., Quiroz, A., Diez, M. C., & Parra, L. (2013c). Using volatile organic compounds to enhance atrazine biodegradation in a biobed system. Biodegradation, 24, 711–720.
Verhagen, P., De Gelder, L., & Boon, N. (2013). Inoculation with a mixed degrading culture improves the pesticide removal of an on-farm biopurification system. Current Microbiology, 67, 466–471.
Vischetti, C., Monaci, E., Cardinali, A., Casucci, C., & Perucci, P. (2008). The effect of initial concentration, co-application and repeated applications on pesticide degradation in a biobed mixture. Chemosphere, 72, 1739–1743.
von Wirén-Lehr, S., del Pilar Castillo, M., Torstensson, L., & Scheunert, I. (2001). Degradation of isoproturon in biobeds. Biology and Fertility of Soils, 33, 535–540.
Yu, Y., Chu, X., Pang, G., Xiang, Y., & Fang, H. (2009). Effects of repeated applications of fungicide carbendazim on its persistence and microbial community in soil. Journal of Environmental Sciences, 21, 179–185.
Zablotowicz, R. M., Weaver, M. A., & Locke, M. A. (2006). Microbial adaptation for accelerated atrazine mineralization/degradation in Mississippi Delta soils. Weed Science, 54, 538–547.
Acknowledgements
The authors acknowledge Vicerrectoría de Investigación, Universidad de Costa Rica (projects 802-B4-503 and 802-B6-137), and the Costa Rican Ministry of Science, Technology and Telecommunications, MICITT (project FI-093-13).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 1125 kb)
Rights and permissions
About this article
Cite this article
Castro-Gutiérrez, V., Masís-Mora, M., Carazo-Rojas, E. et al. Fungal and Bacterial Co-Bioaugmentation of a Pesticide-Degrading Biomixture: Pesticide Removal and Community Structure Variations during Different Treatments. Water Air Soil Pollut 230, 247 (2019). https://doi.org/10.1007/s11270-019-4282-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11270-019-4282-y