Abstract
The biomass characteristics, the process performance, and the microbial community for a sequencing batch reactor (SBR) and a submerged membrane SBR (MSBR) were evaluated. A synthetic wastewater containing only 4-chlorophenol (4CP) was used as the sole source of carbon and energy. Degradation efficiencies of 4CP were higher than 99% for both reactors, and no significant differences on the 4CP degradation rates were observed for the SBR (116.9 ± 0.9 mg 4CP g VSS−1 h−1) as well as for the MSBR (117.3 ± 0.5 mg 4CP g VSS−1 h−1). Despite the similar results obtained for the physicochemical parameters, it was found that the biomass characteristics were different considering the sludge volumetric index, settling velocity, protein content in the mixer liquor, and total suspended solids in the effluent. The settling velocity was three times higher in the SBR than in the MSBR; however, a better quality, considering suspended solids, was observed for the MSBR. The protein concentration in the mixed liquor was higher in the MSBR than in the SBR, generating foaming problems in the MSBR. A similarity analysis was made with the Ochiai–Barkman index. Even though the reactors were inoculated with the same biomass, significant differences in the composition and populations dynamics were observed.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Sequencing batch reactor (SBR) processes have demonstrated their efficiency and flexibility in the treatment of wastewaters with high concentrations of nutrients, nitrogen, phosphorous, and toxic compounds from domestic and industrial sources (Wilderer et al. 2001). The SBR operates under five well-defined phases: fill, react, settle, draw, and idle (Mace and Mata-Alvarez 2002). Most of the advantages of SBR processes may be attributed to their single-tank design and the flexibility allowing them to meet many different treatment objectives, which derives from the possibility of adjusting the duration of the different phases. These batch systems can easily be adapted for continuous variations of pollutant concentrations, which is a general problem found in industrial wastewater treatment plants. For instance, toxic compounds can cause inhibition of microorganisms, which is reflected in a decrease in bacterial activity or even death (Coello Oviedo et al. 2003). To avoid this problem, several strategies, using the discontinuous processes, have been explored in order to increase the biotreatment efficiencies of industrial wastewaters (Wilderer et al. 2001), as in the case of phenolic compounds (Moreno-Andrade et al. 2006a).
Phenolic compounds are commonly found in industrial wastewaters. Typical phenol concentrations for oil refining, pharmacy, electroplanting, paper making, cooking, and iron smelting industries are in the range of 35–400 mg/L (Chen et al. 1997). Chlorophenols are common environmental pollutants that arise from extensive use of wood preservatives, pesticides, herbicides, and fungicides, and they are found frequently in pulp and paper effluents and industrial wastewater. Monochlorophenols can be formed during wastewater chlorination and as a result of the breakdown of pesticides and chlorinated aromatic compounds (Pritchard et al. 1987). Due to its hazardous properties, intensive attention has been paid to the biological degradation of chlorophenols.
On the other hand, membrane technologies have been used recently in order to obtain high-quality effluent in wastewater treatment plants (Tao et al. 2005; Holbrook et al. 2005; Le-Clech et al. 2006). The main goal of the membranes is to separate biomass from the mixed liquor in an integrated step. Due to this separation, it is possible to have a cellular retention time that is independent of the hydraulic retention time needed in the process (Le-Clech et al. 2006; Cicek et al. 2001).
Combining a membrane process with SBR technology may provide advantages for both processes (McAdam et al. 2005). The use of membranes can reduce the SBR cycle length since the settling phase is no longer required and clear water can be extracted during the reaction time. In addition, the separation of biological sludge by means of a membrane leads to complete retention of biomass resulting in a high mixed liquor suspended solids concentration. This allows a very high treatment capacity for a submerged membrane sequencing batch reactor (MSBR) (Kang et al. 2003). The MSBR has been applied for different wastewater including domestic and industrial wastewater, gray wastewater, nutrient-polluted water, reuse, removal of coliform bacteria, and landfill leachate treatment (Bae et al. 2003; Arrojo et al. 2005; Tsilogeorgis et al. 2008; Scheumann and Kraume 2009; Thanh et al. 2010).
However, a common problem in such systems is the membrane fouling. This problem reduces significantly the overall membrane performance, increasing operating cost and shortening membrane life (Fan et al. 2006). New strategies can be applied to avoid this problem; for example, the use of a controlled backwashing during the filtration phase (Vargas et al. 2008).
Other parameters that interact in the membrane performance and fouling are the sludge characteristics and the microbial community (Liang et al. 2007). Carucci et al. (2010) compared different discontinuous technologies to biodegrade a mixture of acetate and 4-chlorophenol (4CP). They found that SBR was the most suitable technology for the acetate and chlorophenol removal; however, the membrane bioreactor prevailed over the SBR regarding the daily waste sludge production and the effluent quality. Thus, it appears that different microbial communities can be developed. In this sense, the objective of this research was to compare the biomass characteristics, the process performance, and the microbial community between a classical SBR and a MSBR degrading a synthetic wastewater containing only 4CP. In particular, it was evaluated whether there are differences in the biodegradation process for an SBR and the MSBR, beyond the inherent filtration properties of the MSBR.
2 Materials and Methods
2.1 Experimental Setup
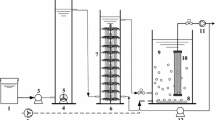
Two reactors with 6 L of useful volume and an exchange volume of 33% (3 L) were used. Figure 1 presents the experimental setup of both reactors. The airflow rate was 2.5 L min−1, and the temperature was maintained at 20°C inside the reactor. The two reactors were made of acrylic with a double wall for the recirculation of water at the controlled temperature. One reactor was operated as a classical SBR, whereas the other SBR was equipped with a submerged membrane (MSBR). The SBR presented a cylindrical shape with a conical bottom (23 cm diameter and 38 cm high). The MSBR was a rectangular prism (10 cm wide, 45 cm large, and 30 cm high). In the MSBR, the membrane was a tubular module of polyvinylidene fluoride with a molecular weight cutoff of 250 kDa and a total area of 0.04 m2. A diaphragm pump coupled to a pressure transducer (C206, Cole-Parmer, USA) and a precision water flow meter (L200CCM, Alicat Scientific, USA) were used to measure on-line the transmembrane pressure and the flux. Both flow and pressure were used to calculate the membrane resistance.
The reactors were inoculated with biomass from a municipal activated sludge wastewater treatment plant [2,000 mg volatile suspended solids (VSS) L−1]. Both reactors were operated using synthetic wastewater containing 400 mg L−1 of 4CP as the sole source of carbon and energy. Due to the exchange volume, the initial concentration inside the reactor was 200 mg L−1. Nutrients (nitrogen and phosphorus) and oligo-elements were added to the feed according to Moreno-Andrade and Buitrón (2004). The 4CP concentration was measured (as total phenols) by taking samples and processing them offline using the colorimetric technique of 4-aminoantipyrine (APHA et al. 2005). Total and volatile suspended solids, sludge volumetric index (SVI), settling velocity (SV), and chemical oxygen demand (COD) were determined according to the standard methods (APHA et al. 2005). Protein quantification was done according to the Lowry technique (Lowry et al. 1951).
The reactors were operated with a filling time of 5 min and a reaction time of 6 h (except for the acclimation, where the reaction was variable and depending on the time needed to reach at least the 97% of the total degradation of the substrate). In the SBR, settling and draw times were 30 and 5 min, respectively, while in the MSBR, permeation lasted approximately 90 min, with no settling. The permeate water was withdrawn at the end of the reaction phase through the membrane modules. The filtration was optimally controlled by an algorithm that used the on-line measured transmembrane pressure and volumetric flow to control the valves and relays using appropriate software according with Vargas et al. (2008). Both filtration and backwashing were done using two diaphragm pumps. The algorithm continually searches for the maximum flux, and whenever there is a loss in this maximal flux (in this case the maximal loss of flux was programmed at 50%), it starts backwashing for a predetermined time period (in this case it was fixed for 10 s). Afterwards, it resumes filtration and searches again for the maximum flux until the exchange volume is complete.
A kinetic analysis of the substrate degradation for the biomass obtained with the SBR and the MSBR was performed feeding the reactors with 200 mgL−1 of 4CP as in the usual operation of the reactors. Each condition represents the average of three kinetics. The Monod model (Eq. 1) was fitted using the Solver tool in Excel to evaluate K s and q max X; q max was obtained considering the average biomass concentration in each reactor.
Where, dS/dt or q represents the 4CP consumption rate in milligrams per hour, q max is the maximal consumption rate in milligrams 4CP liters per hour, X AV is the average VSS concentration in the mixed liquor in grams VSS per liter, S is the 4CP concentration in milligrams per liter, and K s is the saturation constant in milligrams 4CP per liter.
2.2 Comparison of the Microbial Communities
The comparison of the microbial communities was carried out using polymerase chain reaction (PCR) and denaturing gradient gel electrophoresis (DGGE) techniques. Samples of the reactor were taken after the biomass was acclimated (cycle 17th) and the last cycle (cycle 90th). Total DNA was extracted from 2-mL samples of activated sludge using an UltraCleanTM Soil DNA isolation kit (MoBio). PCR amplifications were conformed to a Bio-Rad temperature cycle, with the following program: 15 min of initial activation at 95°C, 35 cycles of denaturation at 94°C (1 min), annealing at 53°C (1 min), and elongation at 72°C (1 min) and a final extension of 72°C (1 min). The universal primers GC-338F and 518R were used for amplification according to Muyzer and Ramsing (1995). PCR products were evaluated by electrophoresis on a 0.2% (w/v) agarose gel. Negative controls, without DNA, were run in all amplifications. PCR products were visualized on a 2% agarose gel electrophoresis in the presence of a 100-bp ladder followed by staining with ethidium bromide. PCR products were separated by DGGE using a DCode universal mutation detection system (Bio-Rad, Hercules, CA) as described by Muyzer and Ramsing (1995). PCR products were separated on a 1.5-mm-thick vertical gel containing 8% (w/v) polyacrylamide (37.5:1 acrylamide/bisacrylamide) and a linear gradient of denaturant urea and formamide, increasing from 42.5% at the top of the gel to 62.5% at the bottom. Here, 100% denaturant is defined as 7 M urea plus 40% v/v formamide. Gels were run for 16 h at 60 V in 1× TAE buffer maintained at 60°C. PCR, DGGE, cloning, and sequencing techniques were used. The gel was dyed with ethidium bromide and analyzed in a Kodak UV transilluminator (302 nm). Specific PCR-DGGE bands were excised from the gel and purified with QIAquick PCR purification Kit (Qiagen, USA). All nucleotide sequences were determined at a DNA sequencing facility at the Instituto de Fisiología Celular–UNAM. Shannon-Wiener diversity index (H′) (Shannon and Weaver 1963) was used to evaluate the structural diversity between microbial communities in SBR and MSBR. DGGE was analyzed employing the PAST software in order to obtain the Ochiai-Barkman’s similarity index (Hammer et al. 2001).
3 Results and Discussion
3.1 MSBR and SBR Operational Parameters
The reactors were operated for 45 days (90 cycles); the biomass was acclimated to degrade the 4CP for 15 degradation cycles. After that time, the reactors showed a stable operation, and there were no significant differences on 4CP removal efficiencies (99.5 ± 0.7% and 99.3 ± 2.2% for SBR and MSBR, respectively). The biomass in both reactors was maintained at 2,259 ± 131 and 2,190 ± 228 mg L−1 for SBR and MSBR, respectively. In the case of the MSBR, there was a punctual decrease to 91% in the removal efficiency in cycle 57th, which occurred because the reaction time was not long enough for the complete degradation of the 4CP. This problem was solved by increasing the reaction phase duration to 12 h during four degradation cycles. After the degradation time was recovered to the normal operational values (around 2 h), the degradation cycle was fixed again in 6 h. This problem was associated with foaming in the MSBR, which caused the attachment of the biomass to the reactor walls. As a consequence, the contact between the substrate and the biomass decreased. Abundance of actinomycetes such as Nocardia or Microthrix is commonly related to foaming in activated sludge plants, and has been identified in membrane bioreactors (MBR) subject to variable organic loading rates. However, foaming in the MBR has also been observed in the absence of foam-forming microorganisms, and in these circumstances, the quantity of foam has been reported as being related to the protein content of extracellular polymeric substances (EPS; Judd 2006).
Figure 2 showed the degradation kinetics after the acclimation phase and during the stable operation of the reactors (cycles 39, 41, and 44 for SBR and 19, 23, and 28 for MSBR). As shown, the Monod model adjusted very well the experimental data. It was found that there were no significant differences between the 4CP degradation for both reactors since the degradation rates, q max, had similar values of 116.9 ± 0.9 and 117.3 ± 0.5 mg 4CP g VSS−1 h−1 for SBR and MSBR, respectively. The values obtained in this work contrast with those reported by Carucci et al. (2010). They found significant differences and a much lower degradation rates for a mixture of 4CP and acetate using a SBR (16.2 mg 4CP g VSS−1 h−1) and a MSBR (3.7 mg 4CP g VSS−1 h−1). The high degradation rates observed in our case can be attributed to the selection and multiplication of specialized microorganisms resulting when 4CP alone is used as the sole source of carbon and energy (Moreno-Andrade and Buitrón 2004), not only in the SBR but also in the MSBR. The K s values were 72 ± 15.6 and 70 ± 8.5 for SBR and MSBR, respectively. The removal efficiencies for COD were higher than 95% for both reactors, and the COD in effluent was 9.4 ± 4.8 and 11.3 ± 5.6 mg/L for the SBR and MSBR, respectively. The results of the COD values in the effluent were analyzed statistically by means of a normal distribution (STATGRAPHICS Plus 5.0, Manugistics Inc.). The statistical analysis determined that the standardized kurtosis values were −0.82 and 0.3 for SBR and MSBR data, determining that the samples come from a normal distribution (the values necessary for applying the normal distribution need to be in the range of −2 to 2 in order to obtain a valid analysis). The normal distribution plot is a descriptive analysis for a single variable to study, in this case the COD in effluent of both reactors, which showed that the effluent quality derived from both process presented excellent characteristics in terms of COD removal since statistically, in 95% of the samples, the COD in the effluent was smaller than 17 and 21.5 mg L−l for SBR and MSBR, respectively (Fig. 3a for SBR and 3b for MSBR). These facts suggested that communities with similar degradation capacities, regarding the biodegradation of 4CP, were acclimated in both reactors.
3.2 Differences Between Biomass Characteristics and Mixed Liquor
Despite the similar results obtained for the physicochemical parameters, it was found that the biomass characteristics were different considering the SVI, SV, total suspended solids (TSS) in the effluent, and protein content in the mixed liquor (Table 1). The settling velocity was three times higher in the SBR than in the MSBR. This behavior can be explained because of the dense flocs formation as a result of the periodical characteristic of the process. In this sense, it has been reported that exposition of the biomass to a feed-starvation regimen in a SBR, and in some cases to a short settling time, induces to a selection of biomass with high settling characteristics and decreases the presence of less dense flocs and free cells (Beun et al. 2002). For this reason, the biomass present in SBR can also aggregate to form granules, producing a high settling velocity rate (de Kreuk and van Loosdrecht 2004; McSwain et al. 2004). A dispersed growth was observed in the MSBR caused by the retention of all the suspended solids by the membrane. That included the microorganisms that are not attached to flocs and thus the production of biomass with a slower settling velocity than the biomass obtained with the SBR, as was also observed by McAdam et al (2005).
It has been reported that during the biological treatment of 4CP, inhibition, loss of viability, and cell lyses occur (Moreno-Andrade et al. 2006b); for this reason, carbohydrates and proteins may remain suspended in the reactor. The results showed that the protein concentration was sixfold higher in the mixed liquor of the MSBR (182.0 ± 17 μg mL−1) than in the SBR (30.2 ± 7.9 μg mL−1). This difference was due to the increase of the soluble microbial products, SMP, composed by the soluble cellular substances that are released during lysis and are retained by the membrane, generating an accumulation in the bulk of the reactor. These SMP, known to cause membrane clogging and foaming phenomenon, have also been correlated with membrane fouling (Lesjean et al. 2005; Judd 2006). In fact, the foaming observed in our MSBR seems to be produced by these SMP, which have the properties of surface-active agents. Di Bella et al. (2011) demonstrated that the foaming in MBR systems can be strongly influenced by a high EPS concentration in the aerobic tank. No foaming was observed in the case of the SBR. In this reactor, the protein content was withdrawn during the draw phase in each cycle. In the SBR’s effluent, the protein content was 25.9 ± 3.5, and this was not detected in the case of the MSBR.
It has been suggested that the excess proteins or EPS in the MSBR that produce fouling problems can be avoided with aerobic granular biomass. In this sense, it has been reported that granular biomass can increase the membrane permeability by 50% compared with a conventional membrane reactor and can reduce the membrane fouling (Li et al. 2005).
3.3 Comparison of the Microbial Communities
For the bacterial community evaluation, the V3 hypervariable region of the 16S rRNA gene was obtained by PCR amplification. The changes in the bacterial communities during operation of MSBR and SBR are shown in Fig. 4. Each band obtained in a DGGE analysis is related to a bacterial species in the tested sample. DGGE revealed only a moderate quantity of bands in both reactors because during the degradation of 4-chlorophenol, there exists a selection of microorganisms (Moreno-Andrade and Buitrón 2004). Nineteen different bands in the DGGE were observed, and only six were possible to excise and amplify for nucleotide sequence analysis.
Left GGE analysis of V3 fragments amplified by PCR of the SBR and MSBR reactors. a SBR, acclimated biomass, cycle 17; b SBR, cycle 90; c MSBR, acclimated biomass, cycle 19; d MSBR, cycle 90. Six bands were identified (GeneBank accession number and percentage of similarity in parenthesis): 1 Xanthomonas sp. (EU604075, 97%), 2 Parvibaculum sp. (EU604074, 97%), 3 Aquificae bacterium (EU604073, 100%), 4 Rhodanobacter sp. (EU604072, 100%), 5 Acinetobacter sp. (EU604071, 100%), 6 Halomonas sp. (EU604070, 96%). Right similarity analysis between the bands observed in DGGE analysis
After the acclimation, more defined bands (bands 1–3, line C in Fig. 4) were found for the MSBR than for the SBR, where only the band 1 clearly appeared (line A). The Shannon-Wiener index was 1.73 and 1.76 for SBR and MSBR, respectively. This demonstrated that there were no differences in the diversity index. However, taking into account the species found with the DGGE analysis, the number of microorganisms is not evenly distributed between all the species. A similarity analysis was made with the Ochiai-Barkman index because this index takes into account the bands present in the samples and also the exclusive bands between the samples (bands that are present only in one line; Fig. 4). It was found that the communities’ composition changed during that time due to the reactors’ operation. For the SBR, the samples demonstrated a dissimilarity of 19% in the microbial diversity, and for MSBR, the difference was 16% (Fig. 4). However, if the communities of the SBR and MSBR are compared, the similarity is only 58%; this demonstrates that the microbial populations are very different between the reactors. This can be attributed to the operation with membrane in MSBR since the inoculum, substrate, and general operation conditions were the same.
Six bands were excised and identified from DGGE analysis. Xanthomonas sp. was consistently present in all the samples, but the intensity of the bands was different in all the samples. Other species were found in both systems as Parvibaculum sp. and Rhodanobacter sp. seem to be involved in the 4-chlorophenol degradation pathway, having the capacity to produce a ring cleavage in aromatic compounds (Sapila et al. 2008). Halomonas sp. was only present in the MSBR at the end of the operation in cycle 90 (line D), indicating that in the MSBR, some species that participate in 4CP degradation were retained. This is important when some slow growth microorganisms must to be retained, as the case for some inhibitory compounds degradation. Acinetobacter sp. was only present in the SBR. The degradation of chlorophenolic compounds by these bacteria was reported by Sharma et al. (2009). It is interesting to note that the communities in each reactor showed differences in the population composition and dynamics, although the inoculum for both reactors was the same. This agrees with the results of other authors (Delbés et al. 2000; Etchebere et al. 2001; Molina-Muñoz et al. 2009) who reported a high microbial population dynamics on anaerobic and anoxic bioreactors, as well as in membrane bioreactors, although the kinetic degradation, removal efficiencies, and other physicochemical parameters remained stable.
4 Conclusions
It was found that there were no significant differences in the performance for the physicochemical parameters between the SBR and MSBR. An excellent performance regarding the 4CP degradation was observed in both reactors, indicating that communities with similar degradation capacities were acclimated in both reactors. Degradation efficiencies of 4CP were higher than 99% for both reactors, and no significant differences on the 4CP degradation rates were observed for the SBR (116.9 ± 0.9 mg 4CP g VSS−1 h−1) as well as for the MSBR (117.3 ± 0.5 mg 4CP g VSS−1 h−1). Despite the similar results obtained for the physicochemical parameters, it was found that the biomass characteristics were different considering the sludge volumetric index, settling velocity, protein content in the mixed liquor, and total suspended solids in the effluent. The settling velocity was three times higher in the SBR than in the MSBR; however, a better quality, considering suspended solids, was observed for the MSBR. The protein concentration in the mixed liquor was sixfold higher in the MSBR than in the SBR, generating foaming problems in the MSBR. Even though the reactors were inoculated with the same biomass, significant differences in the composition and population dynamics were observed. The results showed that MSBR presented a better performance regarding solids removal, but the degradation capabilities were the same as in SBR, in which its implementation may be more economically attractive.
References
APHA, AWWA, & WEF. (2005). Standard methods for the examination of water and wastewater (21st ed.). Baltimore: Port City Press.
Arrojo, B., Mosquera-Corra, A., Garrido, J. M., Méndez, R., Ficara, E., & Malpei, F. (2005). A membrane coupled to a sequencing batch reactor for water reuse and removal of coliform bacteria. Desalination, 179, 109–116.
Bae, T. Y., Han, S.-S., & Tak, T. M. (2003). Membrane sequencing batch reactor system for the treatment of dairy industry wastewater. Process Biochemistry, 39, 221–231.
Beun, J. J., van Loosdrecht, M. C. M., & Heijnen, J. J. (2002). Aerobic granulation in a sequencing batch airlift reactor. Water Research, 36, 702–712.
Carucci, A., Milia, S., Cappai, G., & Muntoni, A. (2010). A direct comparison amongst different technologies (aerobic granular sludge, SBR and MBR) for the treatment of wastewater contaminated by 4-chlorophenol. Journal of Hazardous Materials, 177, 1119–1125.
Chen, J., Rulkens, W. H., & Bruning, H. (1997). Photochemical elimination of phenols and COD in industrial wastewaters. Water Science and Technology, 35, 231–238.
Cicek, N., Macomber, J., Davel, J., Suidan, M. T., Audic, J., & Genestet, P. (2001). Effect of solids retention time on the performance and biological characteristics of a membrane bioreactor. Water Science and Technology, 43(11), 43–50.
Coello Oviedo, M. D., López-Ramírez, J. A., Sales, D., & Quiroga, J. M. (2003). Evolution of an activated sludge system under starvation conditions. Journal of Chemical Engineering, 94, 139–146.
de Kreuk, M. K., & van Loosdrecht, M. C. M. (2004). Selection of slow growing organisms as a means for improving aerobic granular sludge stability. Water Science and Technology, 49(11–12), 9–17.
Delbés, C., Moletta, R., & Godon, J. J. (2000). Monitoring of activity dynamics o fan anaerobic digester bacterial community using 16S rRNA polymerase chain-single-strand conformation polymorphism analysis. Environmental Microbiology, 2, 506–515.
Di Bella, C., Torregrossa, M., & Viviani, G. (2011). The role of EPS concentration in MBR foaming: analysis of a submerged pilot plant. Bioresource Technology, 102, 1628–1635.
Etchebere, C., Errazquin, M. I., Dabert, P., Moletta, R., & Mixí, L. (2001). Evaluation of the denitrifying microbiota of anoxic reactors. FEMS Microbiology Ecology, 35, 259–267.
Fan, F., Zhou, H., & Husain, H. (2006). Identification of wastewater sludge characteristics to predict critical flux for membrane bioreactor processes. Water Research, 40, 205–212.
Hammer, Ø., Harper, D. A. T., & Ryan, P. D. (2001). PAST: Paleontological statistics software package for education and data analysis. Palaeontologia Electronica, 4(1), 1–9. art. 4.
Holbrook, R. D., Massie, K. A., & Novak, J. T. (2005). A comparison of membrane bioreactor and conventional-activated-sludge mixed liquor and biosolids characteristics. Water Environmental Research, 77, 323–330.
Judd, S. (2006). The MBR book. Principles and applications of membrane bioreactors in water and wastewaters treatment. Oxford: Elsevier.
Kang, I.-J., Lee, C.-K., & Kim, K.-J. (2003). Characteristics of microfiltration membranes in a membrane coupled sequencing batch reactor system. Water Research, 37, 1192–1197.
Le-Clech, P., Chen, V., & Fane, A. G. (2006). Fouling in membrane bioreactors used in wastewater treatment. Journal of Membrane Science, 284, 17–53.
Lesjean, B., Rosenberger, S., Laabs, C., Jekel, M., Gnirss, R., & Amy, G. (2005). Correlation between membrane fouling and soluble/collidal organic substances in membrane bioreactors for municipal wastewater treatment. Water Science and Technology, 51(6/7), 1–8.
Li, X., Gao, F., Hua, Z., Du, G., & Chen, J. (2005). Treatment of synthetic wastewater by a novel MBR with granular sludge developed for controlling membrane fouling. Separation and Purification Technology, 46, 19–25.
Liang, S., Liu, C., & Song, L. (2007). Soluble microbial products in membrane bioreactor operation: behaviors, characteristics, and fouling potential. Water Research, 41, 95–101.
Lowry, O. H., Rosebrough, N. J., Farr, A. L., & Randall, R. J. (1951). Protein measurement with the folin phenol reagent. The Journal of Biological Chemistry, 193, 265–275.
Mace, S., & Mata-Alvarez, J. (2002). Utilization of SBR technology for wastewater treatment: an overview. Industrial and Engineering Chemistry Research, 41, 5539–5553.
McAdam, E., Judo, S. J., Glidemeister, R., Drews, A., & Kraume, M. (2005). Critical analysis of submerged membrane sequencing batch reactor conditions. Water Research, 39, 4011–4019.
McSwain, B. S., Irvine, R. L., & Wilderer, P. A. (2004). The influence of settling time on the formation of aerobic granules. Water Science and Technology, 50(10), 195–202.
Molina-Muñoz, M., Poyatos, J. M., Sánchez-Peinado, M., Hontoria, E., González-López, J., & Rodelas, B. (2009). Microbial community structure and dynamics in a pilot-scale submerged membrane bioreactor aerobically treating domestic wastewater under real operation conditions. Science of the Total Environment, 407, 3994–4003.
Moreno-Andrade, I., & Buitrón, G. (2004). Variation of the microbial activity during the acclimation phase of a SBR system degrading 4-chlorophenol. Water Science and Technology, 50(10), 251–258.
Moreno-Andrade, I., Buitrón, G., Betancur, M. J., & Moreno, J. A. (2006a). Control strategy for the optimal degradation of inhibitory wastewater in a discontinuous bioreactor. Journal of Chemical Technology and Biotechnology, 81(4), 713–720.
Moreno-Andrade, I., Lopez-Vidal, Y., & Buitrón, G. (2006b). Effect of the starvation in the activity and viability of Pseudomonas aeruginosa ATCC 10145 acclimated to degrade 4-chlorophenol in a discontinuous reactor. Water Science and Technology, 54(10), 163–168.
Muyzer, G., & Ramsing, N. B. (1995). Molecular methods to study the organization of microbial communities. Archives of Microbiology, 32, 1–9.
Pritchard, P. H., O’Neill, E. J., Spain, C. M., & Ahearn, D. J. (1987). Physical and biological parameters that determine the fate of p-chlorophenol in laboratory test systems. Applied and Environmental Microbiology, 53, 1833–1838.
Sapila, P., Keskinen, A.-K., Akerman, M.-L., Fortelius, C., Haahtela, K., & Yrjälä, K. (2008). High aromatic ring-cleavage diversity in birch rhizosphere: PAH treatment-specific changes of I.E.3 group extradiol dioxygenases and 16S rRNA bacterial communities in soil. ISME Journal, 2, 968–981.
Scheumann, R., & Kraume, M. (2009). Influence of hydraulic retention time on the operation of a submerged membrane sequencing batch reactor (SM-SBR) for the treatment of greywater. Desalination, 246, 444–445.
Shannon, C. R., & Weaver, W. (1963). The mathematical theory of communication. Urbana: University of Illinois Press.
Sharma, A., Thakur, I. S., & Dureja, P. (2009). Enrichment, isolation and characterization of pentachlorophenol degrading bacterium Acinetobacter sp. ISTPCP-3 from effluent discharge site. Biodegradation, 20, 643–650.
Tao, G., Kekre, K., Wei, Z., Lee, T. C., Viswanath, B., & Seah, H. (2005). Membrane bioreactors for water reclamation. Water Science and Technology, 51(6–7), 431–440.
Thanh, B. X., Spérandio, M., Guigui, C., Aim, R. B., Wan, J., & Visvanathan, C. (2010). Coupling sequencing batch airlift reactor (SBAR) and membrane filtration: influence of nitrate removal on sludge characteristics, effluent quality and filterability. Desalination, 250, 850–854.
Tsilogeorgis, J. A., Zouboulis, A., Samaras, P., & Zamboulis, D. (2008). Application of a membrane sequencing batch reactor for landfill leachate treatment. Desalination, 221, 483–493.
Vargas, A., Moreno-Andrade, I., & Buitrón, G. (2008). Controlled backwashing in a membrane sequencing batch reactor used for toxic wastewater treatment. Journal of Membrane Science, 320, 185–190.
Wilderer, P. A., Irvine, R. L., & Goronszy, M. C. (2001). Sequencing batch reactor technolog Scientific and Technical Report Series No. 10 (p. 100). London: IWA Publishing.
Acknowledgments
Financial support was provided by Fondo de Investigación del Instituto de Ingeniería of the Universidad Nacional Autónoma de México (Proyectos internos A2) and Dirección General de Asuntos del Personal Académico, Universidad Nacional Autónoma de México (Project IN104710). Jaime Perez Trevilla and Gloria Moreno are acknowledged for their technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Moreno-Andrade, I., Buitrón, G. Comparison of the Performance of Membrane and Conventional Sequencing Batch Reactors Degrading 4-Chlorophenol. Water Air Soil Pollut 223, 2083–2091 (2012). https://doi.org/10.1007/s11270-011-1006-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11270-011-1006-3