Abstract
A novel clinic protocol of virtual histology using an in vivo cellular imaging and real-time processing system is being developed. Main ideas of photoactivation and miniaturized confocal image scanning are presented. Technical innovation in embedded real-time image processing, feature detection and visualization system is demonstrated.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
The gold standard for detection of mucosal cancers is the ex vivo histological examination of biopsy samples [1]. However, biopsy tissue extraction, especially the excisional biopsy is an invasive procedure. It carries the risk of causing bleeding, infection or perforation. There may also be complications from anesthesia and the biopsies may be non-representative due to the heterogeneity of cells in a single cancer site and the nature of sampling, leading to the underestimation of the severity of the cancer and the tumor invasion. Moreover, the decision to obtain the sample is based on the white light visualization of the tissue, which poses the inability to differentiate flat malignant lesions from normal or inflamed tissue. And finally, the chemical used to preserve the tissue sample causes cell shrinkage, and the slicing of the tissue causes distortion.
Anatomic imaging techniques have been used to locate suspicious lesion site(s), but these techniques only allow cancers to be detected when they are a centimeter or greater in diameter, which is at a point when the site consists of more than 109 cells [2]. Even worse, though the resolution is sufficient for the detection, the screening tends to miss the cancerous site (false negative) or identify non-cancerous sites as suspicious sites (false positive) since pre-cancerous tumors and early stage cancerous tissue often do not have distinctive architectural and morphological features.
In contrast, molecular imaging [3], including optical imaging, radionuclide imaging and magnetic resonance imaging, allows visualization of the expression and activity of specific key molecular targets and host responses associated with early events in carcinogenesis. Notably, optical bioluminescence imaging probes are most commonly used to transfect tumor cell lines which are implanted into animal models for investigation and optical bioluminescence imaging is able to link the activity of luciferase enzyme to particular biological events. Unfortunately, its main disadvantage is the low light penetration. Only a small amount of light produced by the luciferases is able to pass through tissue. Optical coherence tomography (OCT) uses femtosecond infrared pulsed light and optical interference to sense reflections from tissue inhomogeneities. It is a non-invasive imaging technique that offers 2–3 mm penetration and axial resolution up to less than a micrometer through the use of wide bandwidth light sources. It has been employed in the imaging of cancer. Despite the sub-micron resolution, visualization of small (10–15 μm in diameter) cells is still problematic. This could be due to the complicated optical scattering and coherent speckle artifacts from cellular structures and organelles.
Having recognized the problems with biopsy and previous non-invasive imaging modalities, we have proposed and developed a unique virtual histology protocol, based on new molecular imaging probes [4] and fluorescence confocal endomicroscopy imaging instruments [5]. Such an optical fluorescence imaging modality transmits an incident light within the absorption spectrum of the fluorochrome and images the emitted light of a different spectrum. Photosensitizers such as 5-aminolevulinic acid (5-ALA) or 5-ALA esters are used as imaging probes, taking the advantages of their high sensitivity and ability to detect fluorochrome in cancerous cells. Laser light with well-defined wavelength of the photosensitizer’s absorption range is delivered using confocal technology to the tissue through optic fibers. The emitted fluorescence light from the tissue is captured using a photo-multiplier tube.
While the in vivo imaging system allows the capturing and storage of images in real time, processing of the acquired images has become a bottleneck in its clinical application. Hence, we devised a hardware acceleration technique for image computing tasks using Field Programmable Gate Array (FPGA) [6]. FPGA is a type of reconfigurable embedded computing system that allows both the data path and control flow to be manipulated using post-fabrication spatial connections of compute elements. It allows fine-grained parallelism customized to meet design objectives, logic specialization to perform specific functions and hardware-level adaptation to functionality changes as the algorithm changes.
2 Imaging and Processing System
2.1 Photoactivation
The proposed in vivo cellular imaging is based on photoactivation whereby the photosensitizer compound absorbs electromagnetic radiation, becomes energized to a higher energy state, and releases the energy into chemical energy, light energy and heat. The light energy is in the form of fluorescence light emission, and is the energy of interest for detection.
The photosensitizers proposed in our approach include the 5-ALA, fotolon and hypericin. They are approved for human clinically research, selectively accumulate in tumor cells and have a relatively rapid clearance from the body.
Fotolon is a mixture of chlorine e6 and polyvinylpyrrolidone (PVP), while hypericin is an extract from the herb St. John’s wort. Both hypericin and chlorine e6 are new generation photosensitizers, while PVP enhances the penetration of the photosensitizer into the tissue. PVP is a pharmaceutical-grade solvent that is safe for human usage.
5-ALA is a photosensitizer that is a precursor of an endogenous fluorescence metabolite, protoporphyrin IX (PpIX). This metabolite is the second last product in the heme synthesis pathway. Endogenously, ALA is synthesized in the mitochondrion and is transported to the cytosol. As such, no further mitochondria targeting of the instilled 5-ALA is required. The cell absorbs externally instilled 5-ALA into the cytosol. Following which, ALA in the cytosol is converted by enzymes in the pathway and the last product, Coproporphyrinogen III (Coprogen III) synthesized in the cytosol by the heme synthesis pathway is transported back into mitochondria. This product, Coprogen III is converted by enzymes in mitochondria to PpIX. In neoplasm and cancer cells, PpIX is converted at a much lower rate to heme. The resulting accumulation of fluorescence metabolite PpIX in neoplasm and cancer cells, is able to act as an indictor of tumor progression.
2.2 Miniaturized Fluorescence Confocal Scanning
The optical slicing proposed in our approach makes use of a fusion of several instrumentation technologies. The key is to innovate endoscopy technology by incorporating a miniaturized version of the confocal laser scanning microscope (CLSM) into the probe. Amendments are required and new methods of achieving the same concept of CLSM lead to an endomicroscope [7].
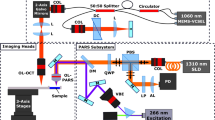
As illustrated in Fig. 1 [7], the optic fiber is used to introduce multi-spectral laser light of wavelengths within a predetermined wavelength range. In order to miniaturize the scanning mirrors and magnifying objective lens into the probe, a collimating lens is adhered at the distal end of the optic fiber. It collimates light emerging from the distal end of the illumination fiber. A dispersing prism then receives the light collimated by the collimating lens and disperses this received collimated light in a predetermined direction, while keeping the light at the same wavelength and collimated. The objective lens then converges the light emerging from the dispersing prism onto the small region of interest (focal spot on sample). As the design is confocal, only the returning rays from the focal spot, return into the optic fiber. The scanning coordinates of the focal spot on the sample are controlled by moving the entire miniaturized objective lens through electrically controlled expansion and contraction of piezoelectric crystal.
The wavelength range of the laser light source is closely coupled to the photosensitizer’s absorption range. This allows the laser light to mainly excite only the photosensitizer, which preferentially accumulates in neoplastic and cancerous cells, and subsequently, these excited unstable photosensitizers decay to emit fluorescence light.
2.3 Real-time Image Processing
A diagram of the proposed image processing system is shown in Fig. 2. A laser optical unit with peak wavelength 488 nm within the blue light electromagnetic wavelength range, is shone through the fiber optic probe. This blue light interacts with the imaging object of interest, the tissues that are instilled with photosensitizers, and the returning light including the reflected incident light and the emitted fluorescence light is filtered by the bandpass filter to remove the incident blue light and enable the emitted fluorescence red light to pass through. Depending on the bandpass filter used, returning green light can be either removed (bandpass filter 550–750 nm) or allowed to pass through (bandpass filter 505–750 nm). This bandpass limited returning fluorescence light intensity for each pixel is captured by the photomultiplier tube to provide information on the tissues, which are the imaging objects of interest.
Processing of the images obtained from the object for feature detection and visualization requires substantial amount of computation cycles. Even at the slowest rate and resolution of digitized PAL analog input, images are generated at a rate of 25 full frames per second with 768 × 576 pixels. As the images are captured from a single channel, there are around 11 million gray-scale inputs per second to be processed and rendered. Thus, computational intensive image processing algorithms will require such substantial computation power that a serial-instruction based computer will not be able to process them in real time.
In our system, the captured images are continuously fed into a FPGA board. FPGA is based on re-configurable logic circuit, with huge computing capacity. It consists of a large array of re-configurable logic elements. These elements allow fine-grained parallel programming. The programmable input–output blocks allow flexible number of inputs and outputs, depending on the design of algorithm. The programmable inter-connections allow efficient routing of hardware circuit path. Hence, the hardware logic circuit of a desired image processing algorithm can be placed and routed onto the FPGA chip. And eventually, the processed images and detected features are visualized on the display device.
3 Embedded Computing System
3.1 Embedded System Development
The embedded system contains a Virtex-II 2V6000 FPGA chip. FPGA, with its reconfigurable logic circuit of large computing capacity [8], allows flexible, high-performance and parallel fine-grained computing, leading to real-time analysis of images [9]. This embedded Virtex-II 2V6000 FPGA chip is programmed using the image processing codes that we developed, such that its programmable connections and resources takes on the hardware circuit specified. The image processing codes are designed using Handel-C language [10]. These Handel-C image processing codes are compiled to Verilog [11] logic blocks using Celoxica DK. The Verilog logic blocks are debugged and optimized to increase performance and reduce resources used. In addition, the performance and resource-optimized logic level circuit are simulated using ModelSim along with the resources’ timing constrain values to ensure that the actual implementation conforms with the simplistic estimates. Following, upon confirmation of conformance, Xilinx ISE is used to generate the configuration file of the FPGA chip specific placed and routed hardware circuit from the optimized Verilog logic blocks.
3.2 A Working Prototype
A full working prototype, as shown in Fig. 3, has been developed with the capability to process captured video signal (25 full fps PAL or 30 full fps NTSC) in real time, with various image processing filters that can be applied concurrently. The prototype is equipped with a touch screen user interface that allows the user to interactively apply various functions and filters with a simple touch on the screen. The touch-screen panel is designed with 16 menu buttons placed on the right. The processed image is displayed on the top left panel. And an informative panel is positioned at the bottom of the screen. The bottom informative panel is used to alternatively display the histograms or to display the minimum, maximum, median and mode measurements, with the selection area being either the entire image or the selected region(s).
The image processing algorithms developed on the FPGA board include arithmetic operation filter (invert intensity filter), neighborhood operation filter (median de-noising filter), convolution operation filters (smoothing filter, sharpening filter and Sobel edge detection filter), segmentation filter (threshold outline filter) and frame difference filter (motion detection filter). In addition, we provide a function to freeze and unfreeze the current video frame, such that the image of interest can be retained for further inspection.
The image measurement functions on the board provide the histograms information and the minimum, maximum, median and mode measurement values. These values are provided for each of the channels, with the selection area being either the entire image or the selected region(s).
Image magnification functions are developed to interactively zoom in, zoom out or reset the video input area to be viewed and processed. This action of digital magnification is interactively reflected on the selection area for image processing and measurement, which is simultaneously being updated accordingly.
3.3 Median De-noising Algorithm
To demonstrate how fine-grained parallelization of these algorithms works for the real-time processing, we present a de-noising filter on the FPGA board, as shown in Fig. 4. It corrects the counting statistic image acquisition defect, which is inherent to all fluorescence images due to the low photon count. It is an optimized incomplete sorting algorithm which obtains the median value.
The logic block is designed in such a way that it is pipelined into eight stages. Each of the stages involves both the basic functions of comparing two input values and copying the input value. The results from the comparisons and copying are stored in nine registers, thus incurring one clock cycle for each stage, and the latency of the filter is only eight clock cycles.
In the first stage, eight of the nine neighborhood pixel values \({\left( {P_{{i - 1,j - 1}} ,P_{{i - 1,j}} ,P_{{i - 1,j + 1}} ,P_{{i,j - 1}} ,P_{{i,j}} ,P_{{i,j + 1}} ,P_{{i + 1,j - 1}} ,P_{{i + 1,j}} ,P_{{i + 1,j + 1}} } \right)}\) undergo comparison through the use of four comparators. This results in four pairs of sorted values being held in registers, which can be represented as follows:
In the second stage, the four pairs of sorted values are further sorted using another four comparators such that the values are grouped into two groups, where the smallest and largest values of each group are found:
In the third stage, the two unsorted middle values in each group are sorted through the use of a total number of two comparators:
In the fourth stage, four comparators are used to combine the sorting of the two groups into one, such that the four smallest values are placed in the first four registers, while the other four largest values are placed in the next four registers:
In the fifth stage, using four comparators, both the first four and next four registers are arranged into two groups of sorted values:
In the sixth stage, using two comparators, the largest value for the first four registers and the smallest value for the next four registers are obtained:
In the seventh stage, we ignore the smallest three values from the first set of four registers, and the largest three values from the next set of four registers. In this way, we optimize the comparison, as un-necessary comparison is not performed. Taking the center two values along with the last pixel value \(\left( {P_{i{\text{ + 1,}}j{\text{ + 1}}} } \right)\), which is copied in the earlier six stages, we sort out the smallest value to the register \({\text{L7\_3}}\):
Finally, in the last stage, using a comparator, the median value is placed into the register L8_8 after comparison between the two register values \({\text{L7\_4}}\) and \({\text{L7\_8}}\). This register value is output as the median value:
The latency of the median de-noising filter is eight clock cycles. Each of the eight levels contributes one clock cycle to the latency. This hardware-based solution’s latency is much shorter than software-based solution. The software-based solution requires 22 clock cycles, as all the comparisons have to be performed in sequential order, instead of being able to capitalize on parallelism.
Furthermore, the output speed for the hardware-based solution is at one output per clock cycle, as the median de-noising circuit is pipelined with registers between each of the levels. In contrast, the software-based solution can only output at the same speed as its latency, which is 22 clock cycles.
3.4 Multiplier-less Parallelized Convolution Algorithm
Likewise, we devise optimally pipelined parallelized multiplier-less convolution filters (as shown in Fig. 5) to smooth, sharpen and detect the edges in the image. The multiplier-less multiplication algorithm significantly reduces resources and clock-cycles, as general resources (adders, wires and sign invertors) are used to replace full multiplier or specific limited multiplier. The parallelizing of the multiplier-less product and adders, allows the convolution to be achieved in two clock cycles, with a through-put rate of one per clock cycle due to the use of pipelining with registers.
In Fig. 5, convolution is illustrated using the Sobel filter. Sobel filter is an edge detector, which consists of performing convolution between the 3-by-3 neighborhood at each pixel location in every image and its vertical and horizontal kernels, respectively. The two convolution kernels are described as follows:
These two convoluted values at every pixel location in the image are combined either as Euclidean norm or as summation of the absolute convoluted values, depending on whether the exact solution algorithm or approximate solution algorithm is used, to form the image consisting of the edges.
The convolution steps consist of firstly, scale multiplication of the pixel values with the kernel values, and secondly, summation of these multiplied values. In the first step (see Fig. 5a), each of the pixel values in the 3-by-3 neighborhood of the selected pixel consisting of unsigned 8 bits is bit extended to signed 16 bits through the use of buffer and wire. It is then multiplied with the convolution kernels. As the convolution kernels (both the horizontal and vertical) contain three zero values, only six multiplications are required. Moreover, as these six kernel values are of either value one or two with either positive or negative sign, the multiplication is calculated without the use of multiplier.
This multiplier-less production is performed for kernel value of positive one using direct wire connection and buffer to the 3-by-3 neighborhood input value. As for kernel value of negative one, the product is obtained by a unary minus connected to the signed 16 bits 3-by-3 neighborhood input value to negate the input value. The kernel value of positive two is a special case, whose product is obtained by shifting the wire connection, which is the equivalent of bit shift. Likewise, for the kernel value of negative two, the product is obtained through the combinatorial use of unary minus and shifting of wire connection. Each of the product values is then connected to a 2-input multiplexer followed by a D flip-flop. This connection of multiplexer with the D flip-flop is to hold the final output value constant until the next clock cycle. The multiplication of the vertical kernel with the 3-by-3 neighborhood input value is similar to the multiplication with the horizontal kernel, and thus will not be further described.
In the second step (see Fig. 5b), the products obtained from step 1 are summed through the use of five 2-input adders connected in a combination of parallel and sequential connections. Using the same method of 2-input multiplexer followed by a D flip-flop, the final summed value is held constant until the next clock cycle. The circuit connections remain the same for the summation of both horizontal and vertical kernel products, with only changes to the input kernel product values.
This convolution filter is optimized for speed, parallelized, and pipelined. It does not rely on the use of specialized limited number of multiplier, and the general logic elements used are parallelized and pipelined to provide a latency of two clock cycles and one output per clock cycle.
3.5 Interactive and Dynamic Region Outlining Algorithm
We devised an algorithm to interactively and dynamically outline regions within a range of intensity based on a user-defined intensity, as shown in Fig. 6. This feature is useful to track the regions of similar color, as the imaging probe is moved within the specimen. The operation of this region outlining filter is such that, upon activation of the threshold outline function in the menu panel, the user is empowered to interactively select any representative point on the touch screen. The outline of the segmentation for every single subsequent frame based on this representative point’s intensities will be displayed in real time. At any time, a new representative point can be selected, and the segmentation outline will be updated accordingly based on the new representative point. This real-time processing of the segmentation and outlining at speed of the analog input video is clearly demonstrated by the ability to segment and outline each new input video frame of different image content without any dropped pixel. In the event that a particular frame is of interest, the user can freeze the frame using the freeze frame function from the menu panel.
Algorithmically, this region outlining filter uses the threshold operation in the first step (see Fig. 6a) where each pixel which falls within the dynamically generated range of intensity based on the user interactively defined intensity is set to a value of one; otherwise, it is set to a value of zero. The resulting one-bit monochrome image consists of dark regions (value one), which represents the regions of interest. In the second step (see Fig. 6b), using optimized morphology operations of an exclusive or of the dilated with the eroded one-bit monochrome threshold image, the outline is obtained.
In the first step, the logic circuit is used to obtain the dynamic upper and lower bounds of the intensity ranges using adder, subtractor and multiplexers. Base on this dynamically calculated upper and lower bound ranges, the input pixel values are compared with these bound values using comparators. This comparison result consisting of either one or zero for each pixel is latched using a multiplexer and a flip-flop, and is updated to the new value at the rising edge of each new clock cycle. The computation time for the thresholding is with a latency time and output rate of one clock cycle, as the bound range calculation and comparison stage are asynchronous (un-clocked), with only the updating of the comparison results being synchronous (clocked).
In the second step, in our context of one-bit monochrome threshold image, the erosion and dilation operations can be described as minimum and maximum operations on each pixel’s 3-by-3 neighborhood. We optimized the minimum and maximum operations, by concatenating the nine neighborhood binary pixel values together and comparing it with constant value 511 and constant value 0 respectively, instead of performing the cascade of parallel comparisons. This optimization reduces both the amount of resources used and the latency time. The computation time for the outline logic circuit has been optimized to one clock cycle for both the latency time and the output rate. Each of the morphological operations—minimum and maximum, only uses a single un-clocked comparison. Whereas, each of the morphological operations—minimum and maximum, using the cascade of parallel comparisons, requires eight clocked comparisons. This leads to a latency of four clock cycles for every morphological operation, instead of one clock cycle as in our optimized case.
4 Conclusion
A significant contribution from this research project is the delivery of a first-of-its-kind in vivo and in situ navigational cellular imaging system, with real-time feedback and dynamic spatial, contrast and temporal optimization while using the miniaturized fluorescence confocal imaging probe. In addition, more complex algorithms, such as the use of wavelet are being developed.
A clinic protocol has accordingly been established, which has a remarkable impact on the medical practice. Specifically, it allows early diagnosis and in vivo imaging of mucous cancer, provides instant assessment and reduces the reliance on invasive physical biopsy. The interventional imaging system has the potential to help better define operative margins for surgery and photodynamic therapy.
References
Koenig, F., Knittel, J., & Stepp, H. (2001). Diagnosing cancer in vivo. Science, 292, 1401–1403.
Weissleder, R. (2006). Molecular imaging in cancer. Science, 312(5777), 1168–1171.
Massoud, T. F., & Gambhir, S. S. (2003). Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes & Development, 17, 545–580.
Zheng, W., Harris, M., Kho, K. W., Thong, P. S., Hibbs, A., Olivo, M., et al. (2004). Confocal endomicroscopic imaging of normal and neoplastic human tongue tissue using ALA-induced-PPIX fluorescence: a preliminary study. Oncology Reports, 12, 397–401.
Thong, P. S. P., Olivo, M., Kho, K. W., Zheng, W., Mancer, K., Harris, M., et al. (2007). Laser confocal endomicroscopy as a novel technique for fluorescence diagnostic imaging of the oral cavity. Journal of Biomedical Optics, 12(1, 014007), 1–8.
“Virtex II Platform FPGA: Complete Data Sheet,” Xilinx Inc, 2005.
Mizuno, R. J. inventor, Confocal probe, U. S. Patent, Patent number 7061673, 13 Jun 2006.
Gokhale, M. B., & Graham, P. S. (2005). Reconfigurable computing: accelerating computation with field-programmable gate arrays. Dordrecht: Springer.
Cheong, L. S., Lin, F., Seah, H. S., Qian, K., Yu, C. W. K., Thong, P. S. P., & Olivo, M. (2007). “Real-time cellular morphologic analysis with fine-grained parallel image processing,” 2nd IAPR Workshop on Pattern Recognition in Bioinformatics, Oct.
“Handel-C language reference manual,” Celoxica Limited, 2005.
Palnitkar, S. (2003). Verilog HDL: a guide to digital design and synthesis. Upper Saddle River: Prentice Hall PTR.
Acknowledgement
This research project is sponsored by a grant from A-Star/BMRC Singapore Bioimaging Consortium (SBIC RP C-010/2006).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cheong, L.S., Lin, F., Seah, H.S. et al. Embedded Computing for Fluorescence Confocal Endomicroscopy Imaging. J Sign Process Syst Sign Image Video Technol 55, 217–228 (2009). https://doi.org/10.1007/s11265-008-0204-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11265-008-0204-8