Abstract
Reticuloendotheliosis virus (REV), classified as a gammaretrovirus, has a variety of hosts, including chickens, ducks, geese, turkeys, and wild birds. REV causes a series of pathological syndromes, especially the immunosuppression of the host, which may lead to an increased susceptibility to other pathogens, thus greatly damaging the poultry industry. Mixed infections of REV and Marek’s disease virus (MDV) have been reported in many countries, including China. Previous reports revealed that MDV vaccines were not efficacious, and even less-virulent MDV strains would cause some losses due to mixed infections with REV. Additionally, contaminants in the MDV vaccine might be the main source of REV. In this study, two clinical samples were collected from two flocks of chickens that were diagnosed with MDV. Subsequently, two REV isolates were obtained from the clinical samples. The isolates, named CY1111 and SY1209, were further confirmed through an indirect immunofluorescence assay and electron microscopy. Complete genome sequences of the two REV strains were determined to test the relationship between them and other REV strains. Phylogenetic trees showed that the two REV strains were closely related to most REV strains that were isolated from a variety of hosts. Therefore, REVs might spread freely among these hosts under natural conditions. Additionally, most REV strains in China were in the same clade. The present work offers some information regarding REV in China.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Reticuloendotheliosis (RE), whose causative agent is the reticuloendotheliosis virus (REV), is characterized by immunosuppression, runting–stunting syndrome, and oncogenicity [1]. Immunosuppression is prone to causing secondary infections that ultimately lead to great losses for the poultry industry. Since the first isolation of REV in the 1950s [2], some reports have described REV infections in different countries and regions [3, 4], including China. Additionally, mixed infections of REV and Marek’s disease virus (MDV) were also reported in China. Yan et al. [5] tested samples from 102 chickens with phymatosis for MDV and REV using a dot-blot hybridization, and the results showed that the rate of mixed infection of MDV and REV was 44.12 %. Wang et al. [6] checked 1126 tumors or suspected tumor samples from 106 flocks for oncogenic virus using PCR and found that 27 flocks had mixed infections of REV and MDV. Furthermore, REV has been reported to drop the effectiveness of the vaccine against MDV. Bulow [7] showed that antibody titers to Turkey herpesvirus (HVT) in chickens infected simultaneously for 3 days with HVT and REV were very low compared with those in chickens inoculated with uncontaminated HVT. Hence, the mixed infection of REV and MDV might bring losses to flocks that have been vaccinated. In addition, a field strain of MDV (GX0101) containing the REV long terminal repeat (LTR) insertion was reported [8]. Subsequently, Sun et al. [9] demonstrated that the deletion of the LTR of GX0101 decreased its capacity for horizontal transmission. Thus, it appeared that REV LTR integration into the MDV genome might increase the potential for MDV transmission, which might harm the poultry industry.
We collected two clinical samples from two flocks in which diseased chickens were diagnosed with MD. Additionally, we isolated two REV isolates, named CY1111 and SY1209, from these samples. In view of the great influence of REV on the MDV vaccine, we analyzed the molecular character of CY1111 and SY1209. Their complete genome sequences were determined to analyze the nucleotide similarity and phylogenetic relationship among other REV isolates.
Materials and methods
Clinical samples and background
The suspected MD in two flocks from Shandong Province in November 2011 and Liaoning Province in September 2012, respectively, emerged, in spite of vaccination with CVI988/Rispens when they were about 1 day old. The clinical symptoms of the chickens from Shandong Province began when they were about 6 weeks old, and the morbidity was about 30 % until 10 weeks. The sick chickens exhibited pathological changes, including swelling of spleens, livers, and kidneys and tumors on organs. In addition, the chickens from Liaoning Province began to get sick when they were 6 weeks old, and the morbidity was 32 % till 10 weeks. The pathological changes of diseased chickens showed swelling and tumors on livers, spleens, ovary, and kidney.
The samples collected from sick chickens of suspected MD were sent to us, and we tested oncogenic viruses [including MDV, REV, and avian leukosis virus (ALV)] and chicken anemia virus (CAV) with PCR assays [10–13]. In the total of 20 sick chickens from Shandong Province, the testing results showed that 15 were MDV positive, six were REV positive, none was detected with ALV and CAV, and six were mixed infected with MDV and REV. Besides, the results of testing 24 diseased chickens from Liaoning Province showed that 18 were MDV positive, eight chickens were REV positive, none was detected with ALV and CAV, and eight were mixed infected with MDV and REV. It was noteworthy that all of the REV-infected chickens from the two flocks were MDV positive.
REV isolation and identification
REVs were isolated using a previously reported method [11]. Briefly, filtered liver and spleen tissue homogenates were inoculated into chicken embryo fibroblasts (CEF) cells and monitored daily for 7 days. To decrease the interference of MDV, we performed the freezing and thawing, followed by centrifugation and incubation with the fresh CEF cells for three passages, which could eliminate the possibility of MDV passage.
Total DNA of CEF cells infected with the two REVs was extracted and checked by PCR using primers mentioned above. CEF cells infected with the REV 07I [14] strain were used as a positive control, and uninfected cells served as a negative control.
DF1 cells in 96-well plates were inoculated with the two isolated viruses. Meanwhile, cells infected with the REV HLJ07I strain were used as the positive control, and uninfected cells served as the negative control. Four days later, cells were fixed with cold absolute ethanol at −20 °C for 30 min. The fixed cells were incubated with an anti-gp90 monoclonal antibody A9E8 [15] in PBST (0.1 M phosphate-buffered saline (PBS) with 0.05 % Tween-20, pH 7.4) for 2 h at room temperature. Then, the cells were washed five times with PBS, followed by incubation with FITC-conjugated goat anti-mouse IgG (Sigma-Aldrich, St. Louis, MO, USA) at a concentration of 1:100 in PBST for 1 h at room temperature. After five washes, the cells were examined by fluorescence microscopy.
REV particles were also attested through electron microscopy, as follows. After 5 days of incubation, the infected CEF cells were scraped off using a cell brush and suspended in PBS. The cells were fixed with glutaraldehyde and pelleted by centrifugation at 3000 rpm for 15 min. The cells were embedded in resin, sectioned, and examined by electron microscopy.
Cloning and Sequencing of proviral genomic DNA
The total DNA of two REV isolate-infected cells was extracted with the TIANamp blood/cell/tissue genomic DNA extraction kit (Tiangen, Beijing, China). Primers for the amplification of the full genome of REV were synthesized based on the report by Barbosa et al. [16]. PCRs were performed as follows: initial denaturation cycle of 5 min at 95 °C, 30 cycles of denaturation for 30 s at 94 °C, annealing for 30 s at 56 °C, and an extension for 2 min at 72 °C, followed by a final extension of 10 min at 72 °C. The PCR products were excised from 1.0 % agarose gel, purified via the gel extraction kit (OMEGA, Guangzhou, China), and then cloned into the vector pMD18-T (TaKaRa, Dalian, China). Three independent clones of each region were sequenced by the Beijing Genomics Institute (Beijing, China).
Proviral DNA sequence and phylogenetic analysis
The overlapping sequences of the PCR products were aligned for preparation of a contiguous sequence of the proviral genome using the Seqman 7.1 function in the DNASTAR sequence analysis software (DNASTAR, Inc., Madison, WI, USA). The percent identity of the LTR, each open reading frame (ORF), and full genome were compared with the reference strain, SNV (GenBank DQ003591), and some other strains isolated in different places, including HA9901 (GenBank AY842951), HLJR0901 (GenBank GQ415646), Chicken/3337/05 (GenBank FJ439120), MD-2 (GenBank JX912710), and APC-566 (GenBank DQ387450), to get a good overview of differences in different places. To analyze SY1209 and CY1111 in more detail, we employed additional REV strains whose full genomes were available, including 1105 (GenBank JQ804915), Goose/3410/06 (GenBank FJ439119), HA1101 (GenBank KF305089), FPV (short for the REV sequence within the genomes of field strains of fowlpox virus, GenBank AF246698), HLJ07I (GenBank GQ375848), GD1210 (GenBank KF709431), DIAV (short for Duck infectious anemia virus, GenBank KF313137), and 104865 (GenBank KJ756349), and their phylogenetic relationships were analyzed using Mega 5.1 with statistical method of neighbor joining, the model of maximum composite likelihood and 1000 bootstrap replicates.
Nucleotide sequence accession numbers
Sequence data have been submitted to the GenBank Nucleotide Sequence Database. CY1111 was listed under Accession No. KJ909531 and SY1209 was listed under Accession No. KJ909530.
Results and discussion
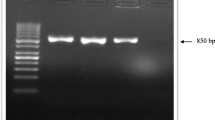
REVs were isolated after three passages of infected cells. The isolate from Shandong Province was named CY1111, and the other one from Liaoning Province was named SY1209. They were confirmed via PCR, IFA, and electron microscopy (Fig. 1). PCRs were preformed with the primers mentioned above to confirm the presence of the REV proviral genome (Fig. 1a). The results, in which REV-specific fragments were produced, demonstrated that the REV proviral genome was present in cells infected with REV CY1111 and REV SY1209. Additionally, REV-infected cells were inspected by an indirect immunofluorescence assay (IFA) using an anti-gp90 monoclonal antibody. Fluorescence was observed in cells infected with CY1111 (Fig. 1b), SY1209 (Fig. 1c), and the positive control 07I (Fig. 1e). There was no fluorescence in the negative control (Fig. 1d). Additionally, the virus particles were detected via electron microscopy (Fig. 1f, g). The round particles were approximately 100 nm in diameter. The enveloped REV particles were observed in the intercellular space of CEF cells infected with CY1111 and SY1209. No REV was detected in CEF cells that were not infected with REV (data not shown).
Identification of REV. a PCR assay tests for reticuloendotheliosis virus (REV). Lanes 1–2, tests of cells infected with REV CY1111 and REV SY1209, respectively, using REV-specific primers; lane 3, negative control; lane 4, positive control (REV 07I strain); lane M, DL2000 Marker. Detecting reticuloendotheliosis virus (REV) CY1111 (b) and SY1209 (c) by an indirect immunofluorescence assay with REV gp90-specific monoclonal antibody. Non-infected cells served as a negative control (d), and cells infected with the REV 07I strain were used as a positive control (e). Electron micrograph of reticuloendotheliosis virus (REV) strains CY1111 (f) and SY1209 (g) with a scale of 500 nm
Many reports have described the existence of REVs. Although there is a fact that REV alone, thus far, has not caused significant losses to the poultry industry, secondary infections resulting from the immunosuppression of REV have been very severe. Many cases of mixed infections, including MDV and REV, have been reported [5, 6]. The interaction between MDV and REV usually leads to losses in the poultry industry. MDV serotype 2 may enhance the development of bursal lymphomas induced by REV [17]. Besides, Witter et al. [18] verified that the protective efficiency of MD vaccine would be depressed due to immunosuppression of REV. Additionally, the REV LTR can integrate into the MDV genome and unexpectedly change the inherent character of MDV. Kim et al. [19] constructed a recombinant MDV rMd5–RM1–LTR virus containing an LTR cloned from the MDV RM1 strain and the virus rMd5–RM1–LTR. The recombinant virus produced a unique 4-kb transcript, which was also detected in RM-1, but not in rMd5 BAC. Subsequently, Mays et al. [20] found that the virus rMd5–RM1–LTR was attenuated at cell culture passage 40, but could still cause severe thymic and bursal atrophy, early immunosuppression, and early cytolytic infection. More importantly, a field MDV strain (GX0101) that contained the REV LTR was isolated, which means that it was possible for the REV LTR to integrate into the MDV genome in the field. Together with the fact that the recombinant forms indeed uncertainly alter the nature of MDV, this reconstruction might accelerate the evolution of virulence of MDV. In our present case, we isolated two REV strains from two clinical samples, confirming the mixed infection of MDV and REV, which might be the reason for losses to the two chicken farms. Mixed infections of REV and MDV are certainly prevalent in China and are very likely to induce losses in flocks.
To test the relationship between them and other REV strains, the complete proviral nucleotide sequences of REV strains CY1111 and SY1209 were determined, and their genome sequences have been submitted to the GenBank Nucleotide Sequence Database. CY1111 was listed under Accession No. KJ909531 and SY1209 was listed under Accession No. KJ909530. Both of their proviral genomes were 8,284 nucleotides long and consisted of a complete viral replication component. Their proviral genomes were divided identically: 5′LTR:1–543, gag:934–2433, pol:2434–6015, env:5952–7712, and 3′LTR:7742–8284. They shared 99.8 % identity within the full-genome sequence and shared the same LTR sequence, and the identities of the gag, pol, and env genes were 99.5, 99.9, and 99.8 %, respectively. The nucleotide percent identities of different genes, compared with representative strain SNV and some other strains isolated in different places (Table 1), showed that the pol genes were more similar to each other than the LTRs and, thus, could be a better target for diagnostic PCRs, which is consistent with view of Barbosa et al. [16].
The phylogenetic relationships among these REV strains, which were isolated from chicken, geese, and wild birds, and integrated fowlpox viruses in different countries and regions were analyzed (Fig. 2). All of the phylogenetic trees, which were based on the LTR sequence, gag gene sequence, pol gene sequence, env gene sequence, and complete genome sequence, showed that most of the REV strains in China were in the same branch, except for GD1210. GD1210, which was isolated in Guangdong Province in southernmost China, was so similar to SNV that they were separated into one small clade, although it has been repeatedly isolated for more than 50 years. Thus, little variation may have taken place in the evolutionary process. These, to some extent, suggested that the majority of REV isolates in China were very similar.
Phylogenetic trees based on a long terminal repeats (LTRs); b gag gene sequences; c pol gene sequences; d env gene sequences; and e complete genome sequences. The solid ball indicates new strains in this study. The scale bars indicate the average number of nucleotide substitution per site, and the numbers at the nodes indicate reliability of each branch
An extraordinary proposition speculated that REVs circulating in poultry and wild birds were derived from the contaminants of vaccines, including the MDV vaccine [21]. Fittingly, the REV MD-2 strain was exactly isolated from a batch of commercial freeze-dried turkey herpesvirus vaccines in China [22]. Therefore, we compared the nucleotide sequences of CY1111, SY1209, and MD-2, and the result showed that CY1111 and SY1209 were very similar to MD-2. Phylogenetic trees also showed that CY1111, SY1209, and MD-2 were in the same clade. Therefore, REV contaminants in vaccines are likely to be reservoirs of isolated strains in farms.
Interestingly, the phylogenetic analysis of CY1111 and SY1209 and other strains showed that the phylogenetic relationship of the HLJ07I gag gene sequence, which was separated into the large clade containing DIAV, GD1210, and SNV, was different from that of LTR, pol, env, and the full genome. This leads to the question of whether recombination events between REV strains led to the evolution of REV. A recent report showed that REV indeed originated from a recombinant retrovirus that circulated in ancestral mammals and was subsequently experimentally introduced into an avian host by contaminant of Plasmodium lophurae [21]. Furthermore, researchers have already identified natural recombinant retroviruses [23]. If recombination is possible, mixed infection of different REV strains might augment the diversity and genome instability of REV.
The phylogenetic trees revealed that CY1111 and SY1209 were more closely related to most of the REV strains that were isolated from various hosts and different places. Therefore, REVs might spread freely among these hosts and places under natural conditions and might not be host specific. Therefore, we should pay more attention to the testing of REV when introducing chickens from other regions and mixed farming of birds. In conclusion, these data provided some information that might help prevent and control REV.
References
A.M. Fadly, G. Zavala, R.L. Witter, in Diseases of Poultry, ed. by Y.M. Saif, A.M. Fadly, J.R. Glisson, L.R. McDougald, L.K. Nolan, D.E. Swayne (Blackwell Publishing, Hoboken, 2008), pp. 568–588
F.R. Robinson, M.J. Twiehaus, Avian Dis. 18, 278–288 (1974)
H.W. Seong, S.J. Kim, J.H. Kim, RDA J. Agric. Sci. Vet. 38, 707–715 (1996)
Celina Buscaglia, Avian Dis. 57, 569–571 (2013)
Y. Yan, Y. Diao, H. Wu, J. Li, J. Sun, Chin. J. Vet. Sci. 30, 1049–1051 (2010)
G. Wang, P. Wei, X. He, China Poult. 24, 13–15 (2002)
V.V. Bulow, Avian Pathol. 6, 383–393 (1977)
Z. Cui, G. Zhuang, X. Xu, A. Sun, S. Su, Virus Genes 40, 236–243 (2010)
A.J. Sun, X.Y. Xu, L. Petherbridge, Y.G. Zhao, V. Nair, Z.Z. Cui, Virology 397, 270–276 (2010)
Y. Zhang, C. Liu, F. Zhang, W. Shi, J. Li, Virus Genes 43, 353–357 (2011)
L. Jiang, X. Qi, Y. Gao, Y. Hua, K. Li, X. Deng, Q. Wang, L. Zhang, H. Chai, Y. Chen, C. Yin, H. Gao, L. Qin, Y. Wang, Y. Qu, Q. Chen, Z. Fan, X. Wang, Vet. Microbiol. 166, 68–75 (2013)
S. Gopal, P. Manoharan, K. Kathaperumal, B. Chidambaram, K.C. Divya, J. Clin. Microbiol. 50, 2668–2673 (2012)
L. Qin, Y. Gao, W. Pan, X. Deng, F. Sun, K. Li, X. Qi, H. Gao, C. Liu, X. Wang, China J. Prev. Vet. Med. 32, 91–93 (2010)
M. Xue, X. Shi, Y. Zhao, H. Cui, S. Hu, X. Cui, Y. Wang, PLoS One 8, e83918 (2013)
M. Xue, X. Shi, J. Zhang, Y. Zhao, H. Cui, S. Hu, H. Gao, X. Cui, Y. Wang, PLoS One 7, e49842 (2012)
T. Barbosa, G. Zavala, S. Cheng, P. Villegas, Virus Res. 124, 68–77 (2007)
M.M. Aly, R.L. Witter, A.M. Fadly, Avian Pathol. 25, 81–94 (1996)
R.L. Witter, L.F. Lee, L.D. Bacon, E.J. Smith, Infect. Immun. 26, 90–95 (1979)
T. Kim, J. Mays, A. Fadly, R.F. Silva, Virus Genes 42, 369–376 (2011)
J.K. Mays, R.F. Silva, T. Kim, A. Fadly, Avian Pathol. 41, 259–265 (2012)
A.M. Niewiadomska, R.J. Gifford, PLoS Biol. 11, e1001642 (2013)
J. Li, C. Yang, Q. Li, H. Li, Y. Xia, D. Liu, K. Yu, H. Yang, Genome Announc. 1, e00555 (2013)
L. Cai, Y. Shen, G. Wang, H. Guo, J. Liu, Z. Cheng, J. Gen. Virol. 94, 2278–2286 (2013)
Acknowledgments
This investigation was supported by Grants from the National Natural Science Foundation of China (No. 31201924), the Special Fund of the State Key Laboratory of China (No. SKLVBP201406), and the Earmarked Fund for the Modern Agro-industry Technology Research System (No. nycytx-42-G3-01).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Edited by Keizo Tomonaga.
Rights and permissions
About this article
Cite this article
Bao, Ky., Zhang, Yp., Zheng, Hw. et al. Isolation and full-genome sequence of two reticuloendotheliosis virus strains from mixed infections with Marek’s disease virus in China. Virus Genes 50, 418–424 (2015). https://doi.org/10.1007/s11262-015-1191-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-015-1191-z