Abstract
Hepatitis C virus (HCV) is a hepatotropic single-stranded RNA virus. HCV infection is causally linked with development of liver cirrhosis and hepatocellular carcinoma. Enhanced production of reactive oxygen species by HCV has been implicated to play an important role in HCV-induced pathogenesis. Mangosteen has been widely used as a traditional medicine as well as a dietary supplement ,thanks to its powerful anti-oxidant effect. In the present study, we demonstrated that the ethanol extract from mangosteen fruit peels (MG-EtOH) is able to block HCV genome replication using HCV genotype 1b Bart79I subgenomic (EC50 5.1 μg/mL) and genotype 2a J6/JFH-1 infectious replicon systems (EC50 3.8 μg/mL). We found that inhibition of HCV replication by MG-EtOH led to subsequent down-regulation of expression of HCV proteins. Interestingly, MG-EtOH exhibited a modest inhibitory effect on in vitro RNA polymerase activity of NS5B. Among a number of xanthones compounds identified within this MG-EtOH, we discovered α-MG (EC50 6.3 μM) and γ-MG (EC50 2.7 μM) as two major single molecules responsible for suppression of HCV replication. This finding will provide a valuable molecular basis to further develop mangosteen as an important dietary supplement to combat HCV-induced liver diseases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatitis C virus (HCV) is a hepatotropic single-stranded RNA virus. HCV infection is responsible for a variety of inflammatory chronic liver diseases including liver cirrhosis and hepatocellular carcinoma [1, 2]. The number of people suffering from HCV infection is estimated to be around 170 million worldwide [3]. In addition, HCV infection is the number one cause for liver transplantation in the United States, accounting for approximately 40–45 % of all liver transplants [4]. However, a prophylactic HCV vaccine is not available so far. Therefore, morbidity and mortality associated with HCV infection are imposing a great challenge on health care system of affected countries. In recent years, a number of anti-HCV drugs including telaprevir, boceprevir, simeprevir, and sofosbuvir have been approved by FDA. However, in spite of their improved efficacy and safety profiles, their high price tags have hampered most of HCV patients in developing countries from enjoying their therapeutic benefits. Thus, a new class of anti-HCV therapeutics with a novel mechanism of action should be developed to provide a more affordable and cost-effective treatment for HCV patients in the future.
Oxidative stress is characterized by enhanced formation of reactive oxygen species (ROS) inside the cell. ROS generally include a broad set of molecules and radicals such as hydrogen peroxide (H2O2), superoxide anion (O •−2 ), and hydroxyl radical (HO•) [5, 6]. Although they are generated as by-products via various cellular metabolisms catalyzed by many different enzymes in mitochondria, endoplasmic reticulum, peroxisomes, and other cellular compartments, they are also thought to play an important role in a variety of biological processes including cell signaling, regulation of cytokine, growth factor and hormone action, transcription, ion transport, neuromodulation, immunomodulation, and apoptosis [7, 8]. Nonetheless, due to their cytotoxic effects, eukaryotic cells have a special defense system to protect themselves against these ROS.
Since the middle of 1990s, researchers have found a positive correlation between oxidative stress and HCV infection [9.] In a good agreement with this observation, most of HCV proteins turned out to be strong inducers of oxidative stress. They include viral structural proteins such as core, E1, and E2 [10] as well as viral nonstructural proteins such as NS3/4A [11], NS4B, and NS5A [10]. Previous researchers even suggested ROS-induced heterogeneity in viral genome could help HCV to evade the immune system for its own survival [12]. However, detailed molecular mechanism of how HCV-induced oxidative stress contributes to progression of inflammatory liver diseases is largely unknown yet.
Garcinia mangostana L. (Clusiaceae), cultivated principally in Indonesia, Malaysia, Philippines, and Thailand, is commonly known as mangosteen and “mangkhut”. The fruits of mangosteen have been used as a traditional medicine in Southeastern Asia for the treatment of diarrhea, dysentery, inflammation, ulcers as well as wound healing [13, 14.] A broad array of biological experiments especially relevant to infectious diseases, cancer chemotherapy and chemoprevention, diabetes, and neurological disorders have been performed using mangosteen extracts and its purified constituents [15–18]. Among 16 xanthones tested in a hydroxyl radical-scavenging assay, α-mangostin turned out to be the most active anti-oxidant compound [19.] In addition, activity-guided fractionation of mangosteen using an HIV-1 protease inhibition assay led to discovery of α- and γ-mangostins as potential antiviral candidates [20.]
Based on these anti-oxidant and antiviral activities of mangosteen described above, we hypothesized that mangosteen may possess therapeutic potential against HCV infection. In order to test this hypothesis, we investigated whether mangosteen has any impact on HCV replication by testing effects of its various extracts and single compounds on HCV replicon cells. Our results suggested that the mangosteen ethanol extract (MG-EtOH) possesses the most potent anti-HCV replication activity. Subsequent HPLC analysis of this MG-EtOH fraction identified α- and γ-mangostins as two major single molecules responsible for its antiviral effect. Finally, we also found that MG-EtOH was able to restore the increased levels of ROS production by HCV to an uninfected normal level in HCV replicon cells, suggesting ROS-scavenging activity of MG-EtOH may play a role in its inhibition of HCV replication.
Materials and methods
Drug
Methyl [(2S)-1-{(2S)-2-[4-(4′-{2-[(2S)-1-{(2S)-2-[(methoxycarbonyl)amino]-3-methylbutanoyl}-2-pyrrolidinyl]-1H-imidazol-4-yl}-4-biphenylyl)-1H-imidazol-2-yl]-1-pyrrolidinyl}-3-methyl-1-oxo-2-butanyl] carbamate which we refer to here as an NS5A inhibitor (BMS-790052) was purchased from Selleck USA. NS5A inhibitor was solubilized in DMSO, and aliquots of 10 mM stocks were maintained at −20 °C. Immediately prior to use, these stocks were diluted to the desired final concentration, and this solution was added directly to the dishes.
Cell culture
The Human hepatoma cell lines Huh7.5 were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Hyclone) supplemented with 1 % l-glutamine (Hyclone), 1 % penicillin/streptomycin (Hyclone), 1 % non-essential amino acids, and 10 % fetal bovine serum (JR Scientific) at 37 °C with 5 % CO2 supplement.
Plasmids
Rluc-J6/JFH1 (FL-J6/JFH-5′C19Rluc2AUbi) [21] is a monocistronic, full-length HCV genome that expresses a renilla luciferase and was derived from the previously described infectious genotype 2a HCV genome J6/JFH1 [22]. Bart79I is a high-efficiency bicistronic subgenomic replicon of HCV derived from an HCV genotype 1b Con1 sequence that harbors the neomycin phosphotransferase gene in the first cistron and the HCV nonstructural proteins in the second cistron under the translational control of an EMCV IRES [23]. This plasmid also has an adaptive mutation (S2204I) in NS5A, which increases replication efficiency. Bart79I-YFP was made by removal of amino acids 2209 to 2254 in Bart79I and insertion of PCR-amplified YFP sequence from pEFYP-C1 (Clonetech, USA) using flanking a NotI site by direct cloning into NS5A of Bart79I. FL-J6/JFH-5′C19Rluc2AUbi and Bart79I were gifts from Dr. Charles Rice at Rockefeller University.
In vitro transcription for production of HCV RNA genomes
In order to obtain HCV RNA genomes, in vitro transcription was performed as previously described [24]. Briefly, plasmids (Bart79I, Bart79I-YFP; genotype 1b, J6/JFH1, or RLuc-J6/JFH1; genotype 2a) were linearized by ScaI or XbaI (NEB) digestion, respectively. The T7 promoter-driven in vitro transcription was performed on the digested plasmids to produce the HCV RNA genomes using a MEGAscript kit (Ambion) according to the manufacturer’s protocol.
Generation of stable HCV replicon cell lines
In order to establish stable HCV replicon cells, in vitro transcribed subgenomic replicon Bart79I, Bart79I-YFP, infectious clone J6/JFH1, or Rluc-J6/JFH1 RNAs were transfected into the Huh7.5 cells using lipofectamine 2000 (Invitrogen). Bart79I and Bart79I-YFP were selected through G418 to a final concentration of 125 μg/mL and the media were replaced every 3 days for 3 weeks. The establishment of Huh7.5 cells stably maintaining a J6/JFH1 infectious clone was performed by transfecting in vitro transcribed J6/JFH1 RNAs into Huh7.5 cells using a lipofectamine 2000 transfection reagent. The establishment of Huh7.5 cells stably maintaining an Rluc-J6/JFH1 clone was also performed similarly.
Cell viability and anti-HCV replication analysis using a luciferase assay
Transfected-Huh7.5 with in vitro transcribed Rluc-J6/JFH1 RNAs through lipofectamine 2000 transfection reagent (Invitrogen) as described by the manufacturer were plated onto a white 96-well plate (Costar 3610) and at next day supplemented with an increasing concentration of MG-EtOH, α- or γ-mangostins. At 3 days after incubation, cells were incubated for 3 h at 37 °C in the presence of EZ-CYTOX (10 % tetrazolium salt; Dogen) reagent to assess cytotoxicity and renilla luciferase activities were measured using a luciferase reagent (1 mM coelenerazine in Methanol-HCL; Goldbio).
Quantitative real-time RT-PCR (qRT-PCR) analysis
Total cellular RNA was extracted using the RNeasy® mini kit (Qiagen) according to the manufacturer’s protocol. The expression of HCV subgenomic RNA and cellular RNA was quantified through quantitative real-time reverse-transcription polymerase chain reaction (qRT-PCR) analysis as previously described. Each sample was normalized by the endogenous reference gene glyceraldehydes-3-phosphate dehydrogenase (GAPDH). The cDNA quantification was performed by the CFX384 real-time PCR detection system (Bio-Rad, US). The used primers were as follows. FW-J6/JFH1-CTCCGCCATGAATCACTC, RV-J6/JFH1-ACGACACTCATACTAACGC, FW-Bart79I-AGAGCCATAGTGGTCT, RV-Bart79I-CCAAATCTCCAGGCATTGAGC, FW-GAPDH-TGGTCTCCTCTGACTTCA, and RV-GAPDH-CGTTGTCATACCAGGAAATG.
HCV pseudoparticle entry assay
In order to generate HCV pseudo virus particle, 293T cells were transfected with following expression vectors encoding the adaptable viral components [25]; pCon-E1E2 is genotype 1b HCV glycoproteins, pH77-E1E2 is genotype 2a HCV glycoproteins, pVSV-G encodes the vesicular stomatitis virus (VSV) G protein, pHIV-gag-pol, and pLN4-1 Fluc RE, using lipofectamine 2000 (Invitrogen). The medium was replaced 24 h later, and then the soup was taken at 48 h post transfection for obtaining virus particle. Target cells were seeded before infection onto a 24-well plate. Following treatment of MG-EtOH at concentrations of 0, 1, 2, 5, 8, and 10 μg/mL, HCV pseudo virus particles were applied to infect target cell in the presence of 10 μg/mL polybrene (Sigma). After 3 days, infected or uninfected cells were lysed. Lysates were laid onto a 96 well white plate and then firefly luciferase activities were measured using a luciferase reagent (Goldbio).
Infection and transfection assay
Infection and transfection were performed as previously described [26]. Briefly, a vaccinia virus encoding T7 RNA polymerase was used to infect Huh7.5 cells. At 1 h after infection, J6/JFH1 plasmid was transfected into Huh7.5 cells. Then, an increasing concentration of MG-EtOH was treated onto infected Huh7.5 cells. At 8 h after incubation, cells were lysed in RIPA buffer (150 mM NaCl, 1 % SDS, 1 % deoxicholic acid sodium salt, 0.1 % sodium dodecyl sulfate, 50 mM Tris–HCl (pH 7.5), 2 mM EDTA; Thermo) containing a cocktail of protease inhibitors (Complete, Roche Diagnostic at final concentration of 1 tablet per 50 mL RIPA buffer). Western blot analysis was performed to detect NS3 protease.
MG-EtOH and NS5A inhibitor combination assay
Hu7.5-Rluc-J6/JFH1 cells were treated with serially diluted MG-EtOH (0, 1, 2, 5, 8, 10 μg/mL) in combination with serially diluted NS5A inhibitor (0.05 and 0.1 pM). At 3 days after incubation, cells were treated with EZ-CYTOX (10 % tetrazolium salt; Dogen) reagent to assess cytotoxicity for 3 h at 37 °C. Renilla luciferase activities were measured using a luciferase reagent (1 mM coelenerazine in Methanol-HCl; Goldbio).
In vitro HCV RNA polymerase assay
Recombinant HCV NS5B protein with an N-terminal hexa-histidine tag was expressed in Escherichia coli and purified, as described previously [27]. In vitro RNA polymerase activity assays were performed with 50 ng of recombinant HCV NS5B lacking the C-terminal 21 hydrophobic amino acids (NS5B∆21), as described previously [27–29]. Briefly, the reaction was performed in 25 μl of RNA polymerase reaction buffer [50 mM Tris–HCl, pH 7.5, 50 mM NaCl, 100 mM potassium glutamate, 2 mM MgCl2, 1 mM dithiothreitol (DTT), 20 units of RNase inhibitor (Promega, Madison, WI, USA)] containing 1 μg of poly(A) RNA template and 10 pmol oligo(U)20, 6 μM UTP, and 5 μCi [α-32P]UTP (3,000 Ci/mmol, Amersham Pharmacia Biotech, Piscataway, NJ, USA). The reaction mixture was incubated for 2 h at 25 °C. Reaction products were resolved on a denaturing 5 % polyacrylamide gel containing 8 M urea. After electrophoresis, gels were dried and then exposed to X-ray film for autoradiography. The amount of radio-labeled RNA was quantified using a Phosphorimager.
Western blot analysis
After incubation of cells (Huh7.5-J6/JFH1 or Bart79I) with followed dose on 5 days, cells were lysed in RIPA buffer (150 mM NaCl, 1 % SDS, 1 % deoxicholic acid sodium salt, 0.1 % sodium dodecyl sulfate, 50 mM Tris–HCl (pH 7.5), 2 mM EDTA; Thermo) containing a cocktail of protease inhibitors (Complete, Roche Diagnostic at final concentration of 1 tablet per 50 mL RIPA buffer). Protein concentration was determined by Bradford assay (Bio-Rad). Equal amounts of protein were electrophoresed on an SDS–polyacrylamide gel. After loaded sample, the samples were transferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore, Bedford). Proteins of interested were detected by following antibodies: anti-Core, anti-NS3, or anti-NS5A (1:1000, 1868 for Core, 1:1000, 1847 NS3, and 1:1000, 1877 for NS5A, Virostat), anti-β-actin (1:5000, Santa Cruz), HRE-conjugated anti-mouse (1:5000, Thermo).
Immunofluorescence
Immunofluorescence analysis was conducted as described previously [30]. Briefly, Bart79I cells were grown on coverslips with indicated dose to 70 % confluency for 3 days. Coverslips were rinsed in phosphate-buffered saline (PBS) three times. Cells were fixed at room temperature for 20 min in 4 % paraformaldehyde, permeabilized in 0.1 % Triton-X in PBS for 5 min, rinsed three times in PBS, and blocked with PBS with 2 % fetal bovine serum (FBS). A mouse anti-NS5A antibody (1:500, 1877, Virostat) was diluted in blocking buffer, and the mixture was incubated for overnight at 4 °C. After three washes in PBS, coverslips were incubated with Alexa 488-conjugated anti-mouse IgG secondary antibodies (Invitrogen, Carlsbad, CA, USA) for 1 h. Following three washes with PBS, coverslips were mounted onto slides using Prolong Gold anti-fade reagent with DAPI (Invitrogen, Carlsbad, CA, USA) and sealed. Fluorescent signals were examined and captured by Nikon confocal laser scanning microscope.
FACS analysis
Huh7.5-Bart79I-YFP cells were plated onto a 6-well plate (Costar 3516) and supplemented with DMSO or mangosteen extracts with indicated concentrations. At 5 days after incubation, cells were washed at several times using 1× PBS (Hyclone). FACS analysis was performed by the FACSAria 3 system (BD Bioscience, US).
Isolation of α- and γ-mangostins
The fruits of G. mangostana were collected from Indonesia. The pericarps of G. mangostana (1.3 kg) were pulverized and extracted with ethanol at the room temperature three times (each 8 L) to obtain 401 g of solid extract. A portion of the methanol extract (200 g) was suspended in H2O and then partitioned with n-hexane, chloroform, ethyl acetate, and n-butanol, resulting in n-hexane, chloroform, ethyl acetate, n-butanol, and aqueous fractions. The chloroform-soluble extract (37.9 g) was subjected to a column chromatography using silica gel (593 g) eluting with gradient mixtures of n-hexane:ethyl acetate (50:1 to 0:100) and ethyl acetate: methanol (20:1 to 0:100) to give 49 fractions (GMC 1 to GMC 49). The α-mangostin (2.25 g) was precipitated from GMC 24 and 25 fractions. GMC35 fraction (698 mg) was chromatographed on a medium pressure liquid chromatography system (MPLC) using MPLC column of silica gel (100 g) eluted with a gradient mixture of chloroform: acetone (2:98 to 25:75) to give 23 fractions (GMC35-1 to GMC35-23). From the GMC35-8 fraction, γ-mangostin (232 mg) was precipitated in methanol and H2O. The NMR spectra of α- and γ-mangostin were identical to those reported in the literature [19]. All the solvents used for extraction were evaporated and extracted materials were precipitated and dissolved in DMSO for all the experiments.
HPLC analysis
As chemicals and reagents, acetonitrile (HPLC grade) and methanol (HPLC grade) were purchased from SK chemical (Seoul, KOREA), and formic acid (HPLC grade) was obtained from Sigma-Aldrich (MO, USA). Water was purified using a Milli-Q system (Milford, MA). α- and γ-mangostins and ethanolic extract (MG-EtOH) stock solutions were prepared by dissolving each sample (1 mg) in methanol (1 mL) and dilution with methanol for each solution was conducted to obtain appropriate concentration of working solution. These solutions were then serially diluted with methanol to provide calibration solutions of 1.0, 5.0, 15.0, 20.0, and 100.0 μg/mL. The solutions were filtered through a 0.5 μm hydrophobic PTFE filter. Analytical method was performed according to the previous report with a slight modification [31]. HPLC system (Shimadzu, Kyoto, Japan) consisted of a quaternary pump (CC-20AD), auto sampler (SIC-20A), column oven (CTO-20A) ,and UV detector (SPD-20A). The wavelength was set at 230 nm for quantification. Chromatographic separation was performed using a TOSOH Tsk-gel ODS-80Ts column (4.6 × 150 mm, 5 μm) with a C18 guard column. The mobile phase consisted of 0.1 % (v/v) formic acid-acetonitrile (A) and 0.1 % (v/v) formic acid-H2O (B) was delivered at a flow rate of 1.0 mL/min by applying the following programed gradient elution: 38 % (A) isocratic for 35 min, 38–100 % (A) in 1 min, 100 % (A) isocratic for 10 min, 100–35 % (A) in 1 min, and 35 % (A) isocratic for 6 min as post-run for reconditioning. The stock solution injection volume was adjusted to 20 μL. In this analytical HPLC method, α- and γ-MG appeared at 29.08 and 16.71 min. In the ranges of 1.0–100 μg/mL of α- and γ-MG, linear regression coefficients of α- and γ-MG were calculated as 0.9988 and 0.9999, respectively.
Measurement of reactive oxygen species
After incubation of cells (Huh7.5, Huh7.5-J6/JFH1, or Huh7.5-Bart79I) with an increasing concentration of MG-EtOH for 72 h, intracellular ROS production was measured by dichlorofluorescein diacetate (DCFH-DA). DCFH-DA was diluted at a final concentration of 20 μM in 2 % FBS in PBS. After incubation for 30 min at 37 °C, the level of ROS was measured by SpectraMax M2e microplate reader (Molecular Devices, USA).
Results
MG-EtOH inhibits HCV genome replication
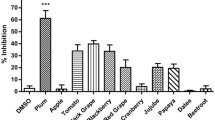
In order to evaluate a potential effect of mangosteen on HCV replication, we prepared a variety of extracts from mangosteen fruit peels using a number of different solvents including methanol, water, hexane, butanol, ethyl acetate, and ethanol. After Huh7.5 hepatocarcinoma cells were transfected with renilla luciferase-linked J6/JFH1 genotype 2a HCV RNAs (Huh7.5-Rluc-J6/JFH1) [21], these cells were incubated with 0.1 % of DMSO, 100 nM of NS5A inhibitor (BMS-790052) [32], or six different mangosteen extracts for 72 h at 10 μg/mL. DMSO and NS5A inhibitor (BMS-790052) were used as negative and positive controls, respectively. Among various mangosteen extracts, ethanol extract (MG-EtOH) was able to induce the most dramatic decrease in HCV replication without any significant cytotoxicity compared with the DMSO-treated mock control (Fig. 1a). Thus, we decided to focus on antiviral effect of MG-EtOH in the following studies, and measured its dose-dependent and time-dependent effects on HCV replication. As shown in Fig. 1b, EC50 (concentration of compound required to reduce virus replication by 50 %) and CC50 (concentration of compound required to reduce host cell viability by 50 %) values of MG-EtOH were determined to be 5.5 and 12.8 μg/mL, respectively. In addition, treatment of MG-EtOH at 10 μg/mL was able to reduce HCV replication in half within 22.3 h (T1/2) without any major cytotoxicity (Fig. 1c).
MG-EtOH inhibits HCV genome replication. a Effect of various extracted fractions from mangosteen fruit peels by methanol, water, hexane, butanol, ethyl acetate, and ethanol on HCV genome replication and cell viability. Huh7.5 hepatocarcinoma cells were transfected with renilla luciferase-linked J6/JFH1 genotype 2a HCV RNAs (Huh7.5-Rluc-J6/JFH1) [21.] These cells were incubated with extracted fractions from MG fruit peels for 72 h at 10 μg/mL. Renilla luciferase and MTT-based cell viability assays were conducted to assess HCV replication and cytotoxicity. BMS-79052, an NS5A inhibitor was used as a positive control. b A dose–response curve was determined by measuring relative cell viabilities as well as renilla luciferase activities in Rluc-J6/JFH1 RNA-transfected Huh7.5 cells treated with an increasing concentration of MG-EtOH for 72 h. c A time-response curve was determined by measuring relative cell viabilities as well as renilla luciferase activities in Rluc-J6/JFH1 RNA-transfected Huh7.5 cells treated with 10 μg/mL of MG-EtOH for an increasing period of time. A dose–response graph was determined by measuring relative HCV well as GAPDH RNA levels via real-time qRT-PCR analyses of Bart79I RNA-transfected Huh7.5 cells (d) or J6/JFH-1 RNA-transfected Huh7.5 cells (e) treated with an increasing concentration of MG-EtOH for 72 h
In order to confirm suppression of HCV replication by MG-EtOH in a reporter-free HCV replicon system, we decided to perform a quantitative RT-PCR to measure levels of viral genomic RNAs in HCV replicon cells treated with MG-EtOH. For this purpose, Huh7.5 cells were transfected with either genotype 1b subgenomic Bart79I or genotype 2a infectious J6/JFH1 HCV RNAs. In case of Huh7.5-Bart79I cells, they were incubated with G418 for 4 weeks to select for HCV replication-competent stable cells. Then, they were treated with an increasing concentration of MG-EtOH from 0 to 10 μg/mL for 3 days. GAPDH was used as an internal control for normalizing HCV RNAs. In this experiment, we found a significant decline in HCV replication of a genotype 1b subgenomic Bart79I replicon upon treatment of MG-EtOH with an EC50 value of 5.1 μg/mL (Fig. 1d). We also demonstrated a more potent inhibitory effect of MG-EtOH on a genotype 2a J6/JFH1 infectious replicon with an EC50 value of 3.8 μg/mL (Fig. 1e). These results strongly suggest that MG-EtOH inhibits HCV genome replication in a dose- and time-dependent manner.
MG-EtOH down-regulates expression of HCV proteins
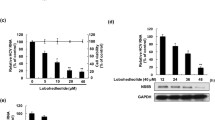
HCV genome replication is coupled to its protein translation. Therefore, blockage of HCV replication naturally leads to disruption of viral protein synthesis. In order to see if suppression of HCV replication by MG-EtOH could result in down-regulation of expression of HCV proteins, we treated Huh7.5-Bart79I or Huh7.5-J6/JFH1 cells with an increasing concentration of MG-EtOH. At 5 days post-treatment, we performed Western blot analyses to compare levels of NS5A protein in Huh7.5-Bart79I cells and core protein in Huh7.5-J6/JFH1 cells. An NS5A inhibitor, BMS-790052 and an anti-β-actin antibody were used as positive and loading controls, respectively. As shown in Fig. 2a, levels of NS5A proteins in Huh7.5-Bart79I cells were gradually reduced upon treatment of an increasing concentration of MG-EtOH with an EC50 value of 6.5 μg/mL. As expected, treatment of an NS5A inhibitor at 100 nM almost completely abrogated expression of NS5A proteins (2.6 %) when compared with untreated Huh7.5-Bart79I cells (100 %). In line with this result, treatment of MG-EtOH at 5 and 10 μg/mL also resulted in a significant drop in the number of NS5A-positive cells as shown in the immunofluorescence analysis of Huh7.5-Bart79I cells using an anti-NS5A antibody (Fig. 2b). As shown in Fig. 2c, similar dose-dependent down-regulation of expression levels of core protein was also observed in MG-EtOH-treated Huh7.5-J6/JFH1 cells with an EC50 value of 6.8 μg/mL.
MG-EtOH down-regulates expression of HCV proteins. A dose-dependent effect of MG-EtOH on expression of HCV proteins was determined by measuring relative levels of NS5A (a) or core (b) as well as β-actin proteins via Western blot analysis of Huh7.5-Bart79I cells (a) or Huh7.5-J6/JFH1 (b) treated with an increasing concentration of MG-EtOH for 120 h. 100 nM of NS5A inhibitor, BMS-790052 was used as a positive control. Numbers shown below two blots represent a relative quantitation of protein levels after normalization to β-actin levels. c Effect of MG-EtOH on expression of HCV proteins was determined by visualizing relative levels of NS5A proteins via immunofluorescence analysis of Huh7.5-Bart79I cells treated with 1 % of DMSO, 5, or 10 μg/mL of MG-EtOH for 120 h. These cells were fixed, blocked with 2 % FBS, and then stained in green by anti-NS5A and Alexa 488 antibodies. Nuclei were stained in blue with DAPI. The images were observed and taken by Nikon confocal laser scanning microscope. Scale bar represents 50 μM. d A dose-dependent effect of MG-EtOH on expression of HCV proteins was determined by measuring relative percentages of HCV NS5A-YFP-positive cells via FACS analysis of Bart79I-YFP RNA-transfected Huh7.5 cells treated with an increasing concentration of MG-EtOH for 120 h. 100 nM of NS5A inhibitor, BMS-790052 was used as a positive control (Color figure online)
In order to further demonstrate reduction of expression of HCV proteins by MG-EtOH, we decided to use Huh7.5-Bart79I-NS5A-YFP cells, which stably maintain genotype 1b subgenomic Bart79I replicons. These cells constitutively express NS5A as an YFP fusion protein to support HCV genome replication in the presence of G418 antibiotic [33]. We were able to confirm stable replication of Bart79I-NS5A-YFP subgenomic replicon by observing a significantly enhanced percentage (44.6 %) of YFP-positive cells when compared with untransfected Huh7.5 cells (0.1 %) by FACS analysis (Fig. 2d). When we treated these cells with an increasing dose of MG-EtOH from 0 to 10 μg/mL for 5 days, we were able to see concentration-dependent reduction in the percentage of YFP-positive cells with an EC50 value of 7.1 μg/mL (Fig. 2d). As expected, treatment of an NS5A inhibitor at 100 nM resulted in complete loss of expression of NS5A-YFP proteins (0.1 %). Therefore, all these results strongly suggested that MG-EtOH is able to down-regulate expression of HCV proteins through inhibition of viral genome replication.
MG-EtOH affects neither HCV entry nor viral protease/polymerase activities
After confirming an inhibitory effect of MG-EtOH on HCV replication, we would like to see if this inhibitory activity of MG-EtOH is due to any defects in HCV entry, NS3-mediated polyprotein processing, or NS5B RNA polymerase activity itself. First, we studied an effect of MG-EtOH on HCV entry using an HCV pseudo particle system (HCVpp) [25]. In this system, pseudo viral particles containing a firefly luciferase-linked retrovirus genome are generated with the help of retroviral transduction system. These particles are studded with HCV E1 and E2 proteins. Therefore, a firefly luciferase activity measured here is directly proportional to a level of E1 and E2-mediated HCV entry. As shown in Fig. 3a, in spite of an increasing dose of MG-EtOH, an efficiency of either genotype 1b or 2a HCV entry remained relatively unchanged. This suggests that MG-EtOH does not affect any steps in HCV entry process. Of note, pseudo virus particles with vesicular stomatitis virus glycoprotein (VSV-G) showed 104-fold higher entry efficiency than HCVpp (Fig. 3a).
MG-EtOH affects neither HCV entry nor viral protease/polymerase activities. a Effect of MG-EtOH on HCV entry was examined by measuring a firefly luciferase activity from Huh7.5 cells infected with pseudo viral particles containing a firefly luciferase-linked retrovirus genome studded with HCV E1 and E2 proteins in the presence of an increasing dose of MG-EtOH. b Effect of MG-EtOH on NS3-mediated polyprotein processing was examined by measuring expression level of NS3 and β-actin proteins in Huh7.5 cells infected with a T7 RNA polymerase-expressing vaccinia virus and transfected with a J6/JFH1 plasmid in the presence of an increasing dose of MG-EtOH by Western blot analyses. c Effect of MG-EtOH on NS5B RdRp activity was examined by measuring in vitro RdRp activity of NS5B in the presence of an increasing dose of MG-EtOH
Viral polyprotein processing is mediated by host signal peptidases as well as viral NS3/4A proteases. In order to test any potential effects of MG-EtOH on this NS3-mediated polyprotein processing, we used a vaccinia infection and transfection system. In this system, transfected HCV replicon plasmids are continuously transcribed into HCV RNAs through a vaccinia virus-encoded T7 polymerase. This system uncouples HCV translation and replication, allowing us to examine effect of MG-EtOH on polyprotein processing without interference on HCV replication. As shown in Fig. 3b, a expected size of NS3 protein was expressed from a newly transcribed genotype 2a HCV RNA. This NS3 protein expression was not affected by addition of MG-EtOH. This suggests that inhibition of HCV replication by MG-EtOH is not dependent on HCV polyprotein processing.
There is another possibility that MG-EtOH might suppress HCV replication by inhibiting the RNA polymerase activity of NS5B itself. In order to test our hypothesis, effect of MG-EtOH on RNA-dependent RNA polymerase (RdRp) activity of HCV NS5B was evaluated by the standard primer-dependent elongation reaction using poly rA/U20 template-primer and recombinant HCV NS5B according to previously described procedures [28]. As shown in Fig. 3c, treatment with MG-EtOH up to a concentration of 1 μg/mL was not able to reduce RNA polymerase activity of NS5B. However, at 10 μg/mL, MG-EtOH showed a modest inhibitory effect on RNA polymerase activity of NS5B (26.4 % inhibition). These data suggest that inhibition of NS5B RNA polymerase by MG-EtOH may play a minor role in suppression of HCV replication.
Additive effect of MG-EtOH and NS5A inhibitor on HCV replication
Recently, a new class of anti-HCV drugs targeting NS5A was identified by a cell-based random high-throughput HCV replicon screen [34]. They were then further optimized through structure–activity relationship analysis to yield BMS-790052. This compound, latter called “daclatasvir” was one of the most potent HCV replication inhibitors described to date, with reported in vitro EC50 values of 9 and 71 pM against HCV genotypes 1b and 2a, respectively [35]. Clinical trial data related with daclatavir have been also very promising so far [36]. Therefore, it would be interesting to use MG-EtOH in combination treatment with daclatavir to see if there is any contraindication for treatment or drug/drug interaction. For this purpose, we determined EC50 values of MG-EtOH in the presence of either 0.05 or 0.1 pM of daclatasvir using Huh7.5-Rluc-J6/JFH1 cells. As shown in Fig. 1b previously, an EC50 value of MG-EtOH was 5.5 μg/mL by itself. When combined with daclatavir at concentration of either 0.05 or 0.1 pM, its EC50 values further decreased to 4.73 and 2.83 μg/mL (Fig. 4). These data suggest that there is at least additive effect of MG-EtOH and NS5A inhibitor on HCV replication.
Additive effect of combined treatment of MG-EtOH and an NS5A inhibitor on HCV replication. Effect of combined treatment of MG-EtOH and daclatasvir on HCV replication was examined by measuring a renilla luciferase activity from Rluc-J6/JFH1 RNA-transfected Huh7.5 cells treated with an increasing concentration of MG-EtOH in the presence of either 0.05 or 0.1 pM of an NS5A inhibitor
Analysis of MG-EtOH and purification of α- and γ-mangostins
After confirming a negative effect of MG-EtOH on HCV replication, we wished to identify single compounds within MG-EtOH responsible for inhibition of HCV replication. For this purpose, we decided to perform HPLC analysis of MG-EtOH. α- and γ-mangostins were known to be the two most abundant constituents present in MG-EtOH based on HPLC chromatogram shown in other literatures (Fig. 5a) [19, 37]. In order to isolate α- and γ-mangostins as single compounds, the percarps of mangostin were pulverized, extracted with ethanol, suspended in H2O, and then partitioned with n-hexane, chloroform, ethyl acetate, and n-butanol. Repeated chromatographic separation of the chloroform-soluble extract led to the isolation of α-mangostin (Fig. 5b) and γ-mangostin (Fig. 5c). The structures of α- and γ-mangostins were confirmed by interpretation of the measured NMR spectroscopic data (data not shown) and comparison with published values in the literature [19]. Furthermore, quantitative analysis was carried out in order to provide the contents of α- and γ-mangostins in MG-EtOH using HPLC–UV method. From this analysis, it was found that α- and γ-mangostins constituted 25 and 4.7 %, respectively, with a ratio of 5:1 approximately of MG-EtOH (w/w) (Fig. 5d).
α- and γ-mangostins inhibit HCV genome replication
After finding α- and γ-mangostins as two major components of MG-EtOH, we wished to examine their effect on HCV replication. For this purpose, we decided to re-utilize Huh7.5-Rluc-J6/JFH1 cells, which were employed previously to measure HCV replication. These cells were treated with an increasing concentration of either α- or γ-mangostins from 0 to 10 μM for 3 days. Both α- and γ-mangostins exhibited antiviral activities with EC50 values of 6.3 μM for α-mangostin and 2.7 μM of γ-mangostin, respectively (Fig. 6a, b). However, both compounds produced a pronounced degree of cytotoxicity when cells were treated with concentration higher than 5 μM, yielding CC50 values of 9.9 μM for α- mangostin and 8.2 μM for γ-mangostin, respectively (Fig. 6a, b). Of note, we found that cell viability remained relatively unaffected up to 10 μM in case of MG-EtOH treatment in a prior experiment (Fig. 2b). According to the result of HPLC analysis, we realized that the ratio of α- and γ-mangostins naturally found in MG-EtOH turned out to be around 5:1 (Fig. 5d). Therefore, we decided to fix the concentration of γ-mangostin at 1 μM and increase the concentration of α-mangostin gradually. Under this condition, α-mangostin was able to block HCV replication with an EC50 value of 2.5 μM without any significant cytotoxicity up to concentration of 8 μM (Fig. 6c). These results indicate that two major constituents of MG-EtOH, α- and γ-mangostins play a crucial role in suppression of HCV replication when they coexist in a naturally occurring ratio.
α- and γ-mangostins inhibit HCV genome replication. Dose–response curves were determined by measuring relative cell viabilities as well as renilla luciferase activities in Rluc-J6/JFH1 RNA-transfected Huh7.5 cells treated with an increasing concentration of either α-mangostin (a) or γ-mangostin (b) for 72 h. c Huh7.5-RLuc-J6/JFH1 cells were incubated with an increasing concentration of α-mangostin in the presence of 1 μM of γ-mangostin for 72 h. Renilla luciferase and MTT-based cell viability assays were conducted to assess HCV replication and cytotoxicity
α- and γ-mangostins down-regulate expression of HCV proteins
In order to see if suppression of HCV replication by α- and γ-mangostins could result in reduced expression of HCV proteins, we treated Huh7.5-Bart79I or Huh7.5-J6/JFH1 cells with an increasing concentration of α-mangostin in the presence of 1 μM of γ-mangostin. At 5 days post-treatment, we performed Western blot analyses using an anti-NS5A for Huh7.5-Bart79I or anti-NS3 antibodies for Huh7.5-J6/JFH1 replicon cells, respectively. Around 6 μM of α-mangostin was required to reduce HCV protein levels by half with 1 μM of γ-MG regardless of genotype 1b or 2a HCV replicons (Fig. 7a, b). Therefore, these results suggested that co-treatment of α- and γ-mangostins lead to down-regulation of expression of HCV proteins.
α- and γ-mangostins down-regulate expression of HCV proteins. a A dose-dependent effect of α- and γ-mangostin on expression of HCV proteins was determined by measuring relative levels of NS5A as well as β-actin proteins via Western blot analysis of Huh7.5-Bart79I cells treated with an increasing concentration of α-mangostin in the presence of 1 μM of γ-mangostin for 120 h. 100 nM of NS5A inhibitor, BMS-790052 was used as a positive control. Numbers shown below two blots represent a relative quantitation of NS5A levels after normalization to β-actin levels. b A dose-dependent effect of α- and γ-mangostins on expression of HCV proteins was determined by measuring relative levels of HCV core as well as host β-actin proteins via Western blot analysis of J6/JFH1 RNA-transfected Huh7.5 cells treated with an increasing concentration of α-mangostin in the presence of 1 μM of γ-mangostin for 120 h. Numbers shown below two blots represent a relative quantitation of core levels after normalization to β-actin levels
MG-EtOH restores increased ROS production by HCV to a normal level
HCV-induced enhancement of oxidative stress and anti-oxidant effect of mangosteen have been well characterized previously [11, 19]. Therefore, we hypothesized that inhibition of HCV replication by MG-EtOH might play a role in restoration of HCV-induced up-regulation of oxidative stress. In order to test this hypothesis, an ROS-sensitive fluorescent dye, DCFH-DA was used to compare ROS levels in the untransfected and HCV RNAs-transfected Huh7.5 cells. As expected, H2O2 treatment gave rise to around 4 and eightfold increases in the levels of ROS in the untransfected and HCV J6/JFH1 RNA-transfected cells, respectively (Fig. 8a). The endogenous level of ROS in the Huh7.5-J6/JFH1 cells was found to be 1.5-fold higher than that in the untransfected cells (Fig. 8a). Interestingly, when Huh7.5-J6/JFH1 cells were treated with an increasing amount of MG-EtOH from 0 to 10 μg/mL, the level of ROS came down to the level equivalent to that found in the untransfected normal Huh7.5 cells. Interestingly, H2O2 itself does not have any effect on HCV replication (Fig. 8b). This result suggests that ROS-scavenging activity of MG-EtOH might play a minor role in disruption of HCV genome replication by MG-EtOH.
MG-EtOH restores increased ROS production by HCV to a normal level. a A dose-dependent effect of MG-EtOH on levels of ROS was determined by measuring relative cell viabilities as well as levels of ROS in the untransfected and Rluc-J6/JFH1 RNA-transfected Huh7.5 cells treated with an increasing concentration of MG-EtOH for 72 h. The ROS level was normalized to cell viability. H2O2 was used as a positive control. b A dose-dependent effect of H2O2 on HCV replication as well as cell viability was determined by measuring a renilla luciferase activity and relative cell viability in Rluc-J6/JFH1 RNA-transfected Huh7.5 cells treated with an increasing concentration of H2O2 for 72 h
Discussion
In this study, we found that MG-EtOH reduces HCV replication in a dose- and time-dependent manner using a renilla luciferase HCV reporter replicon. We also confirmed this MG-EtOH-induced inhibition of HCV replication using a quantitative RT-PCR assay in a reporter-free HCV replicon cells (Fig. 1). Moreover, Western blot, immunofluorescence, and FACS analyses further showed a subsequent down-regulation of levels of HCV proteins including NS5A and core upon treatment of MG-EtOH (Fig. 2). In addition, we also found that MG-EtOH affects neither HCV entry nor viral protease/polymerase activities (Fig. 3). Additive effect of MG-EtOH together with an NS5A inhibitor was also confirmed (Fig. 4). In order to identify active single compounds in MG-EtOH responsible for this abrogation of HCV replication, we performed an HPLC analysis and found α- and γ-mangostins as two major antiviral constituents in MG-EtOH (Fig. 5). A combined treatment of α- and γ-mangostins resulted in a significant decline in both HCV RNA and protein levels, further suggesting α- and γ-mangostins as major players required for disruption of HCV replication (Figs. 6, 7). Finally, we demonstrated restoration of increased production of ROS by HCV to a normal level by treatment of MG-EtOH (Fig. 8a). All these data strongly suggest a direct blockage of HCV replication by mangosteen.
There have been many literatures suggesting an important role of ROS in HCV-induced pathogenesis including liver cirrhosis and hepatocelluar carcinoma (HCC) [10, 11]. However, whether ROS play a beneficial or detrimental role in the life cycle HCV is still controversial. According to a paper published by Choi et al. [38], ROS production by treatment of peroxide was shown to be able to suppress HCV RNA replication in human hepatoma cells. In addition, it was also demonstrated that a widely used anti-malarial drug, artemisinin can act as potent inhibitors of HCV replication through its induction of ROS [39]. Interestingly, they also observed that co-treatment of a well-characterized anti-oxidant, N-acetyl-l-cysteine significantly reduced the antiviral activity of Artemisinin. All these data strongly implies a negative role of ROS in HCV RNA replication. However, other previously published studies also suggested seemingly contradictory results in terms of molecular functions of ROS in HCV infection. For example, beneficial effect of anti-oxidant such as N-acetyl-l-cysteine [40] and vitamin E [41] by decreasing HCV replication was also demonstrated. In addition, another paper also reported that acetylsalicylic acid is able to suppress HCV replication through its anti-oxidant activity by induction of Cu/Zn superoxide dismutase [42]. Our discovery of mangosteen as an inhibitor of HCV replication favors a positive role of ROS production in regulation of HCV RNA genome replication.
Reason for this discrepancy in terms of function of ROS in HCV is still unclear. In our experiment, ROS induced by treatment of H2O2 failed to induce any significant changes in HCV replication (Fig. 8b). It might be due to the fact that HCV may require an optimal level of ROS production in order to fully support its RNA genome replication. In view of this hypothesis, too much of ROS production may result in disruption of architectural structures of subcellular compartments such as Gogi body, endoplasmic reticulum, and lipid droplet, which are necessary to maintain HCV genome replication. On the other hand, too little of ROS production may lead to an inability to stimulate ROS-dependent signaling pathways involving activation of nuclear factor kB and activating protein-1, which are necessary for successful completion of HCV genome replication. Treatment of mangosteen might be able to reset ROS equilibrium from more pronounced oxidant state due to expression of HCV proteins to an ROS-deficient condition. Under this condition, efficient HCV replication may not be able to take place. In this regard, it would be interesting to examine what kind of ROS-dependent signaling pathways fails to activate upon treatment of mangosteen in order to understand an antagonizing effect of mangosteen on HCV replication at molecular level in a more detailed way.
In an effort to identify a potential target viral gene for mangosteen, we tried to generate HCV variant mutants resistant to MG-EtOH by co-culturing Bart79I-transfected genotype 1b replicon cells with MG-EtOH at threefold concentration of EC50 (16.5 μg/mL) for more than 1 month under pressure of G418 selection. However, we failed to isolate any MG-EtOH-resistant HCV variants. Therefore, we were unable to determine a potential viral target for mangosteen. We concluded that this could be due to a relatively narrow therapeutic window between antiviral and cytotoxicity curves (EC50 5.5 μg/mL and CC50 12.8 μg/mL). In order to successfully isolate a variant mutant HCV genome resistant to mangosteen, mangosteen needs to kill all of HCV replicon cells which lost resistance to G418 due to suppression of HCV replication. In parallel, mangosteen should not kill HCV replicon cells which gained resistance to G418 thanks to its mutations resistant to mangosteen. In our several attempts, we also tried 2 (11 μg/mL) and 4 (22 μg/mL) fold EC50 concentrations for selection of resistant variants in vain. We speculate that these two concentrations also killed HCV replicon cells which gained resistance to G418 due to their cytotoxic effects. In addition, we were not able to detect any significant effect of MG-EtOH on HCV IRES-dependent translation of a viral RNA genome (data not shown). These data suggest that mangosteen inhibition of HCV RNA replication does not take place at the translational level. According to our HCV enzymatic assays, MG-EtOH did not affect HCV NS3 protease (Fig. 3b). In case of NS5B polymerase, its RdRd activity was slightly inhibited only when concentration of MG-EtOH was raised up to 10 μg/mL (Fig. 3c). This suggests that inhibition of HCV NS5B polymerase by MG-EtOH may play a minor role in exerting its antiviral activity.
HCV infection correlates cirrhosis and liver cancer development [43.] Unfortunately, the mechanism underlying HCV-induced hepato-carcinogenesis is not fully understood yet. Moriya et al. [44] addressed that the transgenic mice carrying the core gene developed hepatocellular carcinoma (HCC) as well as that oxidative stress in the liver was increased by expression of the core protein. Moreover, not only genetic mutation, but chromosomal modifications also can be induced by ROS, and thus make a contribution to cancer development in multistep carcinogenesis [20, 45.] Therefore, we suppose that mangosteen could be beneficial to treatment of HCV-induced development of HCC due to its dual action in suppression of HCV replication as well as reduction of ROS. In this regard, an HCV-induced liver cancer model using a small animal would be helpful to test a possible dual activity of mangosteen. Furthermore, it would be necessary to investigate potential efficacy of combination treatment of mangosteen with interferon-α and ribavirin in order to test their potential synergism and clinical efficacy of combined therapy.
In conclusion, we demonstrated anti-HCV replication activity of mangosteen. We also identified α- and γ-mangostins as two major single molecules responsible for its antiviral effects. Based on these results, we envisage that consumption of mangosteen as a dietary supplement together with other approved anti-HCV drugs might be considered as an alternative for treatment of HCV patients.
Abbreviations
- HCV:
-
Hepatitis C virus
- ROS:
-
Reactive oxygen species
- HCC:
-
Hepatocellular carcinoma
- MG-EtOH:
-
Magosteen ethanol extract
References
M.J. Alter, D. Kruszon-Moran, O.V. Nainan, G.M. McQuillan, F. Gao, L.A. Moyer, R.A. Kaslow, H.S. Margolis, New Engl. J. Med. 341, 556–562 (1999)
A.M. Di Bisceglie, Hepatology 31, 1014–1018 (2000)
C.W. Shepard, L. Finelli, M.J. Alter, Lancet Infect. Dis 5, 558–567 (2005)
S. Mukherjee, M.F. Sorrell, Gastroenterology 134, 1777–1788 (2008)
S.W. Ryter, H.P. Kim, A. Hoetzel, J.W. Park, K. Nakahira, X. Wang, A.M. Choi, Antioxid. Redox Signal 9, 49–89 (2007)
A. Grakoui, C. Wychowski, C. Lin, S.M. Feinstone, C.M. Rice, J. Virol. 67, 1385–1395 (1993)
G. Gloire, S. Legrand-Poels, J. Piette, Biochem. Pharmacol. 72, 1493–1505 (2006)
H.M. Lander, FASEB J. 11, 118–124 (1997)
A.V. Ivanov, B. Bartosch, O.A. Smirnova, M.G. Isaguliants, S.N. Kochetkov, Viruses 5, 439–469 (2013)
A.V. Ivanov, O.A. Smirnova, O.N. Ivanova, O.V. Masalova, S.N. Kochetkov, M.G. Isaguliants, PLoS ONE 6, e24957 (2011)
H.A. Jung, B.N. Su, W.J. Keller, R.G. Mehta, A.D. Kinghorn, J. Agric. Food Chem. 54, 2077–2082 (2006)
X. Forns, R.H. Purcell, J. Bukh, Trends Microbiol. 7, 402–410 (1999)
K.J. Blight, A.A. Kolykhalov, C.M. Rice, Science 290, 1972–1974 (2000)
P.D. Bryson, N.J. Cho, S. Einav, C. Lee, V. Tai, J. Bechtel, M. Sivaraja, C. Roberts, U. Schmitz, J.S. Glenn, Antiviral Res. 87, 1–8 (2010)
S.X. Chen, M. Wan, B.N. Loh, Planta Med. 62, 381–382 (1996)
Y.W. Chin, H.A. Jung, H. Chai, W.J. Keller, A.D. Kinghorn, Phytochemistry 69, 754–758 (2008)
Y.W. Chin, A.D. Kinghorn, Mini Rev. Org. Chem. 5, 355–364 (2008)
M.T. Chomnawang, S. Surassmo, V.S. Nukoolkarn, W. Gritsanapan, Fitoterapia 78, 401–408 (2007)
S.M. Al-Massarani, A.A. El Gamal, N.M. Al-Musayeib, R.A. Mothana, O.A. Basudan, A.J. Al-Rehaily, M. Farag, M.H. Assaf, K.H. El Tahir, L. Maes, Molecules 18, 10599–10608 (2013)
N. Fujita, R. Sugimoto, N. Ma, H. Tanaka, M. Iwasa, Y. Kobayashi, S. Kawanishi, S. Watanabe, M. Kaito, Y. Takei, J. Viral Hepat. 15, 498–507 (2008)
D.M. Tscherne, C.T. Jones, M.J. Evans, B.D. Lindenbach, J.A. McKeating, C.M. Rice, J. Virol. 80, 1734–1741 (2006)
B.D. Lindenbach, M.J. Evans, A.J. Syder, B. Wolk, T.L. Tellinghuisen, C.C. Liu, T. Maruyama, R.O. Hynes, D.R. Burton, J.A. McKeating, C.M. Rice, Science 309, 623–626 (2005)
K.J. Blight, J.A. McKeating, C.M. Rice, J. Virol. 76, 13001–13014 (2002)
C. Lee, Biomol. Ther. 21, 97–106 (2013)
J. Zhang, G. Randall, A. Higginbottom, P. Monk, C.M. Rice, J.A. McKeating, J. Virol. 78, 1448–1455 (2004)
M. Elazar, K.H. Cheong, P. Liu, H.B. Greenberg, C.M. Rice, J.S. Glenn, J. Virol. 77, 6055–6061 (2003)
S.J. Kim, J.H. Kim, Y.G. Kim, H.S. Lim, J.W. Oh, J. Biol. Chem. 279, 50031–50041 (2004)
S.J. Kim, J.H. Kim, J.M. Sun, M.G. Kim, J.W. Oh, J. Viral Hepat. 16, 697–704 (2009)
D.G. Ahn, S.B. Shim, J.E. Moon, J.H. Kim, S.J. Kim, J.W. Oh, J. Viral Hepat. 18, e298–e306 (2011)
S. Baek, S.M. Kim, S.A. Lee, B.Y. Rhim, S.K. Eo, K. Kim, Biomol. Therap. 21, 42–48 (2013)
S. Yodhnu, A. Sirikatitham, C. Wattanapiromsakul, J. Chromatogr. Sci. 47, 185–189 (2009)
C. Lee, H. Ma, J.Q. Hang, V. Leveque, E.H. Sklan, M. Elazar, K. Klumpp, J.S. Glenn, Virology 414, 10–18 (2011)
G. Jin, S. Lee, M. Choi, S. Son, G. Kim, J. Oh, C. Lee, K. Lee, Eur. J. Med. Chem. 75, 413–425 (2014)
J.A. Lemm, D. O’Boyle 2nd, M. Liu, P.T. Nower, R. Colonno, M.S. Deshpande, L.B. Snyder, S.W. Martin, D.R. St Laurent, M.H. Serrano-Wu, J.L. Romine, N.A. Meanwell, M. Gao, J. Virol. 84, 482–491 (2010)
C. Lee, Arch. Pharmacol. Res. 34, 1403–1407 (2011)
M. Gao, R.E. Nettles, M. Belema, L.B. Snyder, V.N. Nguyen, R.A. Fridell, M.H. Serrano-Wu, D.R. Langley, J.H. Sun, D.R. O’Boyle 2nd, J.A. Lemm, C. Wang, J.O. Knipe, C. Chien, R.J. Colonno, D.M. Grasela, N.A. Meanwell, L.G. Hamann, Nature 465, 96–100 (2010)
Y.H. Choi, S.Y. Han, K. You-Jin, Y.M. Kim, Y.W. Chin, Food Chem. Toxicol. 66, 140–146 (2013)
J. Choi, K.J. Lee, Y. Zheng, A.K. Yamaga, M.M. Lai, J.H. Ou, Hepatology 39, 81–89 (2004)
S. Obeid, J. Alen, V.H. Nguyen, V.C. Pham, P. Meuleman, C. Pannecouque, T.N. Le, J. Neyts, W. Dehaen, J. Paeshuyse, PLoS ONE 8, e81783 (2013)
F. Farinati, R. Cardin, N. De Maria, L.G. Della, C. Marafin, E. Lecis, P. Burra, A. Floreani, A. Cecchetto, R. Naccarato, J. Hepatol. 22, 449–456 (1995)
M.P. Look, A. Gerard, G.S. Rao, T. Sudhop, H.P. Fischer, T. Sauerbruch, U. Spengler, Antiviral Res. 43, 113–122 (1999)
A.M. Rivas-Estilla, O.L. Bryan-Marrugo, K. Trujillo-Murillo, D. Perez-Ibave, C. Charles-Nino, C. Pedroza-Roldan, C. Rios-Ibarra, E. Ramirez-Valles, R. Ortiz-Lopez, M.C. Islas-Carbajal, N. Nieto, A.R. Rincon-Sanchez, Am. J. Physiol. 302, G1264–G1273 (2012)
K. Shirabe, M. Shimada, K. Kajiyama, T. Gion, Y. Ikeda, H. Hasegawa, K. Taguchi, K. Takenaka, K. Sugimachi, Cancer 83, 2312–2316 (1998)
K. Moriya, H. Fujie, Y. Shintani, H. Yotsuyanagi, T. Tsutsumi, K. Ishibashi, Y. Matsuura, S. Kimura, T. Miyamura, K. Koike, Nat. Med. 4, 1065–1067 (1998)
J. Kato, M. Kobune, T. Nakamura, G. Kuroiwa, K. Takada, R. Takimoto, Y. Sato, K. Fujikawa, M. Takahashi, T. Takayama, T. Ikeda, Y. Niitsu, Cancer Res. 61, 8697–8702 (2001)
Acknowledgments
We would like to thank Dr. Jong-Won Oh for providing all the reagents and advices necessary for in vitro HCV NS5B RNA polymerase assay. This work was supported by the GRRC program of Gyeonggi province [(GRRC-DONGGUK2012-A01), Study of control of viral diseases], [(GRRC-DONGGUK2012-B02), Development and discovery of new therapeutic target modulators]. This research was also supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2013R1A1A1011851).
Author information
Authors and Affiliations
Corresponding author
Additional information
Moonju Choi and Young-Mi Kim have contributed equally on this work.
Rights and permissions
About this article
Cite this article
Choi, M., Kim, YM., Lee, S. et al. Mangosteen xanthones suppress hepatitis C virus genome replication. Virus Genes 49, 208–222 (2014). https://doi.org/10.1007/s11262-014-1098-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-014-1098-0