Abstract
The strains of Newcastle disease virus (NDV) were isolated from five suspected outbreaks of ND in broiler (n = 3) and layers (n = 2) poultry farms. The egg-isolated viruses were subjected to biological and genetic characterization. Based on the biological characterization, isolates showed haemagglutination titer ≥log 27, mean death time <55 h, intracerebral pathogenecity index ≤1.8, and egg lethal dose 50 from 10−7.15 to 10−9.31/1 ml. Genetic characterization of the fusion (F) gene revealed that the isolates clustered with NDV strains of genotype VII (VIIf) within class II, which remained predominant genotype in the domestic poultry of Asia. The deduced amino acid sequence of the isolates confirmed virulent motif 112RRQKRF117 at the F protein cleavage site. A bioinformatics and pairwise comparison approach was applied to estimate the synonymous and non-synonymous substitution rates (dN/dS) and selective evolutionary pressure for the F protein. The dN/dS calculated for genotype VII indicate purifying selection, which resulted in a low evolution rate in F gene. The F protein shows a strong negative pressure throughout the length of the protein and no recombination event was determined. Collectively, the results suggest that very similar virulent strains of NDV are involved during current wave of disease outbreak throughout the country. From these results, in conjunction with our recent reports of NDV from Pakistan, it is possible to conclude that emergence of novel group may require revisiting the diagnostics and vaccine control strategies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Newcastle disease (ND) is a highly contagious disease of the birds caused by avian paramyxovirus type 1 (APMV-1) [1]. All avian species are susceptible; however, disease consequences are significantly higher in domestic poultry due to susceptibility to virulent strains [2]. Therefore, it is known to have serious economic losses to the poultry industry around the globe than any other viral diseases of poultry [1].
The NDV genome is approximately 15 kb long that encodes six proteins namely nucleocapsid protein (NP), phosphoprotein (P), matrix protein (M), fusion protein (F), hemagglutinin–neuraminidase (HN), and large RNA-dependent polymerase protein (L) in the order 3′-NP-P-M-F-HN-L-5′ [1]. Within a single serotype, the NDV exist in two distinct classes, class I and class II [3]. Class II strains of NDV, on the basis of partial hypervariable nucleotide sequence of the F gene, are classified in several genotypes (I–XI) [4–6]. Based on pathogenicity, the NDV strains are categorized as velogenic (highly pathogenic), mesogenic (intermediate), and lentogenic (mildly pathogenic) [7]. A specific motif at the cleavage site of the F protein is considered a crucial determinant of NDV pathogenicity. The presence of glutamine at the amino acid position 114 in both mesogenic and velogenic strains determine its pathogenicity, whereas lentogenic strains lack this virulent motif [8].
In Pakistan, both live and killed vaccines are available and are currently being used in both broiler and layer poultry farms. Despite of these extensive vaccination plans, outbreaks are continued to appear of which disease in vaccinated flocks is of special concern. Having these concerns in mind, it is of particular interest to gain insight in the circulating NDV strains and their epidemiological pattern throughout the country. In contrast to our previous study in which samples from northern part of the country were analyzed [5, 9, 10], the viruses were isolated and characterized from the central part, which is an active pole of the poultry but remained largely ignored. In this study, the nucleotide sequence and subsequent F gene-based phylogenetic and evolutionary relationship of the circulating NDVs in Pakistan were determined.
Materials and methods
Sampling history
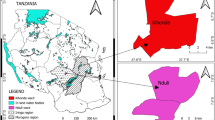
During an epidemic of NDV in 2010/2011, 130 samples (trachea, lung, spleen, cloaca, and blood) were collected from five clinically suspected outbreaks of NDV including commercial broiler (n = 3) and layer farms (n = 2) (Table 1). A mortality rate of 10–15 % per day was observed in broiler flocks, whereas 5–8 % per day was noticed in layer flocks. Collectively, the birds shown dullness, depression, difficult respiration, torticollis, and head tremor, which are believed to be typical signs of neurotropic NDV. All the infected flocks were vaccinated with commercially available lentogenic strain (LaSota) in drinking water.
Biological characterization
The tissue homogenates were inoculated in specific pathogen-free (SPF) eggs in order to isolate the viruses as described earlier [5]. Mean death time (MDT) and intracerebral pathogenicity indices (ICPI) were calculated as reported [1, 11]. The egg lethal dose 50 (ELD50) was calculated by Reed and Muench [12] method.
F gene amplification, sequencing, and phylogeny
The allantoic fluid from eggs, inoculated with clinical material, was impregnated on FTA QiaCard and shipped to Swedish University of Agricultural Sciences (SLU), Uppsala Sweden for further processing. One isolate from each outbreak was selected for analysis due to limited resources. The viral genome was eluted from these FTA QiaCard and run for real-time PCR assay targeting both matrix (M) and fusion (F) genes as we described before [5]. The positive samples were subjected to conventional PCR in order to amplify full-length F gene of NDV, as described before [5], using primers mentioned in Table 2. The amplified PCR products were gel purified with Wizard SV Gel and PCR Clean-Up System (Promega, Co., Madison, WI) according to the manufacturer’s instructions and directly sequenced using ABI PRISM BigDye Terminator version 3.1 (Applied Biosystems, Foster City, CA) as recommended by the manufacturer. Sequencing reactions were run on a 3100 DNA analyzer (Applied Biosystems) both in forward and backward directions. These sequences were assembled and edited in DNAstar Lasergene v8. The consensus sequence was used for the phylogenetic analysis by applying the Neighbor-joining method (Kimura 2-parameter) with 2,000 bootstrap replicates in Molecular Evolutionary Genetics Analysis (MEGA) version 5. The resultant nucleotide data were submitted to the GenBank and are available under accession numbers from JX436340 to JX436344.
Evolutionary study
Selective pressure for the putative F protein was determined using synonymous non-synonymous analysis program (SNAP) services (http://hcv.lanl.gov/content/sequence/SNAP/SNAP.html) [13]. The ratio of non-synonymous to synonymous substitutions (K a/K s ratio) for each amino acid site in the F-coding region was used to scan for evidence of positive or negative selection. To estimate the recombination events, the sequences were analyzed for a recombination detection program, RDP V.3.44. The window size was adjusted to 30 with the highest P value 0.05. The detection of recombination events were applied between sequences sharing 0 and 100 % identity. The NetNGlyc 1.0 Server was used to predict the potential N-glycosylation sites in the F protein (http://www.cbs.dtu.dk/services/NetNGlyc/).
Results
Isolation and pathogenicity assessment of NDV
Exudates from trachea, lungs, spleen, and caecal tonsils, collected aseptically from broilers (n = 3) and layers (n = 2) farms, were inoculated into the SPF egg. The allantoic fluid from individual egg was used for biological characterization including hemagglutination (HA) activity and in vivo pathogenicity tests. The HA titer were determined with 1 % washed chicken RBCs in V- shaped 96-well microtitre plates [14]. All the isolates had an HA titer ≥128 HA U/25 μl (Table 1). All the isolates exhibited MDT <55 h and ICPI <1.8 which are typical velogenic strains of NDV [6] (Table 1). The EID50 was found to be 10−7.15 to 10−9.31/1 ml.
Approximately 300 μl of allantoic fluid from isolates with high pathogenicity indices was used to impregnate FTA QiaCard Indicator (Qiagen, USA) to transport the samples to SLU, Uppsala, Sweden. This method served as an appropriate system for transportation of allantoic fluids. The RNA was eluted using defined protocol [5] and used for the screening of NDV genome in both M and F genes-based real-time PCR [15, 16]. All the samples showed Ct values of less than 20 in both assays indicate high viral genomic load. The complete F gene from each isolate was successfully amplified using the small volume of eluted RNA (4 μl).
Phylogenetic analysis
The robustness of the genetic grouping and topology of the phylogenetic tree was confirmed by comparing F gene sequences of current isolates and published sequences belonging to class I and class II of NDVs. The phylogenetic tree was constructed using selective sequences from Pakistan and other parts of the world representing all lineages and genotypes. The NDV strains were divided into two classes: I and II (Fig. 1). All the Pakistani strains of NDV, characterized in this study, clustered within class II of NDV along with previously reported strains from other parts of the country. Collectively, these strains were branched close to that of Swedish and Russian strains of NDV. As the NDV strains belonging to class I are primarily the strains of wild waterfowls, these strains were withdrawn from this analysis. One strain (DE-R49/99) was included in the tree as an out-group for the phylogenetic interpretation.
Phylogenetic tree based on full length F gene, constructed by the neighbor-joining method. Numbers indicates the bootstrap values (2,000 replicates). Horizontal distances are proportional to sequence distances. Isolates characterized in this study are marked with filled square, whereas isolates reported earlier from Pakistan are marked with empty square
In order to determine the sub-grouping pattern of these Pakistani strains of genotype VII, a phylogenetic analysis was conducted based on the previously reported strains of NDV belonging to all sub-groups of genotype VII. The strain of genotypes VI and V were taken as out-group (Fig. 2). The overall topology of the tree indicated the division into six clear sub-groups which as marked from VIIa to VIIf. It is interesting to observe that all the Pakistani NDV strains, either characterized in this study or reported previously, clustered in a separate sub-genotype (VIIf). However, the reported isolates, though similar in genotype to our previously characterized strains from Northern area of Punjab [5], are distinctly different in sub-genotype in central Punjab. Within this sub-genotype, these Pakistan NDV strains can be divided into two clusters, mainly composed of previously characterized NDV strains and the isolates reported in this study.
Phylogenetic tree based on full length F gene of genotype VII, constructed by the neighbor-joining method. Numbers indicates the bootstrap values (2,000 replicates). Horizontal distances are proportional to sequence distances. Isolates characterized in this study are marked with filled square, whereas isolates reported earlier from Pakistan are marked with empty square
F protein analysis
As a rule of thumb, the Class II isolates can be classified as velogenic viruses on the basis a pair of dibasic amino acids at the cleavage site motif. A consensus amino acid sequence of 112R/K-R-Q-R/K-R-F117 is required for the velogenic (pathogenic) strains of NDV and 112G/E-K/R-Q-G/E-R-L117 is present in lentogenic (apathogenic) NDV isolates. Analysis of the putative F protein indicated that isolates possess three different types of protease cleavage site motifs with significant geographical restrictions. A motif of 112RRQKRF117 was observed in Iranian, Indian, Israelis, Pakistanis, and most of Chinese isolates, whereas 112RRRKRF117 in some of the Pakistani and South Korean isolates. However, the NDV isolates of Class I, primarily from China, contain a unique motif (EQQERL) in the cleavage site of F protein. As was expected from the MDT and ICPI values, a motif 112R-R-Q-K-R-F117, which corresponds to the cleavage site of velogenic viruses, was observed in all the Pakistani isolates reported in this study.
The number and location of predicted N-linked glycosylation sites for the F protein of Pakistani NDV isolates were predicted and found to be conserved. The majority of the isolates had six potential N-glycosylation sites (Asn-Xaa-Ser/Thr) located at positions 85NRTL, 191NNTA, 366NTSA, 447NISI, 471NNSI, and 541NNT. However, the isolates presented in this study, contain substitution at position 541, which results in lost of the N-glycosylation in this site. Such phenomenon has also been observed in some of the Chinese NDV isolates (China/TZ060107/2011 and China ND-XX08/2009).
Evolutionary analysis
A pairwise comparison bioinformatics approach (SNAP) was applied to determine the synonymous and non-synonymous substitution rates and selective evolutionary pressure for the F protein. The results are presented in the Fig. 3, in which numbers above one indicate the positive selection, around one show the neutral selection, whereas below one indicate the negative or purifying selection. Analysis of the Pakistani NDV isolates studied in this report along with other all-available full-length F protein indicated a clear purifying pattern, as it is obvious from the graph (Fig. 3). Analysis of the ratio of synonymous and non-synonymous substitution rates in the isolates also demonstrated the presence of purifying (negative) selection in VII genotype. The calculated K a/K s ratios (0.517) for the F genes predicted a low evolution rate. Furthermore, no recombination events were revealed in the F gene of Pakistani NDV strains.
Discussion
In countries where NDV is endemic, emerging of different genotypes in both classes I and II represents a diverse and continually evolving group of NDV. As the first appearance of NDV in Southeast Asia (Indonesia) in 1926 and the start of organized poultry sector in Pakistan during 1964, the diseases remains endemic throughout the year [5, 9, 10, 15, 17]. The sequence analysis of the F gene appears to be an appropriate tool in understanding the origin of ND outbreaks and continuous surveillance of field isolates. The phylogenetic analysis, based on the F gene, has clearly predicted the circulation of multiple velogenic genotypes in Europe and Asia. Among them, genotype VII representing viruses that emerged in the 1990s in the Far East were prevalent and mainly responsible for ND outbreaks in Far East [18–20], Europe [21–23], and South Africa [24]. Despite the fact that NDV has been reported from selective regions of Pakistan, it is imperative to screen and characterize the NDV throughout the country. Such understanding is of special concern when emergence of novel genotype has recently been reported from Pakistan [5].
In this study, isolates (n = 5) were biologically and genotypically characterized. The tests for pathogenicity assessment showed that Pakistani isolates of NDV are velogenic, which was confirmed by the sequence analysis of the F protein cleavage site. Based on entire ORF sequence (nt 1–1,662), the sequence comparison and phylogenic analysis of the isolates grouped them in Class II and genotype VII. Genotype VI has also been reported in Pakistan [17]; however, it remain to be determined that when genotype VII started to emerge in Pakistan. Among the prevalent NDV genotypes worldwide, VII is considered to be the one involved in outbreaks in Europe [21], Far East [18–20], and South Africa [24]. The isolates from Far East and Israel have been categorized as VIId and from Iran and Indian sub-continent as sub-genotype VIIb. Apart from this, Aldous et al. [4] did report VIIa from the filed outbreaks in the Middle East, Europe, Taiwan, and India. However, in a recent studies, sub-genotype VIIf and a distinct lineage 5i has been reported from Pakistan [5] attributing it mainly due to high genetic difference between vaccinal and field strains of NDV and the broad genetic diversity. It is also not less likely that importation and use of non-recommended vaccines may likely contribute to evolve the local strains into more divergent strains, which demand extensive future investigations. The recent findings of lack of clinical outcome but continuous viral shedding of virulent NDV even from vaccinated birds [25] raised the need of revising the vaccination strains and their schedule accordingly. The continuous trend in genetic mutations may also lead to escape in diagnosis as has been reported in Pakistan [15, 17]. Moreover, wild captive birds and backyard poultry has been suggested to impart an important role in the epizootiology and evolution of NDV [5, 6, 26], which warrant further investigations.
The study provided valuable data on the evolution and circulation of NDVs in central Punjab in the face of current wave of NDV outbreaks in the country. Furthermore, the results collectively indicate that genotype VII of NDV still predominant in the domestic poultry of Asia, and these viruses are not evolutionarily widely different from the isolates reported in 1990s. The results urge the authorities to monitor poultry continuously and report immediately to OIE for its proper diagnosis and control through possible vaccination strategies in future. It is to suggest the use of homologous strains for immunization against NDV and to optimize the vaccination schedules according to diverse environmental conditions of the countries. Most importantly, the study underscores the need to investigate the nature of prevailing NDVs throughout Pakistan.
References
D.J. Alexander, in Disease of poultry, ed. by Y.M. Saif (Iowa State University Press, Ames, 2003)
M. van Boven, A. Bouma, T.H. Fabri, E. Katsma, L. Hartog, G. Koch, Avian Path. 37, 1–5 (2008)
K.S. Choi, E.K. Lee, W.J. Jeon, J.J. Nah, Y.J. Kim, M.Y. Lee, H. Lee, J.H. Kwon, J. Wildl. Dis. 44, 193–198 (2008)
E.W. Aldous, J.K. Mynn, J. Banks, D.J. Alexander, Avian Pathol. 32, 239–256 (2003)
M. Munir, M. Cortey, M. Abbas, Z.U. Qureshi, F. Afzal, M.Z. Shabbir, M.T. Khan, S. Ahmed, S. Ahmad, C. Baule, K. Stahl, S. Zohari, M. Berg, Infect. Genet. Evol. 12, 1010–1019 (2012)
S. Zhang, X. Wang, C. Zhao, D. Liu, Y. Hu, J. Zhao, G. Zhang, PLoS one 6, e25000 (2011)
R.P. Hanson, Newcastle Disease (American Association of Avian Pathologists, Kennett Square, 1980)
B.S. Seal, D.J. King, R.J. Meinersmann, Virus Res. 66, 1–11 (2000)
M. Munir, M. Abbas, M.T. Khan, S. Zohari, M. Berg, Virology J. 9, 46 (2012)
M. Munir, S. Zohari, M. Abbas, M. Berg, Arch. Virol. 157, 765–768 (2012)
OIE Terrestrial Manual (2009). http://www.oie.int/eng/normes/mmanual/2008/pdf/2.03.14_NEWCASTLE_DIS.pdf. Accessed 5 Dec 2012
L.J. Reed, L.H. Muench, Am. J. Hyg. 27, 493–497 (1938)
B. Korber, HIV Signature and Sequence Variation Analysis. Computational Analysis of HIV Molecular Sequences (Kluwer Academic Publishers, Dordrecht, 2000)
OIE. OIE Terrestrial Manual in OIE (ed), 2009
M. Munir, S. Zohari, M. Berg, Indian J. Virol. (2012). doi:10.1007/s13337-012-0073-4
M.G. Wise, D.L. Suarez, B.S. Seal, J.C. Pedersen, D.A. Senne, D.J. King, D.R. Kapczynski, E. Spackman, J. Clin. Microbiol. 42, 329–338 (2004)
T.A. Khan, C.A. Rue, S.F. Rehmani, A. Ahmed, J.L. Wasilenko, P.J. Miller, C.L. Afonso, J. Clin. Microbiol. 48, 1892–1894 (2010)
M. Mase, K. Imai, Y. Sanada, N. Sanada, N. Yuasa, T. Imada, K. Tsukamoto, S. Yamaguchi, J. Clin. Microbiol. 40, 3826–3830 (2002)
Z.M. Qin, L.T. Tan, H.Y. Xu, B.C. Ma, Y.L. Wang, X.Y. Yuan, W.J. Liu, J. Clin. Microbiol. 46, 601–611 (2008)
L.T. Tan, H.Y. Xu, Y.L. Wang, Z.M. Qin, L. Sun, W.J. Liu, Z.Z. Cui, J. Clin. Microbiol. 46, 750–753 (2008)
J. Herczeg, E. Wehmann, R.R. Bragg, P.M. Travassos Dias, G. Hadjiev, O. Werner, B. Lomniczi, Arch. Virol. 144, 2087–2099 (1999)
M. Munir, A.M. Linde, S. Zohari, K. Stahl, C. Baule, B. Engstrom, L.H. M Renström, Virus Genes 43, 261–271 (2011)
M. Munir, A.M. Linde, S. Zohari, K. Stahl, C. Baule, K. Holm, B. Engstrom, M. Berg, Avian Dis. 54, 923–930 (2010)
C. Abolnik, R.F. Horner, S.P. Bisschop, M.E. Parker, M. Romito, G.J. Viljoen, Arch. Virol. 149, 603–619 (2004)
S. Hu, H. Ma, Y. Wu, W. Liu, X. Wang, Y. Liu, X. Liu, Vaccine 27, 904–910 (2009)
N. Jindal, Y. Chander, A.K. Chockalingam, M. de Abin, P.T. Redig, S.M. Goyal, Virol. J. 6, 191 (2009)
Acknowledgments
The authors would like to acknowledge field veterinarians and poultry farmers for their help in sample collection. Additionally, help of Muhammad Arshad and Imran Shafique in sample processing and Martí Cortéy in evolutionary analysis is greatly acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Muhammad Zubair Shabbir and Muhammad Abbas equally contributed as first authors.
Rights and permissions
About this article
Cite this article
Shabbir, M.Z., Abbas, M., Yaqub, T. et al. Genetic analysis of Newcastle disease virus from Punjab, Pakistan. Virus Genes 46, 309–315 (2013). https://doi.org/10.1007/s11262-012-0862-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-012-0862-2