Abstract
The hemagglutinin (HA) gene of novel Swine Origin Influenza A/California/04/2009 (H1N1) was engineered for expression in Pichia pastoris as a soluble secreted protein. The full length HA-synthetic gene having α secretory tag under the control of AOX1 promoter was integrated into P. pastoris genome through homologous recombination. The resultant Pichia clones having single and multiple copy integrants of the expression cassettes were screened for the expression of full length HA protein in the culture supernatant. In order to completely exploit the expression potential of the P. pastoris expression system, a systematic investigation on the influence of gene copy number on the expression of the recombinant protein was made. A panel of Pichia clones carrying increasing copies of the heterologous gene was selected based on Geneticin resistance and SYBR green-based quantitative real-time PCR approach. Using these strategies, recombinant Pichia transformants carrying up to a maximum of four to six copies of the transgene were identified. After optimising the expression conditions for shaker flask culture, the resultant clones demonstrated that the increase in copy number results in a proportional elevation in the expression level of H1N1HA recombinant protein. Our findings clearly suggest that the gene dosage effect play a vital role in high level expression of the pandemic Influenza HA protein in yeast system.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pichia pastoris is methylotrophic yeast that is widely used as a heterologous expression system. This microorganism is as easy to manipulate as Escherichia coli, but has many advantages of eukaryotic expression (e.g., protein processing, folding and post-translational modifications) and it is faster, easier and cheaper to use than other eukaryotic expression systems, such as baculovirus or mammalian tissue culture. It also generally yields higher expression levels. The methylotropic yeast, P. pastoris is widely used as an expression host to express various recombinant proteins because of its potential for high expression levels, efficient secretion of extracellular protein, post-translational modifications and growth to high cell densities [1]. The factors that drastically influence protein production in this system include: copy number of the expression cassette, site of chromosomal integration of the expression cassette, A+T composition of cDNA, transcriptional and translational blocks, nature of secretion signal, endogenous protease activity, host strain physiology, media, growth conditions and fermentation parameters [2]. All these factors should be considered in designing an optimal production system. Among these factors, selection of a clone with suitable copy number of transgene incorporated is important for reaching a high level of recombinant protein expression. It is often desirable to select transformants containing multiple integration events as such clones potentially express significantly higher levels of the recombinant protein.
Amongst several yeast transfer vectors, pPIC9K containing bacterial kanamycin resistance gene (kan from Tn903) that confers resistance to Geneticin (G-418) in Pichia is popular. Because of the genetic linkage between the kanamycin gene and the “expression cassette” one can infer from Geneticin hyper-resistance that the clone in question contains multiple copies of the transgene. The level of Geneticin resistance roughly depends on the number of kanamycin genes integrated. As previously reported [3, 4], an increase in the recombinant protein is anticipated with high copy number transformants that could be attributed to gene dosage effect. However, it should be note that if the recombinant protein produced is toxic to the host cell, the selection of high copy number transformant would have detrimental effects that lead to decreased expression rather than increased expression with increased copy number of the transgene. Hence, it is very critical to experimentally determine the co-relation of expression level with respect to the copy number for any viral structural protein. In case of pPICK9K transfer vector, the expression cassette also contains kan R gene, and hence the antibiotic (Geneticin) tolerance shown by a particular Pichia transformant can be used as a tool to detect transformants that harbour multiple copies of the transgene. This study is aimed to establish a co-relation on the influence of copy number on the expression levels of pandemic influenza recombinant protein in P. pastoris. In addition to the conventional antibiotic-based screening of transformants, we have employed normal genomic DNA PCR and SYBR Green-based real-time quantitative PCR methods for selecting the transformants with different copy numbers. The real-time quantitative PCR used here not only gives precise information on the gene copy number but also re-confirms the transgene. Further the co-relation of gene copy number and expression level is established based on protein expression study and their relative quantitation.
Swine origin Influenza A virus belonging to the Orthomyxoviridae family is an enveloped virus with segmented negative sense RNA genome surrounded by a helical symmetry shell. The 2009 H1N1 novel virus derived its genes from viruses circulating in the pig population [5–7]. Current influenza vaccines protect against homologous viruses but are less effective against antigenic variants and provide little protection against a different subtype. In the event of a pandemic, existing vaccines may be ineffective because the manufacturing process requires at least 6 months from identification of the pandemic strain to distribution which is insufficient time to prevent wide-scale morbidity or mortality. New vaccine strategies are therefore needed that can both accelerate production and provide broader spectrum of protection. Subunit vaccines like recombinant hemagglutinin (HA) protein offer an alternative over conventional vaccine strategies that could save several months of manufacturing time, since the HA gene of the newly circulating strain is available shortly after virus isolation or nucleotide sequencing of HA gene. In contrast to conventional approaches, there is no need for live influenza virus or large quantities of eggs, and subunit vaccines could be deployed earlier in the pandemic for effective reduction of morbidity and mortality. Moreover, it is also economical to produce these vaccines capable of inducing antibody that can neutralise the circulating strain of influenza virus. As it is very important to produce the antigenic protein in its native soluble form, prokaryotic system like bacteria may not be ideal for making this vaccine protein. From the previous studies on bacterially expressed HA proteins of H5N1 avian influenza virus (AIV) reported, it has been found that in the absence of glycosylation, newly synthesised HA proteins are not likely to fold properly or trimerize-like native HA molecules [8]. These HA molecules may not present native conformational epitopes which are important for generation of an effective protective immune response. Indeed the majority of the previous studies did not demonstrate proper folding and/or oligomerization of the HA proteins produced in prokaryotic systems [9–11].
The expression of HA in insect cells and mammalian cells are under development and/or clinical trials [12–15]. The H3N2 HA protein has also been expressed using P. pastoris but in its monomeric form [16]. More recently, the trimeric form of H3N2 HA subunit has been successfully produced using baculovirus system [17]. Both HA and NA proteins of H1N1 have been expressed using Pichia methalonica [18]. The HA monomer is synthesised as a single polypeptide chain which undergoes post-translational cleavages at two sites: the N-terminal signal sequence is removed and depending on the host cell and virus strain, the molecule is cleaved, with the removal of one or more intervening residues, resulting in two polypeptides chains called HA1 (36 KDa) and HA2 (27 KDa), linked via a disulfide bridge [19–21]. The carboxyl terminal stretch of hydrophobic amino acids anchors HA to the viral membrane and though not essential for secretion, this sequence plays a major role in the trimerization process [22]. With the above background, it is practically important to have an alternate heterologous system that can make pandemic influenza HA protein and can also overcome the limitations associated with already reported systems. As Pichia offers several advantages including rapid and economical production of recombinant proteins, an effective influenza HA subunit vaccine can be made with minimal notice for pandemic variants using P. pastoris. We have successfully employed the yeast expression platform for making the HA recombinant protein of Pandemic H1N1 Influenza virus. Further in this study, we have demonstrated the influence of copy number on the expression levels of pandemic H1N1 influenza recombinant HA protein in P. pastoris system.
Materials and methods
Strains, plasmid and media
Pichia pastoris (GS115) and yeast expression plasmid pPIC9K were from Invitrogen, USA. Escherichia coli strain DH5 alpha, the host strain used for plasmid construction was grown in LB (1 % tryptone, 0.5 % NaCl, 0.5 % yeast extract; pH 7.5). For P. pastoris cultivation YPGy (1 % yeast extract, 2 % bacto peptone and 1 % glycerol buffered with 100 mM potassium phosphate buffer, pH 8.2) and for induction, YPM (1 % yeast extract, 2 % bacto peptone and 2 % methanol, buffered with 100 mM potassium phosphate buffer, pH 8.2) media were used.
H1N1 HA gene synthesis and cloning into yeast transfer vector
The DNA sequence of the wild-type HA gene of novel H1N1 A/California/04/2009 (Genbank accession no: FJ966082.1) was synthesised from Biotech desk (Hyderabad, India). The full length HA-encoding synthetic gene was PCR amplified from the holding vector using high fidelity Pfu DNA polymerase (Fermentas, USA) and sub-cloned into pPIC9K yeast transfer vector in frame with the alpha pre-pro secretory tag on the amino terminal end as previously published by us [23].
Pichia pastoris transformation and selection of transformants
The Sal I linearised recombinant plasmid pPIC9KH1N1HA DNA (submitted to NCBI Genbank wide accession number HQ398363.1) was integrated at His4 locus on the Pichia genome through electroporation and HIS+, Mut+ Pichia pastoris GS115 transformants were obtained [23]. The transformants carrying the plasmid-borne Kanamycin resistance gene that confers resistance to Geneticin was used as a selective marker for selecting the putative transformants and also to differentiate the copy numbers integrated. The transformants were streaked on three different replica YPD agar plates containing different concentrations of Geneticin viz 0.25, 0.5 and 0.75 mg/ml and incubated at 30 °C for 3 days and the growth obtained was scored with plus (+) and minus (−) for the presence or absence of growth, respectively.
Expression analysis of H1N1 HA protein from recombinant yeast
The culture conditions including the type of medium, temperature, pH, methanol concentrations and post-induction harvest time were optimised as previously published [23] for better expression of the recombinant HA protein and the optimised conditions were used for analysing the expression of recombinant H1N1HA protein as follows:
The glycerol stock of recombinant P. pastoris clone (Genetically modified P. pastoris culture deposited at International Microbial repository, IMTECH, Chandigarh, India wide accession number MTCC 5607) carrying the transgene was inoculated into 50 ml of YPGy (1 % yeast extract, 2 % bacto peptone and 1 % glycerol buffered with 100 mM potassium phosphate buffer, pH 8.2) taken in 500-ml conical flasks along with negative control (Pichia transformed with pPICK9K without insert) and were incubated at 28 °C in a shaker incubator at 250 rpm until the culture reached OD (A600) of 6–8. The cells were harvested by centrifugation at 3,000g for 10 min at room temperature and the cell pellet was resuspended in required volume of fresh YPM induction medium (1 % yeast extract, 2 % bacto peptone and 2 % methanol, buffered with 100 mM potassium phosphate buffer, pH 8.2) so that the OD (A600) was 5. Incubation was continued at 29 °C on an orbitary shaker (250 rpm) for 2 days. To sustain induction, required volume of 100 % methanol was added once in every 24 h to reach a final concentration of 2 %. Culture supernatant was harvested after 24 and 48 h of post-induction and analysed on SDS-PAGE gel for expression of the recombinant protein. The expressed HA protein was confirmed through western blotting using rabbit anti HA polyclonal antibodies (Genscript, USA) and goat anti-rabbit Horse radish peroxidase conjugated IgG (Sigma, USA) as primary and secondary antibodies, respectively. The colour development was done using H2O2/DAB substrate/chromogen solution. The recombinant protein was further confirmed for its authenticity using MALDI-TOF/MS study.
MALDI-TOF/MS analysis of yeast-derived recombinant HA protein
The HA protein expressed from of Pichia cells was further confirmed using MALDI-TOF/MS study. The coomassie-stained recombinant HA recombinant protein was excised from the gel, and the gel slices were washed thrice with 200 μl destaining solvent (50 % v/v ACN in 25 mM NH4HCO3) with constant vortexing of 10 min. The washed gel pieces were dehydrated with 200 μl of 100 % ACN and dried in speed-vac centrifuge (Thermo, USA). The protein in the gel slices were subjected to digestion using 25 μl (20 μg/100 μl) of trypsin in 50 mM NH4HCO3 at 37 °C for overnight in a shaker incubator at 100 rpm. The peptides were extracted twice from the gel using 200 μl of extraction solvent (50 % ACN, 5 % TFA). This solvent was lyophilised for complete removal of the solvent. The pellets obtained after lyophilisation were reconstituted in 8 μl milli Q water and was subsequently eluted using Mini Tip C18 micro tips (Sigma,USA). Finally, 1 μl of the purified peptide along with 1 μl of HCCA matrix was individually spotted on the MALDI plate in a pre-determined order. Mass spectrometric analysis was performed using MALDI-TOF instrument (Bruker Microflex LRF-20, Flex Control Workstation, Bermen, Germany) equipped with delayed extraction (150 ns) and a UV ionisation laser (N2, 337 nm) with a 3-ns pulse width. The accelerating voltage was 20 kV and the grid voltage was set to 19 kV and laser repetition rate was 20 Hz. For the peptide mass fingerprinting, the instrument was operated in the reflector mode using α-cyano-4-hydroxy cinnamic acid (HCCA) as matrix and hundred shots were averaged per spectra and seven hundred laser shots were accumulated. The spectra were evaluated using the Flex Analysis Software (Bruker Daltonics). The MS spectra obtained was submitted to MASCOT search via Bio tools versions 3.1. The search parameters used were partial methionine oxidation, one missed cleavage, Peptide mass Tolerance 200 ppm and the database selected was NCBI.
Genomic DNA PCR and quantitative real-time PCR (qRT-PCR) based screening of transformants to determine transgene copy number
For preliminary screening of recombinant clones having differential copy numbers of the transgene, a simple PCR was performed using equal concentrations of genomic DNA. A total of 20 clones showing resistance to different concentrations of Geneticin viz, 0.25, 0.5 and 0.75 mg/ml were randomly selected and subjected to genomic DNA PCR using AOX1 forward and HA reverse primers as described previously [23].
One representative PCR positive clone from each Geneticin tolerance category (0.25, 0.5 and 0.75 mg/ml) was further selected for differentiating the copy numbers using qRT-PCR. The genomic template DNA concentrations in all cases were equally adjusted to 50 ng in the PCR reactions to check its sensitivity to screen low, moderate and high copy number integrants along with appropriate negative control. Quantitative real-time PCR was performed using Swine H1 forward and reverse primers (SW H1 Fwd: 5′-AGA CTG GCC ACA GGA TTG AGG AAT-3′; SW H1 Rev: 5′-CGT CAA TGG CAT TCT GTG TGC TCT-3′). The real-time RT-PCR was carried out with SYBR green chemistry-based reagents (Invitrogen, USA) as per the manufacturer’s protocol. Briefly, 12.5 l of 2× SYBR green RT-PCR Master mixture, 0.5 μl each of H1 forward and H1 reverse primers (100 pM), 0.5 l of Superscript III platinum Taq mix, 50 ng of template DNA (2 l) and 9 l nuclease free water in 25 l reaction volume. The Stratagene Mx 3005p system was used to monitor the SYBR Green signal at the end of each extension period for 40 cycles. The thermal profile consist of 10 min of polymerase activation at 95 °C, followed by 40 cycles of PCR at denaturation at 95 °C for 30 s, annealing at 55 °C for 45 s and extension at 72 °C for 30 s. Following amplification, a melting curve analysis was performed to verify the authenticity of the amplified product by its specific melting temperature (T m) using melting curve analysis software of the Mx3005p. This assay resulted in 184 bp amplified product of H1 gene.The threshold cycle (C t) of the amplified H1 gene and the difference between their C t values with respect to the copy numbers was determined. The pPIC9KH1N1HA plasmid purified using plasmid miniprep kit (Qiagen, USA) was used as template DNA to establish the standard curve for determining the copy numbers. The DNA copy numbers of the plasmid pPIC9KH1N1HA was calculated using the following formula:
Thus, we calculated that each microlitre of pPPIC9KH1N1HA (10,967 base pair long) with concentration of 50 ng had 4.1 × 109 copies. A pPIC9KH1N1HA plasmid standard curve with different DNA copy numbers was constructed as described above.
Determination of the relationship of the number of copies and the level of expression
In order to find out the relationship between the copy number of transgene and the HA expression level, six P. pastoris clones each carrying different copy numbers of the transgene based on Geneticin sensitivity and PCR experiments were selected. The glycerol stocks of the above clones were inoculated separately into 50 ml of YPGy taken in 500 ml conical flasks along with negative control (Pichia transformed with pPICK9K without insert) and analysis of recombinant HA protein was done as previously described. Supernatants from all cultures were harvested after 48 h of post-induction and concentrated to 1/10 of its original volume using Molecular weight cut off (MWCO) filters with pore diameter of 10 kDa (Millipore Corporation, USA) by centrifuging at 4,000g for 20–30 min at 4 °C. Protease inhibitor cocktail (Amersco, USA) was added to the concentrated samples and the total protein concentrations were determined through Bradford assay against BSA standards [24]. The concentrated protein samples obtained from all clones were further analysed by running them on 10 % discontinuous PAGE and further staining with Coomassie Brilliant Blue R-250 (Sigma, USA) as per the methods described earlier [25]. The expressed HA protein was confirmed through western blotting using rabbit anti HA polyclonal antibodies (Genscript, USA) as mentioned earlier.
Results and discussion
Pichia pastoris transformation and selection of transformants
Our initial goal was to determine whether there exists any relationship between the copy number of the integrated vector that included the HA gene of swine Influenza A and the expression of recombinant HA. In order to achieve this goal, first the putative transformants were screened on Geneticin plate. After transformation, a total of 145 putative transformants showing resistance to different concentrations of Geneticin were obtained in this study. Out of 145 clones, there were 81, 43 and 21 clones that appeared on antibiotic plate with 0.25, 0.5 and 0.75 mg/ml Geneticin, respectively (Fig. 1). As both Kan R and H1N1HA gene are integrated together, resistance to Geneticin would give a rough idea on the copy number of the integrated H1N1 HA gene. Resistance to higher concentrations of Geneticin is directly proportional to the copy numbers of Kanamycin resistance gene (Kan R) integrated which confers resistance to Geneticin in yeast cells. As a reference, clones that grow well on selection plate with Geneticin concentration of 0.25, 0.5 and 0.75 mg/ml were considered to have low, moderate and high copy integrants, respectively. As reported earlier by Joan Lin Cereghino [26], high copy integrants are due to the multiple gene insertion events at a single locus in Pichia cells that happens spontaneously with a low but at detectable frequency between 1 and 10 % of all selected His+ transformants. The observed less number of high copy integrants is thus justified. In addition, the poor transformation efficiency (365 colonies/30 μg) may also have contributed significantly in obtaining less number of transformants with high copy number integrants. The large size of the plasmid also partly contributed to the observed poor transformation efficiency. In light of the above, we presume that, in order to obtain more number of clones with very high copy number integrants, one has to screen thousands of transformants.
Expression analysis of H1N1 HA protein from recombinant Yeast
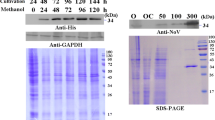
The selected methanol-induced positive Pichia transformant was checked after 24 h and 48 h for the expression of recombinant HA protein. As expected a distinct band of ~80 KDa was observed only in case of 48 h post-induced culture supernatant. Negative control samples derived from induced pPIC9K transformant (having no HA insert) and uninduced positive transformant did not show such band as seen in Fig. 2a. As seen in Fig. 2a, the yeast-derived H1N1 HA protein had an increase in its molecular weight (~80 KDa) which is due to the glycosylation of the expressed protein that happens within Pichia cells. This has a reference from the previous report wherein the similar sized H3N2 HA (75–78 KDa) has been expressed by Saelens et al. [16]. Weldon et al. [17] have reported the size of bacterially expressed protein to be ~60 KDa in its non-glycosylated form. The protein from the positive transformants that showed a distinct band of ~80 KDa through SDS-PAGE were further analysed through western blotting to confirm the authenticity of the expressed protein. Appropriate positive signal was obtained only in case of the culture supernatant of positive yeast transformant collected after 48 h of methanol induction (Fig. 2b).
SDS-PAGE and western blot analysis of the culture supernatants of P. pastoris transformants for confirming the expression of recombinant HA protein. a SDS-PAGE profile of P. pastoris culture supernatants after methanol induction. Lane 1 Pichia transformed from pPIC9K DNA carrying no HA insert after 48 h of post-induction (negative control). Lane 2 Pichia transformed from pPIC9KH1N1HA DNA carrying HA gene (pre-induced negative control). Lane 3 Pichia transformed from pPIC9KH1N1HA DNA carrying HA gene after 24 h of post-induction. Lane 4 Pichia transformed from pPIC9KH1N1HA DNA carrying HA gene after 48 h of post-induction. b Western blot analysis of P. pastoris culture supernatants after methanol induction: Lane 1 Pichia transformed from pPIC9K DNA carrying no HA insert after 48 h of post-induction (negative control). Lane 2 Pichia transformed from pPIC9KH1N1HA DNA carrying HA gene (pre-induced negative control). Lane 3 Pichia transformed from pPIC9KH1N1HA DNA carrying HA gene after 24 h of post-induction. Lane 4 Pichia transformed from pPIC9KH1N1HA DNA carrying HA gene after 48 h of post-induction
MALDI-TOF/MS analysis of yeast-derived recombinant HA protein
The authenticity of the yeast expressed recombinant HA protein was further confirmed via MALDI-TOF/MS analysis. The 80 KDa protein band corresponding to recombinant HA was enzymatically digested as described under methodology section. As shown in Fig. 3, peptide mass profiles were obtained from the destained band. Sixteen peptides (range 800–3,000 Da) were used for peptide mass database search which identified HA protein with 43 % sequence coverage of the protein sequence (Table 1).
Genomic DNA PCR and quantitative real-time PCR (qRT-PCR)-based screening of transformants to determine transgene copy number
Before establishing the relationship between expression level and gene copy number, it is essential to determine the copy number of the integrated transgene. For preliminary screening, a simple genomic DNA PCR was performed as described under materials and methods. Genomic DNA PCR from 20 selected yeast transformants using AOX1 forward and HA gene-specific reverse primer resulted in amplification of ~1.96 Kb DNA in 14 genomic DNA samples along with known positive control. The DNA from non recombinants did not show any amplification (Fig. 4). With genomic DNA PCR, a single band of 1.96 kb was observed in 16 DNA samples. Six of the transformants failed to amplify the transgene which may be due to the poor quality of DNA in these cases as observed from A260/280 ratio. As visualised on the DNA gel (Fig. 3), variation in the amplicon band intensity was noticed with respect to different clones that can be partially attributed to the difference in the transgene copy numbers. Although the DNA template used in PCR reaction is same in all samples, the difference in the copy numbers with multiple insertions of the expression cassette into the Pichia genome have resulted in detectable change in the band intensities with PCR amplification. These findings were more or less matched the Geneticin resistance data also (Table 2). Thus genomic DNA PCR data in combination with the Geneticin resistance data are useful in giving clue for selecting clones with different copy numbers. However, the genomic DNA PCR is not necessarily titratable, hence qRT-PCR was performed to determine the transgene copy numbers present within the selected Pichia clones.
Agarose gel electrophoresis of genomic DNA PCR from selected transformants with different copy numbers of the expression cassette: Lanes 1–20 Genomic DNA PCR amplicons from Pichia clones 1 to 20. Lane 21 PCR amplicon from pPICK9KH1N1HA plasmid DNA (+ve control). Lane 22 Genomic DNA PCR amplicon from Pichia cells transformed with pPICK9K (negative control)
Real-time PCR technique is known for producing linear response over a wide range of starting DNA concentrations [27]. In order to establish the differences in transgene copy number among the selected transformants, SYBR green-based quantitative real-time PCR was optimised. The standard curve generated in this study using pPICK9KH1N1HA plasmid DNA demonstrated the linearity of C t values with respect to the H1 DNA copy numbers that ranged between 4 × 107 to 4 copies (Fig. 5a). Upon melt curve analysis, it was noticed that the all the amplified PCR products had a single T m of 79 °C, thus indicating the specific amplification as expected (Fig. 5b). The standard curves showed high efficiency (R 2 = 0.96) and was sensitive even up to four copies as seen in Fig. 6. Based on the Geneticin sensitivity assay and genomic DNA PCR data, single clone each from low copy, moderate and high copy number transformants were selected for real-time PCR analysis. Similarly the recombinant Pichia strains harbouring different transgene copy number delivered consistent results showing different C t values (Fig. 5c) but same T m values (Fig. 5d). A clear-cut increase in the C t value with decreased copy number was observed in this study, thus demonstrating that selected transformants had a varied number of H1 gene on their chromosomal DNA. Although it is difficult to indicate the exact copy number of the transgene present in each of the Pichia transformants at this stage, the concordance in the results obtained from all the three different experiments viz., Geneticin sensitivity assay, genomic DNA PCR and qRT-PCR have confirmed the selection of recombinant Pichia transformants with different copy numbers for HA.
Quantitative real-time PCR (qRT-PCR) to differentiate the transgene copy numbers within the genome of Pichia transformants: a Amplification plots of showing the C t values with respect to the copy number derived from known concentration of 10-fold serially diluted pPIC9KH1N1HA plasmid; b melt curve analysis of the amplified PCR product from pPICK9KH1N1 HA plasmid DNA. c Amplification plots of showing the C t values with respect to the copy number derived from equal concentration of genomic DNA obtained from different Pichia transformants; d Melt curve analysis of the amplified PCR products from Pichia transformants
Determination of the relationship of the number of copies and the level of expression
The correlation between the numbers of HA copies integrated into the Pichia genome and the respective HA protein expression level play an important role in choosing the best clone that can be used for economical bulk protein production. Hence, to determine the influence of HA copy numbers on the expression of HA protein, six Pichia clones were selected (two each from low, moderate and high copy number integrants) and further studied for the differences in their expression levels of the recombinant protein. Figure 7a–c illustrates the expression profile of the proteins secreted from different Pichia clones with different copy numbers of the transgene. As observed on the gel, there was no or very little expression of HA protein in case of pPIC9KH1N1HA low copy integrants. Whereas the moderate copy integrants had reasonably good amount of HA protein produced with an approximate size of 80 KDa (Fig. 7b). Amongst all transformants tested, only the high copy integrants showed high level expression of H1N1HA protein in secreted form (Fig. 7c).
SDS-PAGE analysis of the culture supernatants of different P. pastoris transformants after methanol induction: a Expression profile of pPIC9KH1N1HA low copy integrants (lanes 1–6). b Expression profile of pPIC9KH1N1HA moderate copy integrants (lanes 1–7 non-recombinant transformant). c Expression profile pPIC9KH1N1HA high copy integrants (lanes 1–7 non-recombinant transformant)
The western blot data of the expressed HA protein revealed intense signal only with moderate and high copy integrants (Fig. 8, lanes 5 and 6), along with the HA protein derived from H1N1 purified virus (Fig 8, lane 1), whereas the protein sample transferred from negative controls, i.e. non-recombinant Pichia transformed with pPICK9K (having no HA insert), positive recombinant Pichia un-induced and low copy number transformant did not develop any signal on the membrane as seen in Fig. 8, Lanes 2, 3, 4, respectively. This data clearly suggest that there is no expression of the recombinant HA protein from low copy integrants. The average amount of protein secreted in case of low copy, moderate copy and high copy integrants was 0.03, 0.07 and 0.16 mg/ml of the culture supernatant, respectively, as estimated through Bradford assay. This accounts for at least fivefold increase in the expression level of recombinant HA protein from high copy number transformants in comparison to single copy integrants. It is also noteworthy that the HA protein is non-toxic to Pichia cells and hence increased copy numbers have no detrimental effect on the host and results in increased expression level of the recombinant protein. This is also supported by the growth kinetics study of the different copy number cultures studied (data not shown). At this stage, we have no clear reasons to justify the observed no expression of HA gene from low copy integrant. However, this needs further investigation for any possible mutation on the gene in the selected low copy transformants. Further screening of more number of low copy integrants for HA expression may also give conclusive results. But these studies were beyond the scope of our study. However, we feel that in case of Pichia when expressing protein from a single recombinant gene copy, the transcript level is most likely to be limiting factor in the HA protein production. Another possible explanation is that the HA protein produced is retained in the endoplasmic reticulum of the host, and this accumulation of recombinant HA protein in the ER limits the yield of HA secreted into the extracellular environment. In this study, we show that there is a proportional elevation in the HA expression with the increased transgene copy number and the poor expression of HA by some clones can be overcome by elevating the gene dosage. Considering the practical utility of the Pandemic Swine flu HA protein, there is a huge advantage of selecting multi-copy transformants for maximal HA protein yield. It is also important to note that if there is a mutation in one particular copy of the expression cassette, arising from the integration process, then the protein that results from such mutant copy may not contribute as significantly to the total amount of protein expressed.
Western blot analysis of Pichia expressed H1N1 HA protein using commercial antibodies (Genescript, USA): lane M pre-stained protein marker. Lane 1 MDCK derived H1N1 purified virus. Lane 2 Pichia culture supernatant from non-recombinant negative clone (48 h post-induced). Lane 3 culture supernatant from positive Pichia clone (uninduced). Lane 4 Pichia culture supernatant from pPIC9KH1N1HA low copy integrant. Lane 5 Pichia culture supernatant from pPIC9KH1N1HA moderate copy integrant. Lane 6 Pichia culture supernatant from pPIC9KH1N1HA high copy integrant
With the ability to replicate certain human glycosylation patterns, yeast-based expression platforms offer an attractive alternative to current mammalian or insect cell culture processes due to a variety of advantages including faster growth rates (and hence shorter culture times), cheaper operating costs, simple chemically defined media, no viral contamination and shorter development times from gene to protein. Unlike bacteria, as the target gene is integrated within the pichia genome, it is difficult to lose the target gene when the recombinant yeast is cultured and passaged. Therefore, P. pastoris has been used successfully to express a wide range of heterologous proteins [28, 29], and the influence of copy numbers on the protein expression level has been well established for several proteins [4, 30–32], this is the first report on establishing the influence of copy number on the expression level of H1N1 virus HA protein. Considering the economical and easy method of producing influenza recombinant protein, practical advantages of Pichia expression system, the present findings are significant and are invaluable towards exploring Pichia expression platform making HA protein of pandemic H1N1 virus in secreted form. This study also has an immense industrial application as the yeast-derived HA protein has got both prophylactic and diagnostic potential.
Conclusions
The expression of glycosylated H1N1 HA protein in P. pastoris required high copy number integrants. Increase in the copy number of the expression cassettes integrated within the Pichia genome number was proportional to the expression level of the recombinant HA protein of pandemic H1N1 virus. The low copy number transformants showed no expression compared to the moderate and high copy number integrants that could be attributed to the gene dosage effect. The observed no expression protein from a single recombinant gene copy is most likely to be associated with the limiting transcription and translational factors that requires further investigation. It is noteworthy that the HA protein of H1N1 is non-toxic to the Pichia host cells, and hence the increased copy number has no detrimental effects on the expression level of the target protein. Selection of positive clones that gives maximum recombinant protein yield based on conventional DNA quantification and Geneticin tolerance methods are less precise, and hence we have performed additional experiments like genomic DNA PCR, qRT-PCR, gene expression analysis and western blotting that are more precise and conclusive. As this is the first study establishing the co-relation of the transgene copy number and expression level of HA protein of pandemic H1N1 virus in P. pastoris expression system, the multi-copy recombinant Pichia transformant obtained in this study has an immense industrial application for large scale production of H1N1 HA recombinant protein for either prophylactic or diagnostic applications.
References
J.L. Cereghino, J.M. Cregg, FEMS Microb. Rev. 24, 45–66 (2000)
K. Sreekrishna, R.G. Brankamp, K.E. Kropp, D.T. Blankenship, J.-T. Tsay, P.L. Smith, P.L. Wierschke, A. Subramaniam, A. Birkenberger, Gene 190(1), 55–62 (1997)
H.-Y. Luo, H.-Q. Huang, Y.-G. Bai, Y.-R. Wang, P.-L. Yang, K. Meng, T.-Z. Yuan, Y.A.O. Bin, Chin. J. Biotechnol. 22(4), 528–533 (2006)
C.A. Scorer, J.J. Clare, W.R. McCombie, M.A. Romanos, K. Sreekrishna, Bio Technol. 12, 181–184 (1994)
G.J. Smith, J. Bahl, D. Vijaykrishna, J. Zhang, L.L. Poon, Proc. Natl. Acad. Sci. USA 106, 11709–11712 (2009)
G.J. Smith, D. Vijaykrishna, J. Bahl, S.J. Lycett, M. Worobey, Nature 459, 1122–1125 (2009)
V. Shinde, C.B. Bridges, T.M. Uyeki, B. Shu, A. Balish, N. Engl. J. Med. 360, 2616–2625 (2009)
S. Shen, G. Mahadevappa, H.L. Oh, B.Y. Wee, Y.W. Choi, J. Med. Virol. 80, 1972–1983 (2008)
F.F. Chiu, N. Venkatesan, C.R. Wu, A.H. Chou, H.W. Chen, Biochem. Biophys. Res. Commun. 383, 27–31 (2009)
Z. Biesova, M.A. Miller, R. Schneerson, J. Shiloach, K.Y. Green, Vaccine 27, 6234–6238 (2009)
J. Curtis-Fisk, R.M. Spencer, D.P. Weliky, Protein Expr. Purif. 61, 212–219 (2008)
J.J. Treanor, R.F. Betts, G.E. Smith, E.L. Anderson, C.S. Hackett, J. Infect. Dis. 173, 1467–1470 (1996)
J.J. Treanor, G.M. Schiff, R.B. Couch, T.R. Cate, R.C. Brady, J. Infect. Dis. 193, 1223–1228 (2006)
C.J. Wei, L. Xu, W.P. Kong, W. Shi, K. Canis, J. Virol. 82, 6200–6208 (2008)
J. Steitz, P.G. Barlow, J. Hossain, E. Kim, K. Okada, T. Kenniston, S. Rea, R.O. Donis, A. Gambotto, PLoS ONE 5(5), e10492 (2010)
X. Saelens, P. Vanlandschoot, W. Martinet, M. Maras, S. Neirynck, R. Contreras, W. Fiers, W.M. Jou, Eur. J. Biochem. 260(1), 166–175 (1999)
W.C. Weldon, B.Z. Wang, M.P. Martin, D.G. Koutsonanos, I. Skountzou, PLoS ONE 5(9), e12466 (2010)
Y. Zhang, Z. Yu, L. Xin, Y. Chen, Q. Tang, Y. Chen, Q. Chen, Y. Shu, Chin. J. Biotechnol. 26(8), 1068–1073 (2010)
H.D. Klenk, R. Rott, M. Orlich, J. Blodorn, Virology 68, 426–439 (1975)
S.G. Lazarowitz, P.W. Choppin, Virology 68, 440–454 (1975)
J.J. Skehel, M.D. Waterfield, Proc. Natl Acad. Sci. USA 79, 968–972 (1982)
I. Singh, R.W. Doms, K.R. Wagner, A. Helenius, EMBO J. 9, 631–639 (1990)
T.N. Athmaram, S. Saraswat, S.R. Santhosh, S.R. Singh, V.V.S. Suryanarayana, R. Priya, N. Gopalan, M. Parida, P.V. Lakshmana Rao, R. Vijayaraghavan, Virol. J. 8, 524 (2011)
M.M. Bradford, Anal. Biochem. 72, 248–254 (1976)
J. Sambrook, D.W. Russell, Molecular cloning: a laboratory manual (Cold Spring Harbor Laboratory Press, New York, 2002)
J. Lin-Cereghino, W.W. Wong, S. Xiong, W. Giang, L.T. Luong, J. Vu, S.D. Johnson, G.P. Lin-Cereghino, Biotechniques 38(1), 44–48 (2005)
S. Abad, K. Kitz, A. Hörmann, U. Schreiner, F.S. Hartner, A. Glieder, Biotechnol. J. 5, 413 (2010)
J.J. Clare, M.A. Romanos, F.B. Rayment, J.E. Rowedder, M.A. Smith, Gene 105, 205–212 (1991)
T.N. Athmaram, G. Bali, G.G. Khang, S. Dwarakanath, Virus Genes 35(2), 265–267 (2007)
R.A. Brierley, R.A. Davis, R.A. Holtz. United States Patent 5,324,639 (1994)
M.A. Romanos, J.J. Clare, K.M. Beesley, F.B. Rayment, S.P. Ballantine, A.J. Makoff, G. Dougan, N.F. Fairweather, I.G. Charles, Vaccine 9, 901–906 (1991)
C.A. Scorer, R.G. Buckholz, J.J. Clare, M.A. Romanos, Gene 136, 111–119 (1993)
Acknowledgments
The authors are thankful to Dr. B. K. Bhattacharya, Head, Div. of Biochemistry for his help on MALDI-TOF/MS study. The authors also acknowledge Director, DRDE, Gwalior, Madhya Pradesh-474 002 for his kind support in executing this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Athmaram, T.N., Saraswat, S., Singh, A.K. et al. Influence of copy number on the expression levels of pandemic influenza hemagglutinin recombinant protein in methylotrophic yeast Pichia pastoris . Virus Genes 45, 440–451 (2012). https://doi.org/10.1007/s11262-012-0809-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-012-0809-7