Abstract
To counteract plant antiviral defense based on RNA silencing, many viruses express proteins that inhibit this mechanism at different levels. The genome of Citrus leaf blotch virus (CLBV) encodes a 227-kDa protein involved in replication, a 40-kDa movement protein (MP), and a 41-kDa coat protein (CP). To determine if any of these proteins might have RNA silencing suppressor activities, we have used Agrobacterium-mediated transient assays in the green fluorescent protein (GFP)-expressing Nicotiana benthamiana line 16c. Only CLBV MP was able to suppress intracellular GFP silencing induced by expression of either single- or double-stranded (ds) GFP RNA, but not cell-to-cell or long distance spread of the silencing signal. The MP suppressor activity was weak compared to other characterized viral suppressor proteins. Overall our data indicate that MP acts as a suppressor of local silencing probably by interfering in the silencing pathway downstream of the steps of dsRNA and small RNAs generation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
RNA silencing is an RNA-based gene regulatory system that plays an essential role in many biological processes [1, 2]. The RNA silencing mechanism is triggered by the presence of double-stranded (ds) RNA molecules in the cell, which are processed into 21–25 nucleotides RNA species termed small RNAs (sRNAs), by enzymes of the RNase III class (DICER-like, DCL). One of the sRNA strands is incorporated into an RNA-induced silencing complex (RISC) and guides cleavage of perfectly complementary mRNAs by an RNase H-like enzyme (Argonaute, AGO). sRNAs can also prime synthesis of new dsRNA molecules by host RNA-dependent RNA polymerases, that will be cleaved by DCLs leading to secondary sRNA accumulation, thus amplifying silencing. In plants, RNA silencing is initially induced at single-cell level, but later a mobile silencing signal is generated that moves cell-to-cell through plasmodesmata and then systemically via the vascular system to cleave target RNAs [1, 3–5].
RNA silencing plays an important antiviral role in plants and animals [6–9]. In plants, ds replicative intermediates of RNA viruses or highly structured single-stranded (ss) viral RNAs may be processed by DCLs and virus specific sRNAs incorporated into RISC target genomic (g) and subgenomic (sg) RNAs for cleavage, thus reducing the virus level in infected cells. To counteract antiviral RNA silencing, most plant viruses have evolved to express silencing suppressor proteins [10, 11]. These proteins do not share common sequences or structural motifs among different viral groups and they interfere with different steps of the RNA silencing pathway. For example, the tombusvirus p19 protein binds sRNAs interfering with their incorporation into RISC [12, 13]. Other viral suppressors bind long dsRNAs [14, 15] or interact with protein components of the host silencing machinery. Thus, the 2b protein encoded by Cucumber mosaic virus (CMV), the p38 protein of Turnip crinkle virus (TCV), the p1 protein of Sweet potato mild mottle virus (SPMMV), and the polerovirus silencing suppressor P0 target the AGO 1 component of RISC [16–20]. RNA silencing suppressors may also block the silencing signal moving cell-to-cell or long distance via the sieve tubes [9]. Most viral suppressors have been identified as pathogenicity determinants [9, 11], some of them affecting the microRNA (miRNA) pathway, that is mainly involved in the developmental regulation of host plants [4, 21]. Therefore, identification and functional analysis of viral silencing suppressors are important for understanding the survival strategy of viruses in their host plants and virulence.
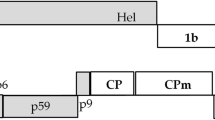
Citrus leaf blotch virus (CLBV), a member of the genus Citrivirus, family Flexiviridae [22–24], has filamentous virions about 960 × 14 nm in size composed of a ss(+)-gRNA of 8747 nt, organized in three open reading frames (ORFs) (Fig. 1a), and a 41-kDa coat protein (CP) [24, 25]. The ORF 1 encodes a ~227-kDa polyprotein that contains the viral replication components with methyl-transferase, AlkB-like, OTu-like peptidase, papain-like protease, helicase and RNA-dependent RNA polymerase (RdRp) domains, and it is translated directly from the gRNA. The other two ORFs encode a ~40-kDa protein with a motif characteristic of cell-to-cell movement proteins (MP) of the 30K superfamily and the CP protein, respectively, and these are translated from two 3′ co-terminal sgRNAs (MP-sgRNA and CP-sgRNA, respectively) (Fig. 1a) [24, 26, 27]. In order to know if any of these proteins might act as an RNA silencing suppressor, we used an Agrobacterium-mediated transient assay in transgenic Nicotiana benthamiana plants expressing the green fluorescent protein (GFP) [28]. Our results show that the CLBV MP protein suppresses the intracellular silencing, but it does not interfere with systemic spread of the silencing signal.
Schematic representation of the CLBV genomic RNA and the constructs used in this study. a An outline of the CLBV genome, with shaded boxes representing the predicted open reading frames (ORFs) and solid lines representing untranslated regions. Proteins potentially encoded by ORFs 2 (MP movement protein) and 3 (CP coat protein), and functional domains in ORF 1 (MT methyl-transferase, AlkB AlkB-like peptidase, OTu OTu-like peptidase, PRO protease, HEL helicase, RdRp RNA-dependent RNA polymerase) are indicated in the boxes. b Schematic representation of binary vector constructs used in this study. The arrow labeled 35S represents a double-enhanced Cauliflower mosaic virus (CaMV) 35S promoter and ellipse labeled Nos-t represents the nopaline synthase terminator. The boxes labeled with MT-AlkB, PRO-HEL-RdRp, MP, and CP indicate proteins encoded by the CLBV genome; GFP green fluorescent protein; the black line between sense and antisense GFP represents an intron, p19 p19 protein of Tomato bushy stunt virus, HC-Pro helper component protein of Tobacco etch virus, p25 p25 protein of Citrus tristeza virus
Materials and methods
Plant growth
The transgenic GFP-expressing N. benthamiana line 16c used in this study has been previously described [29]. Seeds (kindly provided by Professor D. Baulcombe, Sainsbury Laboratory, Norwich, UK) were sown in small pots with an artificial potting mix (50% sand and 50% peat moss) in a plant growth chamber at 20/24°C (night/day), 50% humidity, and a 16/8 h light/dark regime.
Plasmid constructs
All binary plasmids used in this study are outlined in Fig. 1b. The full-length cDNA of the CLBV genome cloned in pBIN19 (IC-CLBV clone) [30] was used as template for PCR amplification of the CLBV MET (methyl-transferase, AlkB-like and OTu-like peptidase domains, positions 74–2925 in the CLBV gRNA of isolate SRA-153, EMBL accession no. AJ318061), REP (protease, helicase and RdRp domains, nt 2927–5962), MP (movement protein, nt 5962–7050), and CP (coat protein, nt 7115–8206) regions using appropriate specific primers (Table 1) and AccuPrime™ Pfx DNA Polymerase (Invitrogen). A nonviral BamHI restriction site was added to the 5′ end of each primer. The BamHI-digested PCR fragments were ligated into BamHI-digested pMOG 180 plasmid (Mogen International), that contains a double-enhanced Cauliflower mosaic virus (CaMV) 35S promoter followed by the BamHI site and the 3′ terminator region of the nopaline synthase gene (Nos-t), flanked by HindIII or EcoRI restriction sites. Then, the complete transcription cassettes excised with HindIII (REP, MP, and CP) or with EcoRI (MET) were inserted into the binary vector pBIN 121 (Clontech) digested with the same enzymes, obtaining constructs pBI-MET, pBI-REP, pBI-MP, and pBI-CP, respectively. Fidelity of all constructs was confirmed by nucleotide sequencing.
GFP mRNA silencing in N. benthamiana 16c plants was induced by expression of the binary plasmids pBI-GFP and pBI-dsGFP (kindly provided by Professor D. Baulcombe) that comprise the CaMV 35S promoter, the mGFP5 sequence (inserted in sense orientation or as an inverted repeat, respectively) and the Nos-t. Binary plasmids pBI-p19, pBI-HC-Pro, and pBI-p25, with the genes encoding suppressor proteins p19 of Tomato bushy stunt virus (TBSV) (kindly provided by Dr. J.A. García, Centro Nacional de Biotecnología, CNB-CSIC, Madrid, Spain), HC-Pro of Tobacco etch virus (TEV) (kindly provided by Dr. F. Tenllado, Centro de Investigaciones Biológicas, CIB-CSIC, Madrid), and p25 of Citrus tristeza virus (CTV) (kindly provided by Dr. L. Peña, Instituto Valenciano de Investigaciones Agrarias, IVIA, Valencia, Spain), respectively, under the control of the CaMV 35S promoter and the Nos-t, were used as positive control for silencing suppression. The empty binary vector (pBI-Ø) containing the CaMV 35S promoter and the Nos-t was used as negative control.
Agro-infiltration of N. benthamiana leaves and GFP imaging
Agrobacterium tumefaciens cells, strain COR 308, carrying the helper plasmid pCH32 (kindly provided by Dr. A. Hamilton, Cornell Research Foundation) were transformed by electroporation with ~10 ng of the corresponding binary plasmid DNA using a Gene Pulser (Bio-Rad) as previously described [30]. Bacterial cells were resuspended in an infiltration buffer (10 mM MES, pH 5.6, 10 mM MgCl2, and 150 μM acetosyringone) to a final OD600 of 1.0 and incubated for at least 3 h at room temperature. In co-infiltration assays, equal volumes of an Agrobacterium suspension carrying the gfp gene and another harboring CLBV-based constructs or positive controls were mixed before infiltration. One-month-old seedlings of GFP-transgenic N. benthamiana 16c were infiltrated with this mix on the third, fourth, and fifth true leaves with a 2 ml syringe without a needle. GFP fluorescence was observed under a long-wavelength UV lamp (Black Ray® model B-100 AP, UV Products). Leaves and whole plants were photographed with a Nikon D-200 digital camera using a yellow filter.
RNA analyses
Total RNA (RNAt) was extracted from 500 mg leaf tissue using a standard protocol with two phenol:chloroform:isoamyl alcohol extractions, followed by ethanol precipitation with sodium acetate, and re-suspended in 25 μl of diethyl pyrocarbonate (DEPC)-treated distilled water [31]. To obtain preparations enriched in sRNAs, RNAt from 250 mg of infected tissue was extracted with TRI-Reagent and 1-bromo-3-chloro-propane (Sigma-Aldrich), precipitated with isopropanol and re-suspended in 150 μl of RNase-free water. High-molecular mass RNAs were precipitated with 1 M NaCl and 10% polyethylene glycol (PEG 8000), and the sRNAs were ethanol precipitated and re-suspended in 25 μl of RNase-free water [32]. RNA content was measured in a NanoDrop™ spectrophotometer (Thermo) and adjusted to the same concentration for electrophoresis. Equal loading was verified by agarose gel electrophoresis and ethidium bromide staining of 5S rRNA or tRNA.
For northern blot analysis of GFP mRNA and CLBV RNAs, 3 μg of RNAt was denatured at 94°C for 5 min in 50% formamide, chilled on ice, separated by electrophoresis in formamide–formaldehyde denaturing 1% agarose gels in MOPS buffer, and electroblotted onto positively charged nylon membranes (Roche) at 250 mA for 1 h and 1 A for 15 h, using 25 mM phosphate buffer, pH 6.45. To analyze gfp-derived sRNAs, 10 μg of sRNAs were mixed with an equal volume of formamide, heated at 95°C for 5 min, and separated in a 15% polyacrylamide Tris–borate-EDTA(TBE)-urea gel. sRNAs were then electroblotted onto positively charged nylon membranes (Roche) at 250 mA for 16 h, using TBE buffer. After UV cross-linking, the membranes were hybridized with digoxigenin (DIG)-labeled RNA probes specific for the GFP mRNA sequence (to detect GFP mRNA or sRNAs) or for the CLBV coat protein gene (to detect viral RNAs), washed and developed as previously reported [27, 30].
Results
The CLBV MP protein suppresses intracellular RNA silencing in agro-infiltrated N. benthamiana leaves
Individual CLBV proteins were screened for silencing suppression ability by co-infiltration of transgenic N. benthamiana 16c plants that express GFP with Agrobacterium cultures transformed with pBIN 121-based binary plasmids carrying the gfp gene (pBI-GFP) or the gene encoding the candidate protein [33, 34]. For this purpose, the different CLBV ORFs were cloned into pBIN 121 under the control of the 35S promoter of CaMV and the nopaline synthase terminator (Nos-t) (Fig. 1b). Because the large size of the ORF 1, it was cloned in two fragments encompassing the methyl-transferase, AlkB-like, and OTu-like peptidase domains (pBI-MET), and the protease, helicase and RNA-dependent RNA polymerase domains (pBI-REP), respectively (Fig. 1b). The empty pBIN 121 vector (pBI-Ø) or plasmids expressing the known silencing suppressor proteins HC-Pro from TEV (pBI-HC-Pro) [35] or p19 from TBSV (pBI-p19) [28] were used as negative and positive controls, respectively (Fig. 1b). All the experiments were repeated at least four times, with six to ten plants being agro-infiltrated in each assay.
Examination of plants at 5 days post-infiltration (dpi) showed a fluorescence decrease in all leaves agro-infiltrated with pBI-GFP and the empty vector as a consequence of RNA silencing activation. In contrast, intense green fluorescence was observed in all leaves co-infiltrated with pBI-GFP and the plasmids pBI-HC-Pro or pBI-p19, expressing silencing suppressors HC-Pro or p19, respectively. This increased fluorescence remained for at least 2 weeks. Among the four CLBV genomic regions tested, only the leaves co-infiltrated with pBI-GFP and pBI-MP showed increased fluorescence at 5 dpi, albeit it was less intense than that observed in leaves infiltrated with positive controls and it remained only until 9 dpi (Fig. 2a). Therefore, we concluded that the CLBV MP protein is a suppressor of RNA silencing weaker than p19 or HC-Pro proteins.
Identification of CLBV MP as suppressor of RNA silencing. a Leaves of N. benthamiana 16c plants infiltrated with a mixture of Agrobacterium tumefaciens cultures harboring binary vectors pBI-GFP and the empty vector (Ø), pBI-p19, pBI-HC-Pro, pBI-MET, pBI-REP, pBI-MP, or pBI-CP. The green fluorescence images of the co-infiltrated leaves were taken at 5 days pos-infiltration (dpi) under a long-wavelength UV lamp. b Northern blot analysis of GFP mRNA and gfp-derived sRNAs extracted from the agro-infiltrated leaf patches shown in a or from non-infiltrated leaves (c) using a DIG-riboprobe specific for the GFP mRNA. Ethidium bromide staining of 5S rRNA and tRNAs are shown as loading controls for mRNA and sRNAs, respectively (Color figure online)
Northern blot analysis of RNAt from the agro-infiltrated leaves using a riboprobe specific for the gfp positive strand showed reduced GFP mRNA accumulation in leaves infiltrated with pBI-GFP and pBI-Ø, pBI-MET, pBI-REP or pBI-CP vectors, in comparison with control leaves non-infiltrated or infiltrated with buffer. However, leaves co-infiltrated with pBI-GFP and pBI-MP, pBI-p19 or pBI-HC-Pro showed higher GFP mRNA accumulation than the control leaves (Fig. 2b).
Since the presence of sRNAs is a hallmark of the RNA-mediated silencing mechanism [36], accumulation of gfp-derived sRNAs in agro-inoculated leaves was analyzed by northern blot. While, gfp-specific sRNAs were readily detected in leaves agro-infiltrated with pBI-GFP and pBI-Ø, pBI-MET, pBI-REP, or pBI-CP constructs, accumulation of these sRNAs was significantly reduced in leaves co-expressing GFP and CLBV MP, or p19 or HC-Pro suppressor proteins used as control (Fig. 2b). Reduced sRNA level correlated with higher GFP mRNA accumulation in these tissues (Fig. 2b), thus confirming that the CLBV MP protein has a suppressor activity that interferes with local RNA silencing in N. benthamiana.
To assess whether the MP protein blocks conversion of GFP mRNA to dsRNA or it inhibits RNA silencing at some step downstream of dsRNA generation, we carried out a dsRNA-triggered silencing assay by co-infiltration of N. benthamiana 16c leaves with A. tumefaciens cultures transformed with pBI-dsGFP (carrying an inverted repeat of the gfp gene that upon transcription would generate GFP dsRNA), and pBI-Ø, pBI-MP, or pBI-p19 (Fig. S1a in Supplementary material). Fluorescence in all leaves agro-infiltrated with pBI-dsGFP plus pBI-Ø dropped at 5 dpi, whereas all leaves co-infiltrated with pBI-dsGFP plus pBI-MP, or pBI-p19 showed green fluorescence (Fig. S1a in Supplementary material). These results were confirmed by northern blot analysis of GFP mRNA and gfp-derived sRNAs, that showed low GFP mRNA and high GFP-specific sRNA levels in leaves co-infiltrated with pBI-dsGFP and empty vector, in comparison with non-inoculated leaves, whereas the opposite effect was observed in leaves co-infiltrated with pBI-dsGFP and pBI-MP, or pBI-p19 (Fig. S1b in Supplementary material). These results indicate that the CLBV MP protein interferes with RNA silencing after dsRNA generation.
The CLBV MP does not prevent cell-to-cell or long distance spread of the RNA silencing signal
To determine whether the CLBV MP protein could interfere cell-to-cell spread of the RNA silencing signal, leaves of N. benthamiana 16c were agro-infiltrated with pBI-GFP and either the empty vector, pBI-MP, or pBI-p19. If the silencing signal is able to move cell-to-cell, a silenced red ring bordering the infiltrated area should be expected, whereas no red ring would appear if the suppressor protein inhibits cell-to-cell movement of the silencing signal [37]. A clear increase of green fluorescence was observed at 5 dpi in leaf areas co-infiltrated with pBI-GFP and pBI-MP or pBI-p19. However, a characteristic silenced red ring bordering the infiltrated area was observed at 11 dpi in leaves infiltrated with pBI-MP but not in those infiltrated with pBI-p19 (Fig. 3a). The red ring was observed at 7 dpi in leaves co-infiltrated with pBI-GFP and the empty vector. These results suggest that the CLBV MP protein does not suppress cell-to-cell spread of the silencing signal.
Effect of CLBV-encoded proteins on the spread of the GFP silencing signal in N. benthamiana 16c plants. a Short-distance silencing spread. Leaves co-infiltrated with A. tumefaciens cultures harboring constructs pBI-GFP and pBI-MP, pBI-p19 or the empty vector (Ø). Photographs were taken at 11 dpi under long-wavelength UV light with a yellow filter. Black arrows show the red border at the edge of the agro-infiltrated leaf area that indicates short-distance intercellular movement of the RNA silencing signal. b Systemic silencing spread. GFP fluorescence at 30 dpi in the upper leaves of plants infiltrated with A. tumefaciens cultures carrying constructs pBI-GFP and either pBI-p25 (positive control), empty vector (Ø, negative control), pBI-MET, pBI-REP, pBI-MP or pBI-CP. c Northern blot analysis of GFP mRNA in upper leaves from plants shown in b or from non-infiltrated control plants (c) using a DIG-riboprobe specific for the GFP mRNA. Ethidium bromide staining of 5S rRNA is shown as loading control (Color figure online)
The mobile silencing signal is thought to spread systemically through the vascular system inducing silencing of homologous sequences in the upper leaves [38]. To examine if CLBV proteins can interfere in systemic silencing, N. benthamiana 16c plants were co-infiltrated with Agrobacterium cultures carrying pBI-GFP and pBI-Ø, pBI-MP, pBI-MET, pBI-REP, or pBI-CP, and the progress of silencing was monitored in the upper leaves. In this analysis, a construct expressing the p25 protein of CTV (pBI-p25) (Fig. 1b), that has systemic silencing-suppressor activity [39], was used as positive control. While, most plants (95%) co-infiltrated with pBI-GFP and pBI-Ø, pBI-MET, pBI-REP, pBI-MP, or pBI-CP, showed reduced green fluorescence in non-inoculated upper leaves at 14 dpi, in plants co-infiltrated with pBI-GFP and pBI-p25 systemic RNA silencing was delayed until 18 dpi and it was observed in only 35% of the plants (Fig. 3b). Northern blot analysis of RNAt from upper leaves of the agro-infiltrated plants using a riboprobe specific for the GFP mRNA showed reduced mRNA accumulation in plants agro-infiltrated with pBI-GFP and pBI-Ø, pBI-MET, pBI-REP, pBI-MP, or pBI-CP constructs, in comparison with control plants infiltrated with buffer, whereas the GFP mRNA level was similar in control plants and in plants co-infiltrated with pBI-GFP and pBI-p25 (Fig. 3c).
To further confirm these results, we conducted a second assay described by Guo and Ding [38], in which the construct that triggers silencing and that expressing the candidate silencing suppressor are agro-infiltrated separately, with the second being expressed in the presumed path of the mobile RNA silencing signal. For this purpose, two basal leaves of N. benthamiana 16c plants were agro-infiltrated with pBI-GFP at the tip (T) and with pBI-Ø, pBI-MET, pBI-REP, pBI-MP, pBI-CP, or pBI-p25 at the leaf base (B). Since, the assay was aimed at examining potential interference of the mobile silencing signal generated at the leaf tip in a source to sink direction, we also performed the opposite infiltrations with the same constructs as control. At 14 dpi, most of the plants (18–19 out of 20) agro-infiltrated with pBI-GFP (T) and empty vector or constructs expressing the CLBV proteins (B) developed systemic silencing in the upper leaves, whereas 12 of the 20 plants agro-infiltrated with pBI-GFP (T) and pBI-p25 (B) and non-infiltrated control plants showed similar levels of fluorescence in the upper leaves (Fig. S2 in Supplementary material). All plants infiltrated with pBI-GFP at the base of the leaves exhibited systemic GFP silencing in the upper leaves regardless the construct agro-infiltrated at the leaf tip (data not shown). Overall, these results suggest that CLBV proteins, including MP, are unable to block long distance spread of the RNA silencing signal.
Expression of the full-length CLBV RNA does not induce detectable silencing suppression in N. benthamiana plants
To assess whether CLBV proteins directly expressed from the full-length CLBV gRNA can elicit silencing suppression, two experiments were performed. In the first, leaves of N. benthamiana 16c plants were co-infiltrated with pBI-GFP and pBI-Ø, pBI-MP, or IC-CLBV, a construct carrying an infectious cDNA clone of the CLBV genome [30]. Leaves agro-infiltrated with pBI-MP showed increased green fluorescence at 5 dpi, whereas no suppression of GFP silencing was observed in plants agro-infiltrated with IC-CLBV or the empty vector, either in the infiltrated areas or in the upper leaves in the four following weeks (Fig. 4a). Virus replication in plants inoculated with IC-CLBV was confirmed in both agro-infiltrated and upper leaves by northern blot analysis using a riboprobe specific to the CLBV CP mRNA (Fig. 4b).
Silencing suppression analysis of CLBV proteins expressed from the viral genome. a Leaves of N. benthamiana 16c plants co-infiltrated with A. tumefaciens cultures carrying constructs pBI-GFP and the empty vector (Ø), pBI-MP or the CLBV infectious clone IC-CLBV (IC). Leaves were photographed at 5 dpi under long-wavelength UV light with a yellow filter. b Northern blot analysis of viral RNAs extracted at 5 and 21 dpi from agro-infiltrated and upper non-infiltrated leaves, respectively, using a DIG-riboprobe specific for the CLBV CP gene. Black arrows indicate the position of gRNA and sgRNAs of the virus
The second experiment was based on a reversion assay on post-transcriptionally GFP silenced N. benthamiana 16c plants [40–42]. The first three leaves of the plants were agro-infiltrated with pBI-GFP, and 20 days later, when post-transcriptional gene silencing affected the whole plant, leaves were agro-infiltrated with pBI-Ø or with the IC-CLBV clone and the plants were observed for green fluorescence appearance along the four following weeks. None of the plants showed GFP silencing suppression in the agro-infiltrated or in the upper leaves, although systemic CLBV infection was achieved in all plants infiltrated with IC-CLBV.
Discussion
To counteract the antiviral plant defense mechanism based on RNA silencing, many viruses have evolved silencing suppressors that inhibit this mechanism at different levels. Analysis of the silencing suppressor activity of CLBV proteins using the A. tumefaciens co-infiltration assay in GFP-expressing N. benthamiana 16c plants [34] showed that only the movement protein (MP) was able to suppress intracellular RNA silencing, albeit its silencing suppressor activity was low compared with other known viral suppressor proteins like p19 from TBSV [28] or HC-Pro from TEV [35]. However, since the protein products expressed by the ORF1 fragments cloned in the pBI-MET and pBI-REP constructs may not be identical to those produced by natural processing of the ~227-kDa polyprotein in CLBV infections, we cannot exclude that some of these products also could have silencing suppressor activity.
Silencing suppressors identified in distinct viruses interfere at different steps of the RNA silencing pathway [10, 43]. The ability of CLBV MP protein to suppress mRNA- and dsRNA-induced silencing in agro-infiltrated N. benthamiana 16c leaves clearly indicates that it must act downstream of dsRNA formation. As previously observed with HC-Pro and p19 suppressor proteins, in leaves co-infiltrated with pBI-GFP and pBI-MP the amount of gfp-specific sRNAs was significantly reduced, but they were not completely eliminated. This was expected in the case of p19 and HC-Pro since they bind sRNAs, and therefore, they do not inhibit DCL activity, in contrast with other suppressors that bind long dsRNAs, thus compromising their DCL-mediated processing [15, 44]. Our results suggest that CLBV MP targets the silencing machinery downstream the sRNA generation step, as do other suppressor proteins of plant viruses [14, 15, 45, 46].
The CLBV MP protein failed to block cell-to-cell or long distance spread of the systemic silencing signal. Long distance suppression was tested using two approaches: (i) simultaneous co-infiltration of the GFP silencing inducer and the suppressor in the same area of N. benthamiana 16c leaves and (ii) concurrent agro-infiltration of both constructs at the tip and the base of single leaves. Therefore, the suppression mechanism of the CLBV MP seems to differ from that of its ortholog in Apple chlorotic leaf spot virus, also a member of the family Flexiviridae [22, 23] that is phylogenetically related to CLBV [24]. The MP protein of ACLSV interferes with systemic spread of the silencing signal without affecting intracellular silencing [32]. Different suppression mechanisms have been also observed in orthologous proteins of two criniviruses: the p22 protein of Tomato chlorosis virus that suppresses intracellular RNA silencing [47] and the p22 protein of Sweet potato chlorotic stunt virus that interferes cell-to-cell and systemic spread of the silencing signal [48], further supporting the notion of independent evolution of silencing suppressors in related viruses [10].
While, N. benthamiana 16c leaves co-infiltrated with GFP- and CLBV MP-expressing constructs showed increased fluorescence, co-infiltration of pBIN-GFP with IC-CLBV, carrying an infectious cDNA clone of the full CLBV genome, did not result in GFP silencing suppression, although systemic CLBV infection was documented. One possibility is that the MP is expressed lower after CLBV systemic infection than after transient expression of the MP gene and it is not enough to cause a detectable silencing suppression in this system. Similarly, the silencing suppressors encoded by TCV (coat protein) and Beet Western yellows virus (P0) were very efficient in suppressing local RNA silencing when they were transiently expressed from binary vectors but not when expressed from the cognate infectious clones [49, 50]. P0 protein was readily detected by immunoblot analysis in N. benthamiana plants agro-infiltrated with pBIN-P0, whereas in leaves agro-inoculated with the full-length viral cDNA it was below the detection threshold [49].
Because CLBV is seed-transmitted [51] and it is difficult to recover CLBV-free plants by shoot-tip grafting in vitro [52], this virus is presumed to overcome the meristem exclusion process that restricts spread of most viruses in infected plants [53]. Previous study with Potato virus X (PVX) and CMV showed that RNA silencing was involved in virus exclusion from meristems [54–56]. Martín-Hernández and Baulcombe [57] observed that the 16K protein of Tobacco rattle virus (TRV), a virus that is seed-transmitted and invades meristems of infected plants, showed a weak suppressor activity in agro-infiltrated N. benthamiana 16c plants, in comparison with the p19 protein of TBSV. Also, TRV mutants obtained by introduction of a stop codon in the 16K protein (TRV-stop) were unable to invade meristems or floral primordia. On the other hand, plants infected with TRV and PVX allowed PVX accumulation in meristems, whereas this virus was excluded from meristems in plants co-infected with TRV-stop and PVX. These data are consistent with TRV 16K protein being the component allowing the virus to invade the meristem. The authors suggested that the weak suppressor activity of the 16K protein might be a crucial factor because a strong suppressor activity would allow high virus accumulation in meristematic cells likely causing severe damage to infected plants. Similarly, the CLBV MP protein that is also a weak silencing suppressor in comparison with p19 or HC-Pro, could be the factor responsible for viral invasion of meristematic cells without causing important symptoms in most citrus hosts [58].
References
D.C. Baulcombe, Nature 431, 356–363 (2004)
E.J. Chapman, J.C. Carrington, Nat. Rev. Genet. 8, 884–896 (2007)
P. Brodersen, O. Voinnet, Trends Genet. 22, 268–280 (2006)
P. Dunoyer, G. Schott, C. Himber, D. Meyer, A. Takeda, J.C. Carrington, O. Voinnet, Science 328, 912–916 (2010)
A. Molnar, C.W. Melnyk, A. Bassett, T.J. Hardcastle, R. Dunn, D.C. Baulcombe, Science 328, 872–875 (2010)
S.W. Ding, O. Voinnet, Cell 130, 413–426 (2007)
I. Krulko, D. Ustyanenko, V. Polischuk, Cytol. Genet. 43, 63–72 (2009)
R. MacDiarmid, Annu. Rev. Phytopathol. 43, 523–544 (2005)
O. Voinnet, Nat. Rev. Genet. 6, 206–220 (2005)
J.A. Díaz-Pendón, S.W. Ding, Annu. Rev. Phytopathol. 46, 303–326 (2008)
F. Li, S.W. Ding, Annu. Rev. Microbiol. 60, 503–531 (2006)
L. Lakatos, G. Szittya, D. Silhavy, J. Burgyán, EMBO J. 23, 876–884 (2004)
D. Silhavy, A. Molnár, A. Lucioli, G. Szittya, C. Hornyik, M. Tavazza, J. Burgyán, EMBO J. 21, 3070–3080 (2002)
L. Lakatos, G. Szittya, V. Pantaleo, E.J. Chapman, J.C. Carrington, Y.P. Liu, V.V. Dolja, L.F. Calvino, J.J. López-Moya, J. Burgyán, EMBO J. 25, 2768–2780 (2006)
Z. Mérai, Z. Kerényi, S. Kertész, M. Magna, L. Lakatos, D. Silhavy, J. Virol. 80, 5747–5756 (2006)
J. Azevedo, D. Garcia, D. Pontier, S. Ohnesorge, A. Yu, S. Garcia, L. Braun, M. Bergdoll, M.A. Hakimi, T. Lagrange, O. Voinnet, Genes Dev. 24, 904–915 (2010)
N. Baumberger, C.H. Tsai, M. Lie, E. Havecker, D.C. Baulcombe, Curr. Biol. 17, 1609–1614 (2007)
D. Bortolamiol, M. Pazhouhandeh, K. Marrocco, P. Genschik, V. Ziegler-Graff, Curr. Biol. 17, 1615–1621 (2007)
A. Giner, L. Lakatos, M. García-Chapa, J.J. López-Moya, J. Burgyán, PLoS Pathog. 6, e1000996 (2010)
X. Zhang, Y.R. Yuan, Y. Pei, S.S. Lin, T. Tuschl, D.J. Patel, N.H. Chua, Genes Dev. 20, 3255–3268 (2006)
E.J. Chapman, A.I. Prokhnevsky, K. Gopinath, V.V. Dolja, J.C. Carrington, Genes Dev. 18, 1179–1186 (2004)
M.J. Adams, G.P. Accotto, A.A. Agranovsky, M. Bar-Joseph, D. Boscia, A.A. Brunt, T. Candresse, R.H.A. Coutts, V.V. Dolja, B.W. Falk, G.D. Foster, D. Gonsalves, W. Jelkmann, A. Karasev, G.P. Martelli, M. Mawassi, R.G.Milne, A. Minafra, S. Namba, A. Rowhani, H.J. Vetten, V.K. Vishnichenko, G.C. Wisler, N. Yoshikawa, S.K. Zavriev, in Family Flexiviridae, ed. By C.M. Fauquet, M.A. Mayo, J. Maniloff, U. Desselberger, L.A. Ball (Elsevier Academic Press, London, 2005) p. 1089
G.P. Martelli, M.J. Adams, J.F. Kreuze, V.V. Dolja, Annu. Rev. Phytopathol. 45, 73–100 (2007)
M.C. Vives, L. Galipienso, L. Navarro, P. Moreno, J. Guerri, Virology 287, 225–233 (2001)
L. Galipienso, M.C. Vives, P. Moreno, R.G. Milne, L. Navarro, J. Guerri, Arch. Virol. 146, 357–368 (2001)
A. Renovell, S. Gago, S. Ruiz-Ruiz, K. Velázquez, L. Navarro, P. Moreno, M.C. Vives, J. Guerri, Virology 406, 360–369 (2010)
M.C. Vives, L. Galipienso, L. Navarro, P. Moreno, J. Guerri, Virology 295, 328–336 (2002)
O. Voinnet, S. Rivas, P. Mestre, D. Baulcombe, Plant J. 33, 949–956 (2003)
M.T. Ruiz, O. Voinnet, D.C. Baulcombe, Plant Cell 10, 937–946 (1998)
M.C. Vives, S. Martín, S. Ambrós, A. Renovell, L. Navarro, J.A. Pina, P. Moreno, J. Guerri, Mol. Plant Pathol. 9, 787–797 (2008)
G. Ancillo, J. Gadea, J. Forment, J. Guerri, L. Navarro, J. Exp. Bot. 58, 1927–1933 (2007)
H. Yaegashi, T. Takahashi, M. Isogai, T. Kobori, S. Ohki, N. Yoshikawa, J. Gen. Virol. 88, 316–324 (2007)
G. Brigneti, O. Voinnet, W.X. Li, L.H. Ji, S.W. Ding, D.C. Baulcombe, EMBO J. 17, 6739–6746 (1998)
O. Voinnet, C. Lederer, D. Baulcombe, Cell 103, 157–167 (2000)
K.D. Kasschau, J.C. Carrington, Cell 95, 461–470 (1998)
A. Hamilton, O. Voinnet, L. Chappell, D. Baulcombe, EMBO J. 21, 4671–4679 (2002)
C. Himber, P. Dunoyer, G. Moissiard, C. Ritzenthaler, O. Voinnet, EMBO J. 22, 4523–4533 (2003)
H.S. Guo, S.W. Ding, EMBO J. 21, 398–407 (2002)
R. Lu, A. Folimonov, M. Shintaku, W.X. Li, B.W. Falk, W.O. Dawson, S.W. Ding, Proc. Natl. Acad. Sci. USA 101, 15742–15747 (2004)
B.M. Roth, G.J. Pruss, V.B. Vance, Virus Res. 102, 97–108 (2004)
O. Voinnet, Y.M. Pinto, D.C. Baulcombe, Proc. Natl. Acad. Sci. USA 96, 14147–14152 (1999)
Z.S. Zhou, M. Dell’Orco, P. Saldarelli, C. Turturo, A. Minafra, G.P. Martelli, J. Gen. Virol. 87, 2387–2395 (2006)
V. Alvarado, H.B. Scholthof, Semin. Cell Dev. Biol. 20, 1032–1040 (2009)
A. Lingel, B. Simon, E. Izaurralde, M. Sattler, EMBO Rep. 6, 1149–1155 (2005)
H. Hemmes, I. Kaaij, D. Lohuis, M. Prins, R. Goldbach, E. Schnettler, J. Gen. Virol. 90, 1762–1766 (2009)
S. Martínez-Turiño, C. Hernández, J. Gen. Virol. 90, 519–525 (2009)
M.C. Cañizares, J. Navas-Castillo, E. Moriones, Virology 379, 168–174 (2008)
J.F. Kreuze, E.I. Savenkov, W. Cuellar, X. Li, J.P.T. Valkonen, J. Virol. 79, 7227–7238 (2005)
S. Pfeffer, P. Dunoyer, F. Heim, K.E. Richards, G. Jonard, V. Ziegler-Graff, J. Virol. 76, 6815–6824 (2002)
F. Qu, T. Ren, T.J. Morris, J. Virol. 77, 511–522 (2003)
J. Guerri, J.A. Pina, M.C. Vives, L. Navarro, P. Moreno, Plant Dis. 88, 906 (2004)
L. Navarro, J.A. Pina, J.F. Ballester-Olmos, P. Moreno, M. Cambra, in Proceedings of the 9th Conference of the International Organization of Citrus Virologists, ed. by S.M. Garnsey, L.W. Timmer, J.A. Dodds (IOCV, Riverside, CA,1984), pp. 234–240
D. Wang, S.A. MacFarlane, A.J. Maule, Virology 234, 112–117 (1997)
T. Mochizuki, S.T. Ohki, J. Gen. Plant Pathol. 70, 363–366 (2004)
F. Qu, X. Ye, G. Hou, S. Sato, T.E. Clemente, T.J. Morris, J. Virol. 79, 15209–15217 (2005)
F. Schwach, F.E. Vaistij, L. Jones, D.C. Baulcombe, Plant Physiol. 138, 1842–1852 (2005)
A.N. Martín-Hernández, D.C. Baulcombe, J. Virol. 82, 4064–4071 (2008)
L. Galipienso, L. Navarro, J.F. Ballester-Olmos, J.A. Pina, P. Moreno, J. Guerri, Plant Pathol. 49, 308–314 (2000)
Acknowledgments
Águeda Renovell was recipient of a doctoral fellowship from the Ministerio de Educación y Ciencia (MEC). Mª Carmen Vives was recipient of a contract from IVIA. This study was supported by grants AGL2006-0316 and AGL2009-08226, co-financed by FEDER funds, and by the MEC and the Ministerio de Ciencia e Innovación (MICINN). We thank Maria Boil for excellent lab assistance and José Juárez for excellent technical support to prepare photographs.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
11262_2011_674_MOESM1_ESM.tif
Fig. S1. Suppression by CLBV MP of RNA silencing triggered by dsGFP. a Leaves of N. benthamiana 16c plants infiltrated with a mixture of A. tumefaciens cultures harbouring binary vectors pBI-dsGFP and pBI-p19, pBI-MP or the empty vector (Ø). The green fluorescence images of the co-infiltrated leaves were taken at 5 days pos-infiltration (dpi) under a long-wavelength UV lamp. b Northern blot analysis of GFP mRNA and gfp-derived sRNAs extracted from the agro-infiltrated leaf patches shown in (a) or from non-infiltrated leaves (c) using a DIG-riboprobe specific for the GFP mRNA. Ethidium bromide staining of 5S rRNA and tRNAs are shown as loading controls for mRNA and sRNAs, respectively. Supplementary material 1 (TIFF 412 kb)
11262_2011_674_MOESM2_ESM.tif
Fig. S2. Effect of CLBV-encoded proteins on systemic spread of the GFP silencing signal in N. benthamiana 16c plants. Basal leaves were agro-infiltrated with pBI-GFP at the leaf tip and with pBI-Ø, pBI-MET, pBI-REP, pBI-MP, pBI-CP or pBI-p25 at the leaf base. Plants were photographed at 14 dpi under long-wavelength UV light with a yellow filter. Only the p25 protein of CTV prevented systemic silencing of the upper leaves. Supplementary material 2 (TIFF 1302 kb)
Rights and permissions
About this article
Cite this article
Renovell, Á., Vives, M.C., Ruiz-Ruiz, S. et al. The Citrus leaf blotch virus movement protein acts as silencing suppressor. Virus Genes 44, 131–140 (2012). https://doi.org/10.1007/s11262-011-0674-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-011-0674-9