Abstract
Human BK virus may cause nephropathy due to viral replication in patients who have undergone renal transplantation. However, the mechanism regulating replication of BKV is still not clear. Previous studies have suggested that epigenetic modifications may play a crucial role in virus replication. In this study, the DNA methylation profiles of five CpG sites located within the promoter/enhancer regions and nine CpG sites located within the early and late coding regions of the replicating BKV genome were investigated. BKV genomic DNA from mature virions and from the early and late phases of replicating BKV were examined for DNA methylation by bisulfite sequencing that covered 14 CpG sites. Our results showed that none of the examined BKV DNA from the various different stages of replication was methylated. This is the first report to analyze the methylation of BKV genomic DNA during viral replication. The results seem to indicate that methylation is not involved in regulation of BKV replication.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Polyomaviruses are small DNA viruses with circular double-stranded viral genome. The virus infects various different mammals that are hosts for replication. There are at least 17 members of the polyomavirus family, of which nine are human polyomaviruses, namely BK virus (BKV) [1], JC virus (JCV) [2], KI virus [3], WU virus [4], Merkel Cell polyomavirus (MCV) [5], Human polyomavirus 6 and Human polyomavirus 7 [6], Trichodysplasia spinulosa polyomavirus (TSV) [7], and Human polyomavirus 9 [8]. BKV has been isolated from the urine of a renal transplant recipient [1] and is now known to be widespread in human populations [9]. During early childhood, asymptomatic primary infection with BKV occurs, and the virus remains latent in human body. Upon immune suppression, BKV may be reactivated and cause lytic infection that can be associated with kidney and urinary tract disorders, such as hemorrhagic cystitis or polyomavirus nephropathy (PVN); these may cause renal dysfunction and graft loss in transplant patients [10–12].
Epigenetic modifications, including DNA methylation, histone modifications, nucleosome positioning, and expression of non-coding RNA influence gene expression without any change in the DNA sequence. The most widely studied epigenetic modifications in humans are cytosine methylation of DNA in CpG dinucleotides (CpG site). It has been found that 60–90% of vertebrate genomic CpG sites are methylated [13, 14]. In contrast, only 3–6% of all cytosines are methylated in normal human DNA [15]. In this context, DNA methylation plays a crucial role in the control of gene activity in higher eukaryotes [16, 17]. For example, DNA methylation within the promoter region of genes has been shown to repress gene transcription, and it has been postulated that such methylation acts as a mechanism that controls gene expression in a tissue-specific manner [18].
There are several lines of evidences showing that DNA methylation can play a role in viral life cycle. For example, evidence shows that integrated adenoviral and retroviral DNA are regulated by methylation [19, 20]. Particularly, hepatitis B virus (HBV) DNA can be methylated in human tissues in both its integrated and nonintegrated forms [21, 22]. Once methylated, HBV DNA undergoes alterations in viral protein production, which suggests that methylation of CpG sites is functionally relevant to viral gene expression [23]. In addition, only some human papillomavirus (HPV) promoters are regulated by methylation [24, 25], but methylation of the HPV E2 promoter hinders E2 protein binding and subsequent inhibition of the viral life cycle [26, 27]. Furthermore, it is thought that DNA methylation changes may switch Epstein–Barr virus (EBV) between reactivation and latency status [28]. Human immunodeficiency virus 1 (HIV-1) silencing seems to be regulated by methylation within the 5’LTR region that contains promoter and enhancer sequences [29, 30]. On the contrary, CpG sites in human adenovirus isolated from purified adenovirus or non-integrated viral DNA extracted from nuclei of infected cells remains unmethylated [19, 20]. Further, studies also demonstrated that CpG sites of several retrovirus including lentivirus are resistant to DNA methylation in mammalian cell [31–33].
Several groups have demonstrated the hypermethylation of tumor suppressor gene promoter in the various human cancers [34–36]; however, the role of CpG methylation of BKV is currently unknown. In this study, we examined the methylation of BKV DNA at the early and late phases of replication as well as in the mature virion.
Materials and methods
Propagation of BKV
BKV [UT genotype; accession no. DQ305492] was propagated in African green monkey kidney or Vero cells [ATCC: CCL-81]. Vero cells infected by BKV were maintained in MEM (Minimum Essential Medium; Invitrogen) containing 10% fetal bovine serum (FBS; Biological Industries), 20 mM l-glutamine (Invitrogen), and 100 U/ml penicillin–streptomycin (Invitrogen). Cells from confluent dishes were trypsinized, and the cells were collected in a 1.5 ml centrifuge tube. The cells were centrifuged at 2500 rpm at 4°C, resuspended in capsid buffer (0.5 mM CaCl2, 10 mM Tris, pH 7.2, 150 mM NaCl, 5% glycerol) and then lysed by freeze/thawing three times. The BKV was partially purified by a 20% sucrose cushion. The BKV was quantified using quantitative real-time PCR with BKV VP2 specific primers (F: 5′-CACTTTTGGGGGACCTAGT-3′; R: 5′-CTCTACAGTAGCAAGGGATGC-3′). Quantitative real-time PCR was performed under the following conditions: 2 min at 50°C, 10 min at 95°C, and 40 cycles of 15 s at 95°C and 1 min at 60°C using 0.5 μl of the viral DNA, 2X SYBR Green Realtime PCR Master Mix (Toyobo) in StepOne (Applied Biosystems). Vero cells were infected by BKV with 100 multiplicity of infection (MOI) and harvested every 5 days to detect DNA methylation and viral protein expression.
Immunofluorescent assay
The BKV infected Vero cells were rehydrated and incubated with normal rabbit serum. The cells were washed with PBS three times and incubated with rabbit anti-BKV VP1 serum (1:1000 dilution) or with monoclonal antibody against SV40 LT (Abcam) for 30 min at room temperature. After washing, the cells were then incubated with FITC-labeled goat anti-rabbit IgG antibody (1:160 dilution) (Sigma). DAKO™ FLUORESCENT Mounting Solution (DaKo) was used for mounting. The cells were observed under a confocal fluorescent microscope, Axioplan2 (ZEISS).
Bisulfite conversion
Sample DNA was bisulfite modified using an EZ DNA methylation kit (ZYMO research) according to the manufacturer’s protocol. In total, 5 μl of M-Dilution buffer was added to 500 ng of DNA in a sample tube, and water was used to adjust the total volume to 50 μl. The mixture was incubated at 37°C for 15 min. Then, 100 μl of CT conversion reagent was added to each sample and mixed thoroughly. The mixtures were then incubated in a thermocycler and with the following program setting: 50°C for 2 h, switch to 95°C for 15 s, back to 50°C for 4 h, and then switch to 95°C for 10 s. Finally, the mixtures were incubated at 50°C for 9 h and 30 min. The mixtures were next placed on ice for 10 min and mixed with 400 μl M-Binding Buffer. The mixture was then loaded into a Zymo-Spin IC Column. The columns were washed with 200 μl of M-Wash Buffer and columns incubated at room temperatures for 20 min with 200 μl M-Desulphonation Buffer. Next, the columns were washed with M-Wash Buffer twice, and finally, the DNA was eluted using 50 μl ddH2O twice. The positive control for methylation was generated by methylation of BKV genome-containing plasmid using CpG methyltransferase, M.SssI (NEB).
Identification of DNA methylation by bisulfite sequencing
CpG sites in the BKV genome and primers for amplification were designed by using MethPrimer software (http://www.urogene.org/methprimer/index1.html). Plasmid, pGEM-Teasy, containing the BKV genomic DNA, was used as a negative control. A total of 10 μl of bisulfite modified BKV DNA was amplified in a 50 μl PCR solution containing 0.25 μM of each forward and reverse primers (Table 1), which were specific for various different regions of the BKV genome. The mixture used contained 250 μM dNTP, 2.5 mM MgCl2, and 0.025 units Platinum Taq DNA polymerase (Invitrogen). A GeneAmp PCR system 2500 thermal cycler (Applied Biosystems) was employed for the amplification. The cycling conditions were 1 cycle of 95°C for 5 min, 35 cycles of 95°C for 30 s, 57°C for 30 s, 72°C for 30 s, and 1 cycle of 72°C for 5 min. The PCR products were cloned into the pGEM-Teasy vector (Promega). Five randomly picked clones were sequenced by Genomics Biotechnology Company (Taipei, Taiwan). Results of the sequencing were analyzed using a web-based tool, namely Biology Workbench (http://workbench.sdsc.edu/).
Results
CpG sites in BKV genome
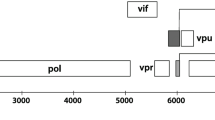
Methylation within a CpG site in a promoter region of a gene may inhibit gene expression [37]. In addition, methylation may also regulate gene expression when it occurs within a coding region [38]. We analyzed the methylation status of 14 CpG dinucleotides within the BKV genome (Fig. 1). With respect to their position, these 14 CpG dinucleotides are located within promoter/enhancer regulatory regions (5 CpGs), the agno protein coding region (3 CpGs), the VP2/3 common region (3 CpGs), and the coding region of the major capsid protein VP1 (1CpG) as well as at the early and common region for the large tumor antigen and small tumor antigen (2 CpGs).
BKV life cycle
DNA methylation may regulate gene expression at various phases of virus life cycle. To investigate DNA methylation of BKV at different stages of propagation, Vero cells were infected with BKV and harvested at 3, 8, 13, 18, 23, and 28 days post-infection. Since a complete life cycle of BKV is approximately 14 days [39], a total of 28 days post infection of BKV covers the early and late phases of BKV replication. Results of immunofluorescent assay (IFA) showed that about 1% of the infected cell expressed LT protein at day 8 post-infection. The LT positive cells gradually increased from 1% at day 8 to 70% at day 28 (Fig. S1a). A similar expression profile was also found for VP1 (Fig. S1b). The percentage of positive cells were quantified and shown in Fig. S1c. These findings indicate that BKV successfully replicated in the Vero cells and completed the virus life cycles.
Detection of methylation on BKV genomic DNA
DNA methylation may be dynamically regulated during different replication stages [40, 41]. To analyze the DNA methylation pattern of BK virus, viral DNA was extracted from mature virion at 18, 23, and 28 days of post-infection corresponding to the early to late phases of the life cycle. Four pairs of primers (Table 1) flanking all CpG sites (Fig. 1) were used for bisulfite-PCR. After bisulfite conversion, PCR was performed to amplify DNA fragments containing the CpG sites. As expected, the fragment sizes of the PCR products using the specific primers (Table 1) were 437, 497, 527, and 282 bp, respectively, after amplification from different BKV genomes (Fig. 2). The DNA fragments were then cloned into pGEM-Teasy replicative plasmid for further sequencing (Fig. 3). The results showed that all of the BKV genomic DNAs, including those from virion particles and from early phase replication to late phase (18, 23, and day 28) were devoid of any methylation. For positive control of methylation, in vitro methylated BKV genomic DNA containing plasmid showed methylation at all CpG sites (Fig. 3). In contrast, plasmid BKV genomic DNA which was extracted from E. coli was essentially unmethylated. The findings suggest that BK viral DNA is not modified by methylation during the virus replication life cycle.
Amplification of various BKV genomic DNA fragments after bisulfite conversion. BKV genomic DNAs extracted from virion particles (BKV), viral plasmid DNA (BKV pl), in vitro methylated viral genome (Me BKV pl) and post-infection on day 18 (d18), day 23 (d23), and day 28 (d28). After bisulfite conversion, the viral DNAs were amplified by PCR using primer pairs 1–4 as listed in Table 1. (−) PCR negative control, M DNA markers
Sequencing of CpG sites within the BKV genomic DNA. CpG sites within the BKV genomic DNA were sequenced after bisulfite conversion (a). The viral genomic DNAs were extracted from BKV virion particles, plasmid BKV DNA, in vitro methylated BKV genome (Me plasmid BKV DNA), and post-infection on day 18 (d18), day 23 (d23), and day 28 (d28). Methylation of viral DNA was summarized in b. Open circle unmethylated, Filled circle methylated
Discussion
In this study, we analyzed the DNA methylation status of BKV genomic DNA from the early to the late phase of infection, as well as the mature virus particles. The results from bisulfite sequencing showed that BKV DNAs from these different sources were devoid of any methylation. This is the first report on the methylation of BKV genomic DNA during various different stages of replication. These results indicate that DNA methylation is not involved in the regulation of BKV replication.
A previous study using murine polyoma virus to infect mouse kidney cells found that both viral and cellular DNA seems to be methylated [42]. Further studies have also demonstrated that polyoma and SV40 DNA are methylated in vitro and when microinjected into cells for early and late gene expression [43, 44]. Furthermore, recent studies demonstrated that DNA methylation in the coding region of several genome of DNA viruses are presented in human cancers and correlates with expression level of viral genes and disease progression [38, 45]. However, the expression of only late genes and not early genes was affected by DNA methylation [43]. Similar results were found for integrated polyomavirus DNA in polyoma-transformed rat cells, where only late genes were methylated [46]. Similarly, LTR of several retroviruses are also found to be unmethylated in human cancer cells [32, 33, 47]. Taking the previous findings and our current results together, it may concluded that methylation does not seem to be involved in polyomavirus gene regulation during productive infection but this leaves open the possibility that methylation is involved in transformation or non-productive infection. However, further experiments are underway to confirm if BK viruses are resistant to DNA methylation in human cells.
DNA viruses, such as Kaposi’s sarcoma-associated herpesvirus (KSAV) [48], HBV [49, 50], and EBV [51], may trigger cellular DNMT expression, and this may subsequently affect promoter methylation of cellular genes and thus their expression. However, in this context, it is important to note that viral DNA can also be methylated after activation of DNMT [52]. Viral DNA methylation may be associated with the innate immune response. For example, unmethylated HBV DNA may stimulate Toll-like receptor expressing cells and subsequently cause the activation of the NF-κB pathway [53]. It is known that the NF-κB pathway may inhibit HBV replication via the innate immune system. A similar mechanism for immune escape has also been found for KSAV infection [54]. During BKV infection, it would seem that LT may activate DNMT1 [55]. However, BKV viral DNA does not seem to be methylated during the viral replication process based on the findings of this study. However, further studies are needed to clarify whether BKV infection can affect the methylation of cellular DNA through activation of DNMT.
In addition to DNA methylation, histone modifications may also be involved in gene regulation. It has been reported that the viral expression of the latent-associated transcripts (LATs) of herpes simplex virus type 1 is regulated by histone acetylation in the area of the promoter region of the LAT genes [56]. Therefore, it is possible that the viral gene expression of BKV may be regulated by histone modification.
It is also possible that microRNA may also play a role in viral gene regulation. Previous reports have shown that microRNA can be detected during polyoma virus infection without affecting viral protein expression [57]. Notwithstanding this, it is still possible that microRNA may play a role in the autoregulation of viral gene expression during polyoma virus infection [57]. During SV40 infection, the presence of viral microRNA does not impair the yield of infectious virus but does reduce the production of LT, which results in the effect of cytotoxic T cells on the virus-infected cells being hindered [58]. More recently, production of viral microRNA has also found to occur during BKV, JCV, and MCV infection [59, 60]. MicroRNA would seem to regulate the amount of early protein present during the late phase of infection to promote late protein expression. Similarly, a “switch” from early to late phases during lytic infection may be regulated by viral transcripts. During the polyoma virus life cycle, it was found that the viral late transcripts annealed with early transcripts to form dsRNA and this results in a switch into late phase [61]. Therefore, viral microRNA and late transcripts may also be involved in the regulation of viral replication and this needs to be investigated. In conclusion, our result showed that the examined BKV CpG sites were devoid of any methylation, which suggests that DNA methylation is not involved in the regulation of BKV replication. Nonetheless, the effect of BKV infection on methylation of cellular genes seems to deserve further investigation.
Abbreviations
- BKV:
-
BK virus
- JCV:
-
JC virus
- MCV:
-
Merkel cell polyomavirus
- TSV:
-
Trichodysplasia spinulosa polyomavirus
- HPyV:
-
Human polyomavirus
- PVN:
-
Polyomavirus nephropathy
- HBV:
-
Hepatitis B virus
- HPV:
-
Human papillomavirus
- EBV:
-
Estein–Barr virus
- HIV:
-
Human immunodeficiency virus
- IFA:
-
Immunofluorescent assay
- KSAV:
-
Kaposi’s Sarcoma-associated herpesvirus
- DNMT:
-
DNA methyltransferase
- LATs:
-
Latent-associated transcripts
References
S.D. Gardner, A.M. Field, D.V. Coleman, B. Hulme, Lancet 1, 1253–1257 (1971)
B.L. Padgett, D.L. Walker, G.M. ZuRhein, R.J. Eckroade, B.H. Dessel, Lancet 1, 1257–1260 (1971)
T. Allander, K. Andreasson, S. Gupta, A. Bjerkner, G. Bogdanovic, M.A. Persson, T. Dalianis, T. Ramqvist, B. Andersson, J. Virol. 81, 4130–4136 (2007)
A.M. Gaynor, M.D. Nissen, D.M. Whiley, I.M. Mackay, S.B. Lambert, G. Wu, D.C. Brennan, G.A. Storch, T.P. Sloots, D. Wang, PLoS Pathog 3, e64 (2007)
H. Feng, M. Shuda, Y. Chang, P.S. Moore, Science 319, 1096–1100 (2008)
R.M. Schowalter, D.V. Pastrana, K.A. Pumphrey, A.L. Moyer, C.B. Buck, Cell Host Microbe 7, 509–515 (2010)
E. van der Meijden, R.W. Janssens, C. Lauber, J.N. Bouwes Bavinck, A.E. Gorbalenya, M.C. eltkamp, PLoS Pathog 6, e1001024 (2010)
N. Scuda, J. Hofmann, S. Calvignac-Spencer, K. Ruprecht, P. Liman, J. Kuhn, H. Hengel, B. Ehlers, J. Virol. 85(9), 4586 (2011)
W.A. Knowles, in Human Polyomaviruses: Molecular and Clinical Perspectives, ed. by K. Khalili (Wiley, New York, 2002), p. 527
H.H. Hirsch, W. Knowles, M. Dickenmann, J. Passweg, T. Klimkait, M.J. Mihatsch, J. Steiger, N. Engl. J. Med. 347, 488–496 (2002)
A. Bedi, C.B. Miller, J.L. Hanson, S. Goodman, R.F. Ambinder, P. Charache, R.R. Arthur, R.J. Jones, J. Clin. Oncol. 13, 1103–1109 (1995)
T. Shinohara, M. Matsuda, S.H. Cheng, J. Marshall, M. Fujita, K. Nagashima, J. Med. Virol. 41, 301–305 (1993)
A.P. Bird, Nature 321, 209–213 (1986)
A. Bird, M. Taggart, M. Frommer, O.J. Miller, D. Macleod, Cell 40, 91–99 (1985)
M. Esteller, Annu. Rev. Pharmacol. Toxicol. 45, 629–656 (2005)
W. Doerfler, J. Gen. Virol. 57, 1–20 (1981)
A. Razin, J. Friedman, Prog. Nucleic Acid Res. Mol. Biol. 25, 33–52 (1981)
E. Li, C. Beard, A.C. Forster, T.H. Bestor, R. Jaenisch, Cold Spring Harb. Symp. Quant. Biol. 58, 297–305 (1993)
U. Gunthert, M. Schweiger, M. Stupp, W. Doerfler, Proc. Natl. Acad. Sci. USA 73, 3923–3927 (1976)
C. Kammer, W. Doerfler, FEBS Lett. 362, 301–305 (1995)
P. Vivekanandan, D. Thomas, M. Torbenson, J. Viral Hepat. 15, 103–107 (2008)
J.Y. Chen, H.C. Hsu, C.S. Lee, D.S. Chen, A.J. Zuckerman, T.J. Harrison, J. Virol. Methods 19, 257–263 (1988)
P. Vivekanandan, D. Thomas, M. Torbenson, J. Infect. Dis. 199, 1286–1291 (2009)
V. Badal, L.S. Chuang, E.H. Tan, S. Badal, L.L. Villa, C.M. Wheeler, B.F. Li, H.U. Bernard, J. Virol. 77, 6227–6234 (2003)
S. Badal, V. Badal, I.E. Calleja-Macias, M. Kalantari, L.S. Chuang, B.F. Li, H.U. Bernard, Virology 324, 483–492 (2004)
K. Kim, P.A. Garner-Hamrick, C. Fisher, D. Lee, P.F. Lambert, J. Virol. 77, 12450–12459 (2003)
A. Thain, O. Jenkins, A.R. Clarke, K. Gaston, J. Virol. 70, 7233–7235 (1996)
R.F. Ambinder, K.D. Robertson, Q. Tao, Semin. Cancer Biol. 9, 369–375 (1999)
T. Ishida, A. Hamano, T. Koiwa, T. Watanabe, Retrovirology 3, 69 (2006)
D.P. Bednarik, J.D. Mosca, N.B. Raj, J. Virol. 61, 1253–1257 (1987)
F. Zhang, A.R. Frost, M.P. Blundell, O. Bales, M.N. Antoniou, A.J. Thrasher, Mol. Ther. 18, 1640–1649 (2010)
J. Hejnar, D. Elleder, P. Hajkova, J. Walter, J. Blazkova, J. Svoboda, Biochem. Biophys. Res. Commun. 311, 641–648 (2003)
J. Gimenez, C. Montgiraud, G. Oriol, J.P. Pichon, K. Ruel, V. Tsatsaris, P. Gerbaud, J.L. Frendo, D. Evain-Brion, F. Mallet, DNA Res. 16, 195–211 (2009)
J.L. Chou, H.Y. Su, L.Y. Chen, Y.P. Liao, C. Hartman-Frey, Y.H. Lai, H.W. Yang, D.E. Deatherage, C.T. Kuo, Y.W. Huang, P.S. Yan, S.H. Hsiao, C.K. Tai, H.J. Lin, R.V. Davuluri, T.K. Chao, K.P. Nephew, T.H. Huang, H.C. Lai, M.W. Chan, Lab. Invest. 90, 414–425 (2010)
E. Lara, V. Calvanese, C. Huidobro, A.F. Fernandez, A. Moncada-Pazos, A.J. Obaya, O. Aguilera, J.M. Gonzalez-Sancho, L. Sanchez, A. Astudillo, A. Munoz, C. Lopez-Otin, M. Esteller, M.F. Fraga, Mol Cancer 9, 170 (2010)
T.L. Lee, W.K. Leung, M.W. Chan, E.K. Ng, J.H. Tong, K.W. Lo, S.C. Chung, J.J. Sung, K.F. To, Clin. Cancer Res. 8, 1761–1766 (2002)
P.A. Jones, D. Takai, Science 293, 1068–1070 (2001)
A.F. Fernandez, C. Rosales, P. Lopez-Nieva, O. Grana, E. Ballestar, S. Ropero, J. Espada, S.A. Melo, A. Lujambio, M.F. Fraga, I. Pino, B. Javierre, F.J. Carmona, F. Acquadro, R.D. Steenbergen, P.J. Snijders, C.J. Meijer, P. Pineau, A. Dejean, B. Lloveras, G. Capella, J. Quer, M. Buti, J.I. Esteban, H. Allende, F. Rodriguez-Frias, X. Castellsague, J. Minarovits, J. Ponce, D. Capello, G. Gaidano, J.C. Cigudosa, G. Gomez-Lopez, D.G. Pisano, A. Valencia, M.A. Piris, F.X. Bosch, E. Cahir-McFarland, E. Kieff, M. Esteller, Genome Res. 19, 438–451 (2009)
A.S. Dugan, S. Eash, W.J. Atwood, J. Virol. 79, 14442–14445 (2005)
S. Ito, A.C. D’Alessio, O.V. Taranova, K. Hong, L.C. Sowers, Y. Zhang, Nature 466, 1129–1133 (2010)
S.C. Wu, Y. Zhang, Nat. Rev. Mol. Cell. Biol. 11, 607–620 (2010)
E. Winocour, A.M. Kaye, V. Stollar, Virology 27, 156–169 (1965)
A. Fradin, J.L. Manley, C.L. Prives, Proc. Natl. Acad. Sci. USA 79, 5142–5146 (1982)
M. Graessmann, A. Graessmann, H. Wagner, E. Werner, D. Simon, Proc. Natl. Acad. Sci. USA 80, 6470–6474 (1983)
C. Sun, L.L. Reimers, R.D. Burk, Gynecol. Oncol. 121, 59–63 (2011)
H. Manor, J. Virol. 56, 734–742 (1985)
J. Blazkova, K. Trejbalova, F. Gondois-Rey, P. Halfon, P. Philibert, A. Guiguen, E. Verdin, D. Olive, C. Van Lint, J. Hejnar, I. Hirsch, PLoS Pathog. 5, e1000554 (2009)
M. Shamay, A. Krithivas, J. Zhang, S.D. Hayward, Proc. Natl. Acad. Sci. USA 103, 14554–14559 (2006)
I.Y. Park, B.H. Sohn, E. Yu, D.J. Suh, Y.H. Chung, J.H. Lee, S.J. Surzycki, Y.I. Lee, Gastroenterology 132, 1476–1494 (2007)
D.L. Zheng, L. Zhang, N. Cheng, X. Xu, Q. Deng, X.M. Teng, K.S. Wang, X. Zhang, J. Huang, Z.G. Han, J. Hepatol. 50, 377–387 (2009)
R. Hino, H. Uozaki, N. Murakami, T. Ushiku, A. Shinozaki, S. Ishikawa, T. Morikawa, T. Nakaya, T. Sakatani, K. Takada, M. Fukayama, Cancer Res. 69, 2766–2774 (2009)
P. Vivekanandan, H.D. Daniel, R. Kannangai, F. Martinez-Murillo, M. Torbenson, J. Virol. 84, 4321–4329 (2010)
X. Liu, Q. Xu, W. Chen, H. Cao, R. Zheng, G. Li, Oncol. Rep. 21, 941–947 (2009)
S.C. Verma, T. Choudhuri, E.S. Robertson, J. Virol. 81, 3402–3413 (2007)
M.T. McCabe, J.A. Low, M.J. Imperiale, M.L. Day, Oncogene 25, 2727–2735 (2006)
N.J. Kubat, R.K. Tran, P. McAnany, D.C. Bloom, J. Virol. 78, 1139–1149 (2004)
C.S. Sullivan, C.K. Sung, C.D. Pack, A. Grundhoff, A.E. Lukacher, T.L. Benjamin, D. Ganem, Virology 387, 157–167 (2009)
C.S. Sullivan, A.T. Grundhoff, S. Tevethia, J.M. Pipas, D. Ganem, Nature 435, 682–686 (2005)
G.J. Seo, C.J. Chen, C.S. Sullivan, Virology 383, 183–187 (2009)
G.J. Seo, L.H. Fink, B. O’Hara, W.J. Atwood, C.S. Sullivan, J. Virol. 82, 9823–9828 (2008)
M. Kumar, G.G. Carmichael, Proc. Natl. Acad. Sci. USA 94, 3542–3547 (1997)
Acknowledgments
This study was supported by research grant from the National Science Council, Taiwan, ROC: NSC97-2320-B-194-002-MY3 to MWYC and NSC99-2321-B-194-001, and NSC97-2320-B-194-001-MY3 to DC.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.

11262_2011_627_MOESM1_ESM.jpg
Fig. S1. Replication of BKV. Vero cells were infected by BKV. LT (a) and VP1 (b) were detected by immunofluorescent assay using anti-LT and anti-VP1 antibodies on day 3, 8, 13, 18, 23, and 28 post-infection. The percentage of positive cells were quantified and this is shown in panel c. (JPEG 817 kb)
Rights and permissions
About this article
Cite this article
Chang, CF., Wang, M., Fang, CY. et al. Analysis of DNA methylation in human BK virus. Virus Genes 43, 201–207 (2011). https://doi.org/10.1007/s11262-011-0627-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-011-0627-3