Abstract
H1 influenza A viruses that were distinct from the classical swine H1 lineage were identified in pigs in Canada in 2003–2004; antigenic and genetic characterization identified the hemagglutinin (HA) as human H1 lineage. The viruses identified in Canadian pigs were human lineage in entirety or double (human–swine) reassortants. Here, we report the whole genome sequence analysis of four human-like H1 viruses isolated from U.S. swine in 2005 and 2007. All four isolates were characterized as triple reassortants with an internal gene constellation similar to contemporary U.S. swine influenza virus (SIV), with HA and neuraminidase (NA) most similar to human influenza virus lineages. A 2007 human-like H1N1 was evaluated in a pathogenesis and transmission model and compared to a 2004 reassortant H1N1 SIV isolate with swine lineage HA and NA. The 2007 isolate induced disease typical of influenza virus and was transmitted to contact pigs; however, the kinetics and magnitude differed from the 2004 H1N1 SIV. This study indicates that the human-like H1 SIV can efficiently replicate and transmit in the swine host and now co-circulates with contemporary SIVs as a distinct genetic cluster of H1 SIV.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Influenza in swine is an acute respiratory disease caused by influenza A viruses of the family Orthomyxoviridae. Orthomyxoviruses have negative-sense single-stranded RNA genomes that are segmented, allowing for reassortment and production of novel viruses. There are two major surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA), that define subtypes and are important for host range, antigenicity, pathogenesis, and diagnostic detection. Other than sporadic transmission to humans [1], classical swine influenza A viruses of the H1N1 subtype were historically distinct from avian and other mammalian influenza viruses based on host specificity, serologic type, and/or genotype. A novel H3N2 virus appeared in the U.S. swine population around 1998 and quickly became endemic. The H3N2 viruses were demonstrated to have acquired HA, NA, and PB1 genes of human virus origin, PA and PB2 genes of avian virus origin, and the remaining internal genes, NP, M, and NS, of swine virus origin, thus giving rise to the triple reassortant designation [2]. The human lineage PB1, avian lineages PB2 and PA, and swine lineages NP, M, and NS found in contemporary swine viruses are referred to as the triple reassortant internal gene (TRIG) constellation [3]. Since 1998, three predominant swine influenza virus (SIV) subtypes have circulated in U.S. swine, H1N1, H1N2, and H3N2. The H1N1 viruses contain the HA and NA from the classical swine H1N1 virus and the TRIG from triple reassortant H3N2 viruses making them reassortant H1N1 (rH1N1); the H1N2 viruses contain the HA from the classical swine virus and the NA and TRIG from the triple reassortant H3N2 viruses [4, 5]. Since the acquisition of the TRIG cassette, an increase in the rate of genetic change in North American swine influenza isolates appears to have occurred in H1 virus subtypes, and distinct genetic and antigenic clusters have evolved [6]. At present, there are a number of reassortant viruses that have been identified in pigs, including further drift variants of H3N2 [5, 7–9], H1N2 [4, 10], rH1N1 [5], and H3N1 viruses [11, 12]. The TRIG was shown to have accepted an avian H2 and N3, producing a novel triple reassortant swine H2N3 in 2006 [13]. Furthermore, the recently emerged human pandemic H1N1 has been demonstrated to contain six gene segments of the North American triple reassortant lineage with the M and NA from the Eurasian lineage H1N1 [14, 15]. The events of spring 2009 provide compelling evidence that the TRIG has an enhanced ability to pick up novel surface glycoprotein gene segments, and these novel viruses can have pandemic potential.
The H3N2, rH1N1, and H1N2 viruses have become endemic and co-circulate in most major swine-producing regions of the U.S. and Canada. In addition, introduction of H1 viruses with the HA gene of human virus origin (hu-like H1) that are genetically and antigenically distinct from the classical swine H1 lineage were reported in pigs in Canada [16]. The viruses identified in Canadian pigs were human lineage in entirety or double (human–swine) reassortants. Since 2005, hu-like H1N1 and H1N2 viruses have emerged in swine herds across the U.S. [17]. Here, we report the whole genome sequence analysis of four hu-like H1 viruses isolated from U.S. swine, two isolated in 2005 and two isolated in 2007. These viruses are compared to the hu-like swine H1 viruses described by Karasin et al. [16]. One of the 2007 hu-like H1N1 SIV isolates utilized in the sequence analysis was selected to be evaluated in an experimental pathogenesis and transmission model and compared to a 2004 swine-lineage rH1N1 SIV isolate.
Materials and methods
Viruses and sequence analysis
Four hu-like H1 viruses isolated from the outbreaks of respiratory disease in pigs submitted to the Minnesota Veterinary Diagnostic Laboratory were selected for full genome sequencing based on partial HA sequence determination of a hu-like origin. The isolates A/Sw/IL/00685/05 (IL05), A/Sw/NC/00573/05 (NC05), A/Sw/IL/07003243/07 (IL07), and A/Sw/MN/07002083/07 (MN07) were grown in MDCK cells. Viral RNA was extracted from MDCK cell fluids with the QIAamp viral RNA Mini Kit (Qiagen Inc., Valencia, CA). For sequencing, all genes were amplified by RT-PCR in their entirety using previously described primer sets [18, 19] with SuperScript III and high fidelity Platinum TaqDNA polymerase (Invitrogen Corporation, Carlsbad, CA), and the fragments were purified by QIAquick® Gel Extraction Kit (Qiagen, Inc.) and sequenced for genetic comparison. In short, PCR products were quantitated using the Pico Green assay for dsDNA (Invitrogen Corporation, Carlsbad, CA). The appropriate quantity of dsDNA PCR product was labeled in each direction using Big Dye terminator chemistries (Applied Biosystems Inc., Foster City, CA) according to the manufacturer’s instructions. The labeled products were sequenced using an ABI 3100 genetic analyzer (Applied Biosystems Inc., Foster City, CA) using the primary PCR primers and a series of internal sequencing primers (available upon request). The sequences were analyzed using SeqMan (DNAStar, Inc., Madison, WI). Phylogenetic analyses, molecular evolutionary analyses, and pairwise analyses of base differences or amino acid differences per site were conducted using MEGA version 4 [20]. Influenza A virus sequences available in GenBank were included in the multiple alignments and phylogenetic analysis and are identified by their accession numbers in Figs. 1 and 2. The evolutionary history was inferred using the Neighbor-Joining method. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) were shown next to the branches. The trees were drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method and reported in the units of the number of base substitutions per site. All positions containing alignment gaps and missing data were eliminated only in pairwise sequence comparisons (Pairwise deletion option). Putative antigenic sites in the HA were identified by alignment with the PR8 H1 reference strain [21], and putative receptor binding and host determinant sites were identified as previously described [22–24].
Phylogenetic trees for each of the HA and NA gene segments based on nucleotide sequences from U.S. hu-like H1 (indicated by open circle), Canadian hu-like H1 (open diamond), and other sequences available from GenBank. a HA phylogenetic tree with four clusters of related viruses, H1α (cH1N1), H1β (rH1N1-like), H1γ (H1N2-like), and H1δ (hu-like H1) as indicated by the bars on the right of the tree. b NA phylogenetic tree with N1 and N2 subtypes and lineages indicated by bars on the right. Phylogenetic analyses were conducted in MEGA4. The reference viruses used in the analysis are abbreviated with their state and year of origin, subtype preceded by host abbreviation (sw swine; hu human; du duck; tu turkey), and GeneBank accession number
Phylogenetic trees for the six internal gene segments based on nucleotide sequences from U.S. hu-like H1 (indicated by open circle), Canadian hu-like H1 (open diamond), and other sequences available from GenBank: a PB2, b PB1, c PA, d NP, e M, f NS. Lineages are indicated by bars to the right of the tree. The triple reassortant internal gene (swTRIG) constellation circulating in swine influenza virus from North America is indicated to the right as well. Phylogenetic analyses were conducted in MEGA4. The reference viruses used in the analysis are abbreviated with their state and year of origin, subtype preceded by host abbreviation (sw swine; hu human; du duck; ch chicken; tu turkey), and GeneBank accession number
In vivo study
Seventy-seven 4-week-old cross-bred pigs from a herd free of SIV and porcine reproductive and respiratory syndrome virus (PRRSV) were divided into three groups. All pigs were treated with ceftiofur crystalline free acid (Pfizer, New York, NY) to reduce bacterial contaminants prior to the start of the study. Groups were housed in individual isolation rooms and cared for in compliance with the Institutional Animal Care and Use Committee of the National Animal Disease Center. Pigs were humanely euthanized with a lethal dose of pentobarbital (Sleepaway, Fort Dodge Animal Health, Fort Dodge, IA) at the appropriate times during the course of the study. Eighteen pigs were inoculated intratracheally with 2 × 105 TCID50/pig of A/Sw/MN/07002083/07 H1N1 (MN07) to represent the recent cluster of hu-like H1 SIVs. Twenty pigs were inoculated intratracheally with 2 × 105 TCID50/pig of A/Sw/IA/00239/04 H1N1 (IA04) to represent the swine lineage H1 SIVs. Twelve control pigs were inoculated with noninfectious cell culture supernatant. The virus and sham inocula were given intratracheally, while the pigs were anesthetized with an intramuscular injection of a cocktail of ketamine (8 mg/kg), xylazine (4 mg/kg), and Telazol (6 mg/kg, Fort Dodge Animal Health, Fort Dodge, IA). Ten contact pigs were comingled with each of the groups of inoculated pigs on 3 days postinfection (dpi) to study transmission efficiency. Pigs were observed daily for clinical signs of disease. Nasal swabs were taken and placed in 2 ml minimal essential medium (MEM) on 0, 3, 5, and 7 dpi or days postcontact (dpc) to evaluate nasal shedding and stored at −80°C until study completion. Five inoculated pigs and three control pigs were euthanized on 3, 5, and 7 dpi to evaluate lung lesions and viral load in the lung. The contact pigs and remaining primary inoculated pigs were euthanized on 24 dpc or 27 dpi.
For virus isolation and titration in nasal swabs, samples were subsequently thawed and vortexed for 15 s, centrifuged for 10 min at 640×g, and the supernatant was passed through 0.45 μm filters to reduce bacterial contaminants. An aliquot of 200 μl of the filtrate was plated onto confluent phosphate-buffered saline (PBS)-washed MDCK cells in 24-well plates. After 1 h of incubation at 37°C, 200 μl serum-free MEM supplemented with 1 μg/ml TPCK trypsin and antibiotics was added. All the wells were evaluated for cytopathic effect (CPE) between 24 and 48 h and subsequently frozen. After euthanasia, each lung was lavaged with 50 ml MEM to obtain broncho-alveolar lavage fluid (BALF) and stored at −80°C for virus isolation and titration. Tenfold serial dilutions in serum-free MEM supplemented with TPCK trypsin and antibiotics were made with each BALF sample and each virus isolation-positive nasal swab filtrate sample. Each dilution was plated in triplicate in 100 μl volumes onto PBS-washed confluent MDCK cells in 96-well plates. Plates were evaluated for CPE between 48 and 72 h postinfection. At 72 h, plates were fixed with 4% phosphate-buffered formalin and stained using immunocytochemistry with an anti-influenza A nucleoprotein monoclonal antibody as previously described [25]. A TCID50 was calculated for each sample using the method of Reed and Muench [26].
Pathologic examination of lungs
At necropsy, lungs were removed and evaluated for the percentage of the lung affected with purple-red consolidation typical of SIV infection. The percentage of the surface affected with pneumonia was visually estimated for each lung lobe, and a total percentage for the entire lung was calculated based on weighted proportions of each lobe to the total lung volume [27]. Tissue samples from the trachea and right cardiac lung lobe and other affected lobes were taken and fixed in 10% buffered formalin for histopathologic examination. Tissues were routinely processed and stained with hematoxylin and eosin. Lung sections were given a score from 0 to 4 to reflect the severity of bronchial epithelial injury based on previously described methods [8]. A single pathologist scored all slides and was blinded to the treatment groups.
Serologic assays
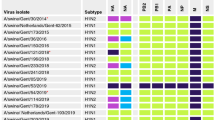
Blood samples were collected by jugular venipuncture prior to challenge, at approximately 14 dpi or dpc, and at 27 dpi or 24 dpc. For use in the hemagglutination inhibition (HI) assay, sera were heat inactivated at 56°C for 30 min, then treated with a 20% suspension of Kaolin (Sigma–Aldrich, St. Louis, MO) to remove nonspecific hemagglutinin inhibitors and natural serum agglutinins and adsorption with 0.5% turkey red blood cells (RBC), respectively. The HI assays were done with the MN07 and IA04 viruses as antigens and turkey RBC using standard techniques [28]. In addition to homologous HI assays, a panel of previously described H1 antisera [6] was evaluated against the MN07 virus and a subset of the MN07 anti-sera was evaluated against the hu-like H1 NC05, IL05, and IL07 viruses. Reciprocal HI titers were log2 transformed for analysis and reported as geometric means.
Statistical analysis
Macroscopic pneumonia scores, microscopic pneumonia scores, log10 transformations of BALF and nasal swab virus titers were analyzed using analysis of variance (ANOVA) with a P value ≤0.05 considered significant (JMP, SAS Institute, Cary, NC). Response variables shown to have a significant effect by treatment group were subjected to comparisons between MN07 and IA04 groups using the Student’s t test.
Results
Sequence analysis
All the eight genes from the IL05, NC05, IL07, and MN07 viruses were sequenced, and the sequences have been deposited in GenBank database under accession numbers FJ611895–FJ611902 and FJ638295–FJ638318. The sequences generated from this study were then compared to the hu-like H1 viruses isolated from pigs in Canada, described by Karasin et al. [16], and other influenza A viruses published in GenBank. Multiple alignment, pairwise analysis, and phylogenetic analysis were performed. Based on the nucleotide analysis, NC05, IL05, and MN07 are H1N1 SIVs and IL07 is an H1N2 SIV, all contain human lineage HA and NA. The HA genes of IL05, NC05, IL07, and MN07 differed by 0–5% at the nucleotide level when compared to the 2003 and 2004 hu-like H1 SIV from Canada. The HA gene of the H1N2 IL07 was genetically more similar to the Canadian hu-like swine isolates with only 2–3% difference at the nucleotide level. Phylogenetic analysis of the HA gene indicated two distinct clusters of hu-like swine H1 viruses, with the Canadian isolates and IL07 forming one cluster and the three remaining U.S. isolates, IL05, NC05, and MN07, forming a separate cluster (Fig. 1a).
Comparison of the amino acid sequences of the hu-like H1 HA proteins with swine-lineage H1 HAs revealed an amino acid deletion at position 130 of the mature hu-like HA. There were numerous amino acid differences between hu-like H1 and swine H1 at putative antigenic sites [21]. A substitution between swine lineage H1 HA (133T) and hu-like H1 HA (133S) was identified at a human host restrictive residue [22], with all the hu-like H1 SIV isolates studied here maintaining the hu-like 133S (H1 numbering with 1 being the first amino acid after the signal sequence). NC05 and the Canadian hu-like HAs contained 183P, whereas IL05, IL07, and MN07 contained 183S. The P183S substitution was reported as a human host-range restrictive residue [22]; however, swine lineage H1 HA contained either an S or P at this position. Position 183 has also been identified as a residue in the phylogenetically important region (PIR) I of H1 viruses [23]. PIRs, designated A–S, differentiate viruses of avian, human, or swine lineage, and amino acid changes within PIRs in the hu-like H1 HA may suggest adaptation. In contrast to the differences at PIRs identified by Karasin and colleagues in the Canadian hu-like SIV [16], amino acid positions 94, 128, and 141 remained hu-like in the HA of the U.S. hu-like H1 viruses. However, an A96T substitution was noted between the human-lineage H1 reference sequences and NC05, IL05, and MN07 in PIR D. Changes in host-restrictive sites and PIR are summarized in Table 1.
The NA genes from NC05, IL05, and MN07 formed a cluster of N1 genes distinct from N1 genes of swine origin and were most similar to human H1N1 from 2002 to 2003, whereas the NA of IL07 had hu-like N2 (Fig. 1b). The N1 genes of NC05, IL05, and MN07 displayed 0–1% nucleotide difference from each other and 1–2% from the closest human N1 genes. When compared to the N1 genes from the human isolates referenced in Fig. 1b, several deduced amino acid changes were noted in the N1 genes of the hu-like SIV, suggesting possible adaptation to swine host cells (Table 1). Two changes (K329R and S367I) were present in all three of the hu-like SIV N1s and were located in previously described PIRs, PIRs I, and L, respectively, of the N1 regions designated A–O [24]. In addition, the MN07 N1 contained G77R and A86V substitutions in PIR C and V389M in PIR M. The NA gene of IL07 H1N2 was 6% divergent when compared to the H1N2 isolates from Canada, and 3–7% difference for contemporary U.S. H1N2 and H3N2 viruses at the nucleotide level. Similar to the N1 genes, there were several amino acid changes in the N2 gene of the IL07 isolate as compared to the reference virus of human origin in Fig. 1b. Three of these changes were found in previously described PIRs for N2, designated A′–L′: M51I in A′, S331R in F′, and R403M in K′.
The internal genes of the four U.S. isolates were similar to the composition of the contemporary swine TRIG (Fig. 2a–f), but differed significantly from the internal gene composition of the Canadian hu-like H1 viruses which were wholly human (A/Sw/ON/52156/03 H1N2) or human–swine double reassortants (A/Sw/ON/48235/04 H1N2 and A/Sw/ON/55383/04 H1N2). The PB2 genes of the U.S. isolates were of avian lineage, whereas the PB2 from the Canadian hu-like H1 viruses were of human or swine lineage (Fig. 2a). The U.S. PB2 genes differed from one another by 0–2% and from the Canadian isolates by 16%. The PB1 genes of the U.S. and Canadian isolates were of H3 human lineage, although the U.S. isolates tended to be more related to one another (0–5% difference) than to the Canadian PB1 viral genes (4–6% difference). Gene segment 2 from IL05, NC05, and MN07 also encoded 57 amino acid truncated forms of the PB1-F2 protein, whereas the IL07 PB1 encoded the full-length 90 amino acid PB1-F2. The PA genes of the U.S. isolates were of avian lineage whereas the Canadian PA genes were of human H3 or swine lineage. The U.S. PA genes differed from one another by 0–3% and from the Canadian isolates by 17–19%. The NP genes of the U.S. isolates were of swine origin and differed by 0–4%. Two of the Canadian isolates also had NP genes of swine origin, but differed by 8–9% from the U.S. NP genes. The wholly human Canadian virus isolate (A/Sw/ON/52156/03) had an NP gene of human H3 origin and differed by 17–18% from all other isolates. The M genes of the U.S. isolates were of swine origin and differed from each other by 0–2%. Two of the Canadian M genes were also of swine virus origin, but differed from the U.S. M genes by 7–8%. The wholly human Canadian virus M gene was of human H3 virus origin and differed by 11–12% from the other human-like swine isolates. The NS genes of the U.S. isolates were of swine virus origin and differed by 0–6% from one another. Two of the Canadian NS genes were also of swine virus origin, but differed from the U.S. NS genes by 8–9%. The wholly human Canadian virus NS gene was of human H3 virus origin and differed by 17–19% from the other isolates under comparison.
In vivo pathogenesis and transmission
All the pigs were seronegative for specific antibodies to SIV by the HI assay and PRRSV by ELISA prior to the start of the study. The kinetics of virus replication, pathogenesis, as well as host humoral immune response differed between the two viruses. Pigs inoculated with MN07 virus demonstrated mild clinical illness similar in effect to the IA04-infected pigs. Necropsy revealed mild macroscopic lung lesions typical of SIV (purple-red colored, consolidated areas) in inoculated pigs, but none in control pigs. Macroscopic lung lesions for pigs infected with MN07 hu-like H1N1 averaged 14% on 3 dpi, 11% on 5 dpi, and 10% on 7 dpi, whereas the averages for pigs infected with IA04 were higher with 18% on 3 dpi, 24% on 5 dpi, and 15% on 7 dpi. Macroscopic lung lesions were significantly different at P < 0.05 between MN07 and IA04 groups on 5 and 7 dpi. Microscopic lung lesion scores (0–4) averaged 1.9 on 3 dpi, 2.5 on 5 dpi, and 2.9 on 7 dpi for MN07, and 3.5 on 3 dpi, 2.8 on 5 dpi, and 3.1 on 7 dpi for IA04, reflecting similar kinetics as the macroscopic lesions but differing in magnitude between the two viruses. No macroscopic or microscopic lung changes were observed in negative control pigs.
Virus titers in BALF averaged 106.4 TCID50/ml on 3 dpi, 104.0 TCID50/ml on 5 dpi, and were negative by 7 dpi in the MN07 inoculated group. In the IA04 inoculated group, virus titers in BALF averaged 106.9 TCID50/ml on 3 dpi, 106.1 TCID50/ml on 5 dpi, and were negative by 7 dpi. BALF titers were statistically different between the groups at 5 dpi. In the MN07 inoculated group, 77.7% of the pigs were shedding virus in nasal swab samples on 3 dpi, with an average titer of 103.2 TCID50/ml. On 5 dpi, 92% of nasal swabs were positive with an average titer of 102.9 TCID50/ml. All of the nasal swab samples from MN07 inoculated pigs were negative on 7 dpi. In comparison, the IA04 inoculated group had significantly more virus in nasal swab samples on 3 and 5 dpi, with average titers of 104.4 and 104.2 TCID50/ml, respectively. In contrast to MN07, 50% of the IA04 primary infected pigs were still shedding on 7 dpi. Nasal shedding is summarized in Table 2.
Contact pigs demonstrated clinical illness similar to the primary challenge group, although somewhat more sporadic. The MN07 hu-like H1N1 virus was transmitted from the primary challenge pigs to all contact pigs, although viral titers were significantly less than in IA04 rH1N1 contact pigs on 3 and 5 dpc. In the MN07 contact group, 60% of the pigs were positive by nasal swab samples on 3 dpc, with an average of 101.0 TCID50/ml. On 5 dpc, 40% of the pigs were shedding virus with an average of 101.3 TCID50/ml. In contrast to the primary inoculated pigs where none were shedding detectable amounts of virus, 80% of the MN07 contact pigs continued to shed virus on 7 dpc, with an average titer of 101.3 TCID50/ml. The IA04 virus demonstrated more rapid transmission and shedding in the contact pigs with 100% shedding on 3 dpc with an average of 103.4 TCID50/ml and 100% shedding on 5 dpc with an average titer of 103.9 TCID50/ml. On 7 dpc, 90% of the IA04 contact pigs continued to shed virus, with an average titer of 101.3 TCID50/ml.
Serologic responses
The kinetics of the humoral immune response also differed between the recently emerged MN07 hu-like H1N1 and the IA04 rH1N1. Three of the three remaining MN07 primary inoculated pigs were seropositive by the HI assay at 14 dpi, with a geometric mean reciprocal titer of 260. In contrast, only three of the five remaining IA04 inoculated pigs were seropositive at 14 dpi with a geometric mean reciprocal titer of 23. All of the MN07 contact pigs (N = 10) were seropositive by 14 dpc, with a geometric mean reciprocal titer of 171, demonstrating efficient transmission by contact, despite the relatively low nasal swab virus titers detected in that group. However, only 80% of the IA04 contact pigs were seropositive by 14 dpc, with a much lower geometric mean reciprocal titer of 30, even though all the contact pigs were shown to have high levels of virus by nasal swab as early as 3 dpc. By 27 dpi for the primary pigs and 24 dpc for the contact pigs, all had seroconverted to the respective challenge virus, but the geometric mean homologous HI titers for the IA04 groups remained six- to ninefold lower than the MN07 groups.
Cross-HI demonstrated little cross-reactivity between swine lineage H1-antisera [6] and the 4 U.S. hu-like H1 viruses (Table 3). In addition, there was limited cross-reactivity between the MN07 antiserum and the other three U.S. swine viruses of human H1 lineage, with an eightfold or greater reduction in HI titer between the homologous and heterologous reactions. The reduction in cross-reactivity was consistent with amino acid changes at putative antigenic determinant sites [21] in the HA peptide demonstrated between the four U.S. hu-like H1 viruses. The reduction in cross-reactivity suggests that not only are there two genetic clusters of hu-like H1 viruses in North American swine, but also antigenic drift within a cluster has likely occurred since introduction into the U.S. swine population.
Discussion
Human H1N2 and human–swine reassortant H1N2 were first isolated from pigs in Canada in 2003 and 2004 [16]. Millions of hogs are imported into the U.S. each year from Canada for pork processing. In addition, millions of hogs are transported within the U.S. due to the integrated market in the North American swine industry. This movement provides a possible route of introduction and/or transmission of newly emerging pathogens to U.S. swine. In 2005, hu-like H1 viruses began to be identified in U.S. swine (M.R. Gramer, unpublished). However, to date, full genome sequence analysis has neither been reported on U.S. isolates, nor have these hu-like H1 SIV isolates been evaluated experimentally in the pig. Full genome sequencing revealed substantial differences between the U.S. isolates and those first isolated in Canada. The four isolates evaluated here are triple reassortant SIV, with genes from human, avian, and swine lineage, with a TRIG constellation similar to that found in contemporary North American SIV [5]. The HA genes of three of the U.S. isolates form a separate phylogenetic cluster from the original three Canadian hu-like H1 isolates, suggesting two introduction events. These same three isolates also differed in that they contain hu-like N1 rather than N2. The hu-like N1 are phylogenetically distinct from N1 of swine origin. The internal gene constellation also differed between the U.S. and Canadian isolates, with clear distinction in lineages for the PA and PB2 genes. And although the PB1, NP, M, and NS genes were of similar lineages, separate clusters divided the U.S. and Canadian hu-like viruses. Further evaluation is required to determine if distinct populations of hu-like viruses continue to co-circulate in geographic regions of the North American swine herds.
Deduced amino acid differences were identified in the HA genes of the four U.S. hu-like isolates when compared to HA of swine lineage, as well as a limited number of changes among viruses within the hu-like HA genetic cluster. Amino acid changes were demonstrated in putative antigenic determinant sites [21], consistent with the loss in cross-reactivity among the hu-like SIV antigens. The limited cross-reactivity with anti-sera from swine lineage H1 viruses suggests that despite circulation of swine lineage H1 in the U.S. swine population, a lack of herd immunity to the hu-like H1 is likely. Reduced cross-reactivity was demonstrated among the four U.S. hu-like isolates, as well, further complicating strategies to protect the swine population from H1 influenza virus infection. To date, only one commercially available SIV vaccine contains antigens from this newly emerged cluster of hu-like H1 SIV. The implementation of this vaccine as a control for hu-like SIV remains largely unknown since it has only recently become available.
The MN07 hu-like H1N1 isolate was chosen to represent the U.S. cluster of hu-like H1 SIVs to be evaluated in an in vivo challenge and transmission model. Our results indicate that the MN07 virus was pathogenic in pigs and was transmissible among pigs. The MN07 virus induced macroscopic and microscopic lesions that are indistinguishable from those induced by contemporary SIV; however, the clinical signs and lesions were reduced in severity as compared to the IA04 rH1N1 challenge in this study. The IA04 chosen to represent the swine-lineage H1 viruses had been previously evaluated in pigs and was shown to induce consistent lung pathology and nasal shedding [6]. The MN07 virus was transmitted from inoculated pigs to contact pigs as demonstrated by nasal swab virus isolation and seroconversion. Contact pigs that were infected via natural transmission routes shed virus longer than the intratracheally inoculated pigs. Contact pigs began to shed virus as early as 3 dpc, and 8 out of 10 continued to shed for as long as 7 dpc. Nasal swabs were not taken after day 7 postcontact; therefore, an endpoint was not reached for nasal shedding in the contact pigs. Both the hu-like MN07 infected and contact pigs shed lower titers of virus than the IA04 rH1N1 groups, suggesting that although this hu-like H1N1 is successful in transmission, it may not replicate as efficiently as contemporary swine-adapted SIV. Likewise, although virus titers in the lung were similar on 3 dpi, by 5 dpi, the MN07 virus had declined significantly, whereas the IA04-infected pigs maintained high titers on 3 and 5 dpi. The humoral responses were inversely related to the differences in virus titers between MN07 and IA04. Pigs in this study mounted a more robust humoral response to the MN07 hu-like H1N1, both in kinetics and magnitude, as compared to the IA04 rH1N1. This is likely related to the interplay between virulence properties of the virus and host factors such as receptor distribution in the respiratory tract and/or early innate immune response. No differences were observed in hemagglutination of the turkey red blood cells by the IA04 or MN07 isolates to suggest the serologic response was an artifact of the HI assay.
A number of determinants have been proposed for barriers against interspecies transmission of influenza A viruses (reviewed in [29]). The binding of the HA protein to the host receptor has been demonstrated to be a major barrier. The putative sites important for receptor recognition have been described, with two amino acid residues having a fundamental role in avian and human species specificity of H1 viruses (E190 and G225 in avian viruses and D190 and D225 in human viruses, H3 numbering) [30]; however, D190 and D225 are conserved between human and swine H1 isolates. As predicted, these positions were demonstrated to be D190 and D225 in the HA of hu-like SIV isolates evaluated in this study. Other amino acid sites in the H1 HA thought to play a role in differential recognition of avian or human receptor types [22] were consistent between the hu-like SIV and the recent H1 SIV included in this evaluation. Changes within PIRs of the HA and NA genes suggest possible adaptation to the swine host; however, future work is required to monitor further changes as the hu-like H1 viruses circulate in the swine population.
The pig has been suggested to be a “mixing vessel” for influenza viruses with pandemic potential due to the presence of both avian and mammalian receptors located on the epithelial cells of the respiratory tract [31]. The 2009 pandemic H1N1 is evidence of the potential for viruses with genes from swine lineages to emerge in the human population [15]. Although the novel H1N1 has not been identified circulating endemically in swine, reassortants between the North American and Eurasian lineage swine viruses have been identified from pigs in China [32]. It is apparent that pigs may be infected at least transiently with wholly avian and/or human viruses, long enough for reassortment to occur and allowing swine viruses to acquire avian and/or human virus gene segments [2, 9, 16, 33–35]. Further supporting the occurrence of such events is our recent identification of a swine and avian reassortant H2N3 virus [13], an HA subtype not seen in the human population since the 1957 H2N2 pandemic virus disappeared in 1968. To our knowledge, no reports of human illness associated with these hu-like H1 SIVs have been made. A limited number of human infections with contemporary triple reassortant SIV with swine lineage H1 have been reported [36], and most of these cases had documented exposure to swine with little evidence for person to person transmission. The ability of the hu-like triple reassortant SIV to infect humans with subsequent person to person transmission is unknown. The limited genetic change in the HA genes between the hu-like SIV and the reference human viruses suggests that these triple reassortant swine viruses would likely react with reference human anti-sera or be detected by molecular diagnostic assays directed against human lineage HA. Thus, these emerging hu-like SIV may be overlooked in routine seasonal influenza monitoring by human health laboratories.
The internal gene cassette from the swine triple reassortant viruses found in North America seems to have lent contemporary SIV an ability to acquire new HA and NA genes as well as an increased rate of antigenic drift. This was not previously observed with classical swine H1N1 viruses. The potential for a successful swine to human jump of contemporary North American SIV is unknown, but the rapid rate at which SIVs in North America are evolving and emerging potentially increases the risk. The 2009 pandemic H1N1 underscores the potential risk to the human population of other influenza virus subtypes and genotypes on the SIV TRIG backbone. Increased surveillance and monitoring of the hu-like H1 as well as other SIV in both the swine and human populations are warranted to better understand this risk.
References
K.P. Myers, C.W. Olsen, G.C. Gray, Clin. Infect. Dis. 44, 1084–1088 (2007)
N.N. Zhou, D.A. Senne, J.S. Landgraf, S.L. Swenson, G. Erickson, K. Rossow, L. Liu, K. Yoon, S. Krauss, R.G. Webster, J. Virol. 73, 8851–8856 (1999)
A.L. Vincent, W. Ma, K.M. Lager, B.H. Janke, J.A. Richt, Adv. Virus Res. 72, 127–154 (2008)
A.I. Karasin, J. Landgraf, S. Swenson, G. Erickson, S. Goyal, M. Woodruff, G. Scherba, G. Anderson, C.W. Olsen, J. Clin. Microbiol. 40, 1073–1079 (2002)
R.J. Webby, K. Rossow, G. Erickson, Y. Sims, R. Webster, Virus Res. 103, 67–73 (2004)
A.L. Vincent, K.M. Lager, W. Ma, P. Lekcharoensuk, M.R. Gramer, C. Loiacono, J.A. Richt, Vet. Microbiol. 118, 212–222 (2006)
R.J. Webby, S.L. Swenson, S.L. Krauss, P.J. Gerrish, S.M. Goyal, R.G. Webster, J. Virol. 74, 8243–8251 (2000)
J.A. Richt, K.M. Lager, B.H. Janke, R.D. Woods, R.G. Webster, R.J. Webby, J. Clin. Microbiol. 41, 3198–3205 (2003)
C.W. Olsen, A.I. Karasin, S. Carman, Y. Li, N. Bastien, D. Ojkic, D. Alves, G. Charbonneau, B.M. Henning, D.E. Low, L. Burton, G. Broukhanski, Emerg. Infect. Dis. 12, 1132–1135 (2006)
Y.K. Choi, S.M. Goyal, M.W. Farnham, H.S. Joo, Virus Res. 87, 173–179 (2002)
W. Ma, M. Gramer, K. Rossow, K.J. Yoon, J. Virol. 80, 5092–5096 (2006)
P. Lekcharoensuk, K.M. Lager, R. Vemulapalli, M. Woodruff, A.L. Vincent, J.A. Richt, Emerg. Infect. Dis. 12, 787–794 (2006)
W. Ma, A.L. Vincent, M.R. Gramer, C.B. Brockwell, K.M. Lager, B.H. Janke, P.C. Gauger, D.P. Patnayak, R.J. Webby, J.A. Richt, Proc. Natl Acad. Sci. USA 104, 20949–20954 (2007)
F.S. Dawood, S. Jain, L. Finelli, M.W. Shaw, S. Lindstrom, R.J. Garten, L.V. Gubareva, X. Xu, C.B. Bridges, T.M. Uyeki, N. Engl. J. Med. 360, 2605–2615 (2009)
R.J. Garten, C.T. Davis, C.A. Russell, B. Shu, S. Lindstrom, A. Balish, W.M. Sessions, X. Xu, E. Skepner, V. Deyde, M. Okomo-Adhiambo, L. Gubareva, J. Barnes, C.B. Smith, S.L. Emery, M.J. Hillman, P. Rivailler, J. Smagala, M. de Graaf, D.F. Burke, R.A. Fouchier, C. Pappas, C.M. Alpuche-Aranda, H. Lopez-Gatell, H. Olivera, I. Lopez, C.A. Myers, D. Faix, P.J. Blair, C. Yu, K.M. Keene, P.D. Dotson Jr., D. Boxrud, A.R. Sambol, S.H. Abid, K. St. George, T. Bannerman, A.L. Moore, D.J. Stringer, P. Blevins, G.J. Demmler-Harrison, M. Ginsberg, P. Kriner, S. Waterman, S. Smole, H.F. Guevara, E.A. Belongia, P.A. Clark, S.T. Beatrice, R. Donis, J. Katz, L. Finelli, C.B. Bridges, M. Shaw, D.B. Jernigan, T.M. Uyeki, D.J. Smith, A.I. Klimov, N.J. Cox, Science (2009). doi:https://doi.org/10.1126/science.1176225
A.I. Karasin, S. Carman, C.W. Olsen, J. Clin. Microbiol. 44, 1123–1126 (2006)
M.R. Gramer, SIV: An update on circulating strains, advances in diagnostic tests and interpretation of test results, in 38th Annual Meeting of the American Association of Swine Veterinarians, Orlando, FL, 2007
C.H. Chan, K.L. Lin, Y. Chan, Y.L. Wang, Y.T. Chi, H.L. Tu, H.K. Shieh, W.T. Liu, J. Virol. Methods 136, 38–43 (2006)
E. Hoffmann, J. Stech, Y. Guan, R.G. Webster, D.R. Perez, Arch. Virol. 146, 2275–2289 (2001)
K. Tamura, J. Dudley, M. Nei, S. Kumar, Mol. Biol. Evol. 24, 1596–1599 (2007)
A.J. Caton, G.G. Brownlee, J.W. Yewdell, W. Gerhard, Cell 31, 417–427 (1982)
M.N. Matrosovich, A.S. Gambaryan, S. Teneberg, V.E. Piskarev, S.S. Yamnikova, D.K. Lvov, J.S. Robertson, K.A. Karlsson, Virology 233, 224–234 (1997)
T.G. Fanning, J.K. Taubenberger, Virus Res. 65, 33–42 (1999)
T.G. Fanning, A.H. Reid, J.K. Taubenberger, Virology 276, 417–423 (2000)
P. Kitikoon, D. Nilubol, B.J. Erickson, B.H. Janke, T. Hoover, S. Sornsen, E.L. Thacker, Vet. Immunol. Immunopathol. 112, 117–128 (2006)
L.J. Reed, H. Muench, Am. J. Hyg. 27, 493–497 (1938)
P.G. Halbur, P.S. Paul, M.L. Frey, J. Landgraf, K. Eernisse, X.J. Meng, M.A. Lum, J.J. Andrews, J.A. Rathje, Vet. Pathol. 32, 648–660 (1995)
D.F. Palmer, M.T. Coleman, W.R. Dowdle, G.C. Schild, Advanced Laboratory Techniques for Influenza Diagnosis. Immunology Series No. 6 (U. S. Department of Health, Education and Welfare, Washington, DC, 1975), pp. 51–52
G.A. Landolt, C.W. Olsen, Anim. Health Res. Rev. 8, 1–21 (2007)
M. Matrosovich, A. Tuzikov, N. Bovin, A. Gambaryan, A. Klimov, M.R. Castrucci, I. Donatelli, Y. Kawaoka, J. Virol. 74, 8502–8512 (2000)
T. Ito, J.N. Couceiro, S. Kelm, L.G. Baum, S. Krauss, M.R. Castrucci, I. Donatelli, H. Kida, J.C. Paulson, R.G. Webster, Y. Kawaoka, J. Virol. 72, 7367–7373 (1998)
G.J.D. Smith, D. Vijaykrishna, J. Bahl, S.J. Lycett, M. Worobey, O.G. Pybus, S.K. Ma, C.L. Cheung, J. Raghwani, S. Bhatt, J.S.M. Peiris, Y. Guan, A. Rambaut, Nature 459, 1122–1125 (2009)
A.I. Karasin, K. West, S. Carman, C.W. Olsen, J. Clin. Microbiol. 42, 4349–4354 (2004)
C.W. Olsen, A. Karasin, G. Erickson, Virus Res. 93, 115–121 (2003)
H. Yu, G.H. Zhang, R.H. Hua, Q. Zhang, T.Q. Liu, M. Liao, G.Z. Tong, Biochem. Biophys. Res. Commun. 356, 91–96 (2007)
M.M.W.R. Morb, Mortal Wkly. Rep. 58, 400–402 (2009)
Acknowledgments
We thank the NADC sequencing facility, Matt Kappes, Michelle Harland, Deb Adolphson, Brian Pottebaum, and Jason Huegel for assistance with laboratory techniques and animal studies and Dr. Brian Brunelle for critical review of the manuscript. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Vincent, A.L., Ma, W., Lager, K.M. et al. Characterization of a newly emerged genetic cluster of H1N1 and H1N2 swine influenza virus in the United States. Virus Genes 39, 176–185 (2009). https://doi.org/10.1007/s11262-009-0386-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-009-0386-6