Abstract
Chicken anaemia virus (CAV) was detected by a Nested-PCR assay in field samples from different regions of Brazil. The 539 bp amplified fragments of vp1 gene from 44 field samples were sequenced and 10 new nucleotide sequences of CAV were observed. These sequences were phylogenetically analysed by Mega2 using neighbour joining distance methods with 1000 bootstrap replications. Phylogenetic analysis did not show correlation between CAV pathology pattern and genetic groups. The 10 nucleotide sequences of the Brazilian samples were also analysed together with 30 sequences of CAV strains previously described from other countries. The genetic variability observed was not related to the geographical distribution. Amino acid substitutions were detected at 9 positions of the Brazilian sequences and two of them had not been observed before, 65R replacing the Q residue and 98F replacing Y residue.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chicken anaemia virus (CAV) is a member of the Circoviridae family, Gyrovirus genus [1] and was first described in 1979 by Yuasa and colleagues in Japan [2]. CAV infections induce either clinical or sub-clinical signs [3] and both result in measurable economical. losses [4–6]. Signs and lesions include stunting, increased mortality, anaemia, bone marrow cell depletion, subcutaneous haemorrhage, and atrophy of secondary lymphatic organs [7]. This infection is often associated with secondary bacterial, viral infections or vaccination failures [8–11]. CAV can be vertically transmitted by either dam or sire to progeny [3, 12] and the transmission is greatly reduced but not entirely eliminated in immune breeders [13]. CAV has been detected worldwide by isolation, serology or DNA amplification in both laying and broiler chickens. The presence of CAV has been investigated in cell culture, sera and tissues by polymerase chain reaction (PCR) [13–17]. The CAV genome structure is composed by a circular ssDNA of about 2.3 kb with three open reading frames (ORFs) coding for proteins of 52 (VP1), 24 (VP2) and 14 (VP3) kDa [18].
CAV was known as a very conserved virus of one serotype [2] with several genetic groups [19], although an additional serotype has been recently reported [20, 21]. Renshaw et al. [22] identified an hypervariable region in VP1 and suggested that certain amino acid changes in this region could influence the rate of virus replication and spread of CAV strains in MDCC-MSB-1 cells. Recently, Chowdhury et al. [23] reported that low and high passage isolates were phylogenetically divergent and differed in their pathogenicities, but it would not accurately predict the phylogeny of the virus and rather it is more indicative of selection of a particular isolate in the cell culture.
Little is known about the phylogenetic analysis of CAV Brazilian samples. In the present work, we report the amplification of a vp1 fragment of the CAV from Brazilian field samples. The amplification products were sequenced and submitted to phylogenetic analysis by neighbour joining distance methods to evaluate genetic diversity and a possible correlation with their induced pathology.
Material and methods
Samples
One hundred and fifty field samples from commercial breeders, broilers and free-range chickens (originated by random crosses between different breeds introduced in Brazil following colonization, around 1500) [24] were analysed. They were selected from different states of Brazil and were collected from the year 2000 until 2004. Tissue samples (80), sera (55) and litter (15) were collected from flocks with or without clinical signs of chicken anaemia. Also, samples from flocks with diagnosis of infectious bursal disease (IBD), Pasturella multocida (PM) infectious laryngotracheitis virus (ILTV) and Ornithobacterium rhinotracheale (ORT) were tested (Table 1). A CAV commercial vaccine (Nobilis CAV P4, Intervet International, Netherlands) was also sequenced.
DNA amplification and sequencing
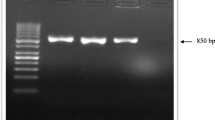
The DNA extraction methods, primers and cycling conditions for the Nested-PCR assay were performed as described previously [25]. The 539 bp amplification products were purified with GFX PCR DNA and Gel Band Purification Kit (Amersham Biosciences, Sweden). Both strands were sequenced using the Big Dye Terminator version 3.1 cycle sequencing RR-100 kit on an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, USA).
Sequence analysis
The obtained nucleotide sequences and the deduced amino acid sequences were aligned using Clustal W [26]. GeneDoc software [27] was used to measure similarity degrees. Kimura 2-parameter pairwise distances [28], calculated for the different nucleotide sequences detected in Brazil were used to construct a phylogenetic unrooted tree by using the neighbour joining distance methods in the Molecular Evolutionary Genetics Analysis software MEGA 2 [29]. The statistical confidence of the tree topologies was performed by 1000 bootstrap replications on the same software.
In order to compare with the Brazilian samples in the phylogenetic studies (using the same analysis method described above), we detected seventy five CAV related sequences in GenBank, comprising the same genome region analysed in the present work and thirty of these sequences were selected to represent the diversity of CAV worldwide.
Results
From the 150 field samples analysed, 135 were Nested-PCR positive and from which 44 samples were selected for nucleotide sequencing representing flocks from different geographical regions. From the 539 bp amplification product sequenced, 481 nucleotides were further analysed. Twenty-three (52%) samples sequenced displayed more than one sequence in the same sample, including all CAV DNA sequences from São Paulo (8) and Minas Gerais (2) States. The remaining unambiguous 21 sequences (48%) were aligned and 10 different sequences (Table 1) with 92.0–99.0% similarity were found. Five samples detected in clinically healthy flocks had identical sequences and are represented here by GenBank accession number AY855079, which showed 100% identity with the amplification product of the commercial vaccine.
Phylogenetic analysis of Brazilian samples was performed to verify a possible relation of CAV induced pathology with a specific genetic group. Phylogenetic tree showed three groups (Fig. 1). The first (I) was formed by four sequences (GenBank accession numbers: AY855079, AY855084, AY855085 and AY855086) detected in clinically healthy flocks and litter. The second group (II) was formed by two sequences (GenBank accession numbers: AY855080 and AY855081) detected in flocks with ORT antibodies. The third group (III) was composed by four sequences (GenBank accession numbers: AY855082, AY855083, AY855087 and AY855088). The sequence AY855082 shared 99% similarity with AY855088 and both were detected in flocks displaying CAV clinical signs. Seven other samples had identical sequence to AY855082 and were detected in flocks with diagnosis attributed to CAV, PM, ILTV or with ORT antibodies. AY855003 was obtained from a clinically healthy flock and AY855087 was from a flock with IBD. These ten CAV Brazilian sequences did not show relation between genetic group and the state of origin.
Brazilian CAV nucleotide sequences were compared with the 30 other CAV sequences deposited in the database to know if there was any geographical relation worldwide (Fig. 2). Phylogenetic trees constructed by using maximum parsimony or maximum likelihood methods also supported the same clustering as well as the bootstrap values (data not shown).
The alignment from the deduced VP1 amino acid sequences of the 10 Brazilian samples showed amino acid substitutions in nine different positions. Two of them presented amino acid substitution 65R replacing the Q residue in AY855088 and 98F replacing the Y residue in AV855085 (Fig. 3). AY855088 and AY855085 sequences were obtained from a free-range chicken with severe chicken anaemia symptoms and a commercial breeder flock with IBD, respectively.
Discussion
The 539 nucleotide sequences of the 2.3 kb CAV genome from commercial broiler and hens and free-range chickens from Brazil were obtained. The DNA samples were extracted from biological specimens without virus isolation in cell culture. This procedure enables us to characterize CAV that might not have replicated in vitro [19] and avoided the potential nucleotide changes that can occur during cell culture passage. The analysis of the nucleotide sequences obtained from the vp1 amplification product of field samples showed that 52% of these had more than one sequence. Probably, these chickens were infected by more than one strain, or the different sequences constitute CAV quasispecies. This finding is in agreement with the observations of van Saten et al. [30] who reported ambiguous sequences in PCR products. Claessens et al. [31] also reported nucleotide differences in two clones obtained from the same sample.
Phylogenetic analysis of the 10 nucleotide sequences described in the present work displayed three groups. It was not observed any relationship between CAV pathology and the genetic group. Indeed, the severity of CAV disease is also influenced by the presence of other infectious pathogens. Interestingly, the AY855079 sequence, detected in clinically healthy flocks showed higher divergence with AY855088, detected in chicken with clinical signs of chicken anaemia in phylogenetic tree. These results were similar to an earlier study that indicated that isolates with different pathogenicities were also phylogenetically divergent [23].
The phylogenetic tree of Brazilian sequences and the 30 CAV described did not show a clear geographical correlation. Islam et al. [19] also reported that an Australian strain did not demonstrate any grouping with strains detected in five different countries. Brazilian sequences are not all found on a separate group. Therefore, these sequences are not more similar to each other than they are to CAV isolates of other countries. This result was similar to the observations of van Santen et al. analysing strains from USA [30]. However, some Brazilian nucleotide sequences displayed greater homology with USA strains what can be explained by the fact that some commercial chicken lines and vaccines are usually imported from USA. Our results suggest that the diversity displayed was probably related to different origins of virus introduction more than its evolution inside Brazil.
Brazilian samples differed in 42 nucleotide positions that resulted in nine predicted amino acid substitutions. The VP1 alignment confirmed previous findings from Renshaw et al. [22] that described a hyper-variable region from amino acid positions 139 to 157. However, another variable region from amino acid positions 75 to 98 was also observed. Scott et al. [32] suggested that an amino acid substitution (T→A) at position 89 in the VP1 could be associated with the non-reactivity of one monoclonal antibody as well as the attenuation of some strains. We observed that the 10 Brazilian sequences had 89T. Interestingly, only two of these were present in broilers with clinical signs of chicken anaemia, indicating that other factors are probably involved in the induction of the clinical disease. This discrepancy could be attributed to the use of clones sequenced from high passage viruses [32].
Seven out of the 10 Brazilian sequences had 75I, 97L, 139Q and 144Q and three had 75V, 97M, 139K, and 144E amino acid profile. These findings are in agreement with an earlier study of 14 CAV strains detected in 10 clinical samples obtained from commercial broiler chickens in Alabama State, USA [30]. The amino acids at positions 139 and 144 did not appear to be independent of each other, since a 139Q is always followed by a 144Q and a 139K is always accompanied by a 144E.
Renshaw et al. [22] suggested that 139Q and/or 144Q can influence the rate of replication or spread of infection in MDCC-MSB-1 cells. Seven out of the 10 Brazilian sequences were 139Q and 144Q, however, only two of them, were detected in flocks with CAV clinical signs. It was also verified that the three sequences that were 75V, 97M, 139K and 144E amino acid positions, were detected in flocks without CAV clinical signs and these residues could be associated with attenuated viruses. The AY855088 sequence, detected in broilers with severe signs of chicken anaemia presented amino acid change Q→R in position 65 not observed before, which could be related to the pathogenicity of CAV strains. These questions will require more investigations to confirm correlations between pathogenicity and genetic determinants. The knowledge about these differences and their pathogenic potential may be useful in the future development of vaccines. However, sequence analysis and biological studies require careful interpretation concerning the mutation in the CAV genome.
In conclusion, when the Brazilian sequences were analysed alone, they formed three genetic groups that were not clustered when these sequences were analysed together with sequences from all over the world. Also, there was no clear relation between pathology and the genetic groups. The results obtained add new and important information about genetic variability of VP1 in Brazil and its putative correlation with CAV pathogenesis.
References
C.R. Pringle (1999) Arch. Virol. 144 2065–2070 Occurrence Handle10550679 Occurrence Handle1:CAS:528:DyaK1MXotFSlurg%3D Occurrence Handle10.1007/s007050050728
N. Yuasa T. Taniguchi I. Yoshida (1979) Avian. Dis. 23 366–385 Occurrence Handle10.2307/1589567
K.A. Schat, in Diseases of Poultry, ed. by Y.M. Saif, H.J. Barnes, A.M. Fadly, J.R. Clisson, L.R. Mcdougald, D.E. Swayne (Iowa State University, 2003), pp.182–202
M.S. McNulty S.G. Mcllroy D.W. Bruce D. Todd (1991) Avian. Dis. 35 263–268 Occurrence Handle1854312 Occurrence Handle1:STN:280:By6A3cjks1E%3D Occurrence Handle10.2307/1591175
S.G. McIlroy M.S. McNulty D.W. Bruce J.A. Smyth E.A. Goodall M.J. Alcorn (1992) Avian. Dis. 36 566–574 Occurrence Handle1417588 Occurrence Handle1:STN:280:ByyD3sfjt1w%3D Occurrence Handle10.2307/1591750
I. Davidson M. Kedem H. Borochovitz N. Kass G. Ayali E. Hamzani B. Perelman B. Smith S. Perk (2004) Avian. Dis. 48 108–118 Occurrence Handle15077804 Occurrence Handle1:STN:280:DC%2BD2c7ot1WltA%3D%3D
B.M. Adair (2000) Dev. Comp. Immunol. 24 247–255 Occurrence Handle10717291 Occurrence Handle1:CAS:528:DC%2BD3cXisleisLc%3D Occurrence Handle10.1016/S0145-305X(99)00076-2
S.S. Cloud J.K. Rosenberger H.S. Lillehoj (1992) Vet. Immunol. Immunop. 34 353–366 Occurrence Handle1:STN:280:ByyD1cflvFM%3D Occurrence Handle10.1016/0165-2427(92)90175-P
S.S. Cloud H.S. Lillehoj J.K. Rosenberger (1992) Vet. Immunol. Immunop. 34 337–352 Occurrence Handle1:STN:280:ByyD1cflvFI%3D Occurrence Handle10.1016/0165-2427(92)90174-O
A.M. Miles S.M. Reddy R.W. Morgan (2001) Avian. Dis. 45 9–18 Occurrence Handle11332504 Occurrence Handle10.2307/1593006
H. Toro O. González C. Escobar L. Cerda M.A. Morales C. Gonzeles (2001) Avian. Dis. 45 215–255 Occurrence Handle11336070 Occurrence Handle1:STN:280:DC%2BD3MrgsV2rug%3D%3D Occurrence Handle10.2307/1593031
R.K. Hoop (1992) Avian. Pathol. 21 493–501
C.J. Cardona W.B. Oswald K.A. Schat (2000) J. Gen. Virol. 81 2067–2075 Occurrence Handle10900046 Occurrence Handle1:CAS:528:DC%2BD3cXlsFOhur8%3D
D. Todd K.A. Mawhinney M.S. McNulty (1992) J. Clin. Microbiol. 30 1661–1666 Occurrence Handle1321165 Occurrence Handle1:CAS:528:DyaK38XkvVCgs7s%3D
K.M. Tham W.L. Stanislawek (1992) Avian. Dis. 36 1000–1006 Occurrence Handle1485846 Occurrence Handle1:STN:280:ByyC3srmvFc%3D Occurrence Handle10.2307/1591561
K. Imai M. Mase S. Yamaguchi N. Yuasa (1998) Res. Vet. Sci. 64 205–208 Occurrence Handle9690604 Occurrence Handle1:CAS:528:DyaK1cXlt1eltr4%3D Occurrence Handle10.1016/S0034-5288(98)90126-6
H. Yilmaz N. Turan N.Y. Ozgur C.R. Helps O. Akay (2001) Avian. Dis. 45 529–533 Occurrence Handle11417840 Occurrence Handle1:STN:280:DC%2BD38%2FhsVCqsA%3D%3D Occurrence Handle10.2307/1593000
M.H.M. Noteborn G.F. Boer ParticleDe D.J. Roozelaar ParticleVan C. Karreran O. Kranenburg J.G. Vos S.H.M. Jeurissen R.C. Hoeben A. Zantema G. Koch G.H. Ormondt Particlevan A.J. Eb Particlevan Der (1991) J. Virol. 65 3131–1139 Occurrence Handle1851873 Occurrence Handle1:CAS:528:DyaK38XjsFantA%3D%3D
M.R. Islam R. Johne R. Raue D. Todd H. Müller (2002) J. Vet. Med. B 49 332–337 Occurrence Handle1:CAS:528:DC%2BD38XpsVCisb0%3D Occurrence Handle10.1046/j.1439-0450.2002.00581.x
E. Spackman S.S. Cloud C.R. Pope J.K. Rosenberger (2002) Avian. Dis. 46 945–955 Occurrence Handle12495056 Occurrence Handle1:STN:280:DC%2BD38jjt1aitA%3D%3D
E. Spackman S.S. Cloud J.K. Rosenberger (2002) Avian. Dis. 46 956–663 Occurrence Handle12495057 Occurrence Handle1:STN:280:DC%2BD38jjt1aitQ%3D%3D
R.W. Renshaw C. Soiné T. Weinkle P.H. O’Connel K. Ohashi S. Watson B. Lucio S. Harrington K.A. Schat (1996) J. Virol. 70 8872–8878 Occurrence Handle8971016 Occurrence Handle1:CAS:528:DyaK28XmvVGqtL8%3D
S.M.Z.H. Chowdhury A.R. Omar I. Aini M. Hair-Bejo A.A. Jamaluddin BM. Md-Zain Y. Kono (2003) Arch. Virol. 148 2437–2448 Occurrence Handle14648297 Occurrence Handle1:CAS:528:DC%2BD3sXptleksrw%3D Occurrence Handle10.1007/s00705-003-0189-3
C.A.V. Lima-Rosa C.W. Canal A.F. Streck L.B. Freitas A. Delgado-Cañedo S.L. Bonatto F.M. Salzano (2004) Anim. Genet. 35 278–284 Occurrence Handle15265066 Occurrence Handle1:CAS:528:DC%2BD2cXntVGqtLo%3D Occurrence Handle10.1111/j.1365-2052.2004.01160.x
S. Simionatto C.A.V. Lima-Rosa L.L. Rubin C.W. Canal (2005) Pes. Vet. Bras. 25 112–118
J.D. Thompson D.G. Higgins T.J. Gibson (1994) Nucl. Acids. Res. 22 4673–4680 Occurrence Handle7984417 Occurrence Handle1:CAS:528:DyaK2MXitlSgu74%3D
K.B. Nicholas H.B. Nicholas SuffixJr. D.W.I.I. Deerfield (1997) Embnew News 4 14
M. Kimura (1980) J. Mol. Evol. 16 111–120 Occurrence Handle7463489 Occurrence Handle1:CAS:528:DyaL3MXmtFSktg%3D%3D Occurrence Handle10.1007/BF01731581
S. Kumar K. Tamura M. Nei (1993) MEGA: Molecular Evolutionary Genetics Analysis Pennsylvania State University University Park, PA
V.L. Saten Particlevan L. Li F.J. Hoerr L.H. Lauerman (2001) Avian. Dis. 45 373–388 Occurrence Handle10.2307/1592977
J.A.J. Claessens C.C. Schrier A.P.A. Mockett E.H.J.M. Jagt P.J.A. Sondermeijer (1991) J. Gen. Virol. 72 2003–2006 Occurrence Handle1908516 Occurrence Handle1:CAS:528:DyaK38XksVWktbY%3D Occurrence Handle10.1099/0022-1317-72-8-2003
A.N.J. Scott T.J. Connor J.L. Creeland M.S. McNulty (1999) Arch. Virol. 144 1961–1975 Occurrence Handle10550669 Occurrence Handle1:CAS:528:DyaK1MXotFSltbg%3D Occurrence Handle10.1007/s007050050718
Acknowledgements
We thank Brazilian chicken rearing companies for help in the collection of biological material. S. Simionatto received a scholarship from. Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). Financial support was provided by Fundação de Amparo a Pesquisa do Estado do Rio Grande do Sul (FAPERGS), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Pró-Reitoria de Pesquisa da Universidade Federal do Rio Grande do Sul (PROPESQ/UFRGS).
Author information
Authors and Affiliations
Corresponding author
Additional information
The nucleotide sequence data reported in this paper have been submitted to the GenBank nucleotide sequence database and have been assigned the accession numbers AY855079 to AY855088.
Rights and permissions
About this article
Cite this article
Simionatto, S., Lima-Rosa, C.A.d.V., Binneck, E. et al. Characterization and Phylogenetic Analysis of Brazilian Chicken Anaemia Virus. Virus Genes 33, 5–10 (2006). https://doi.org/10.1007/s11262-005-0033-9
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s11262-005-0033-9