Abstract
Background and objectives
Long noncoding RNAs (lncRNAs) play key roles in process of cancer cell growth and apoptosis and have received increasing attention. SChLAP1 is a novel lncRNA that is required for development and progression of prostate cancer. We hypothesized that SChLAP1 also has important biological functions in human bladder cancer which is another type of urological cancer.
Methods
The expression of SChLAP1 in bladder cancer was determined using real-time qPCR. Bladder cancer T24 and 5637 cells were transfected with SChLAP1 siRNA or negative control siRNA. Cell proliferation, apoptosis and migration were determined using CCK-8 assay, flow cytometry analysis and wound healing assay, respectively.
Results
SChLAP1 was overexpressed in bladder cancer tissues compared to paired normal bladder tissues. Cell growth arrest, apoptosis induction and migration inhibition were also observed in bladder cancer T24 and 5637 cells after transfection with SChLAP1 siRNA.
Conclusions
Our data suggest that SChLAP1 plays oncogenic roles and can be used as a therapeutic target for treating human bladder cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bladder cancer is one of the most common types of genitourinary tumors, and the major subtype of this cancer is urothelial carcinoma. Although many genes and pathways are found to be correlated with bladder cancer, its carcinogenesis is still far from clear [1, 2].
Long noncoding RNAs (lncRNAs) are a family of nonprotein coding RNAs with length more than 200 nucleotides. With the completion of human genome project, long noncoding RNAs (lncRNAs) have increasingly been discovered to be deregulated in many human cancers, including bladder cancer [3]. For example, MALAT1 [4], TUG1 [5], UCA1 [6] and HOTAIR [7] were reported to have affected the development of bladder cancer. These lncRNAs play critical roles in carcinogenesis and act as potential tumor promoters or tumor suppressors [8]. LncRNA SChLAP1 is a novel biomarker that predicts poor clinical outcome for prostate cancer [9–12]. However, the relationship between SChLAP1 and bladder cancer is largely unknown.
In this study, we found that SChLAP1 was overexpressed in bladder cancer tissues compared to matched normal bladder tissues. Silencing SChLAP1 inhibited bladder cancer growth or migration and induced apoptosis.
Materials and methods
Patient samples
We included 36 bladder cancer patients who received partial or radical cystectomy in the expression analyses, and the Institutional Review Board of The People’s Hospital of Xinjiang Uyghur autonomous region approved this study. We also got written informed consents from all patients. Bladder cancer tissues and paired normal bladder tissues from patients were snap-frozen in liquid nitrogen immediately after resection.
Cell culture
Bladder cancer T24 and 5637 cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). T24 cells were cultured in Dulbecco’s Modified Eagle’s Medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10 % fetal bovine serum (Invitrogen, Carlsbad, CA, USA) at 37 °C in a 5 % CO2 atmosphere. 5637 cells were cultured in RPMI-1640 Medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10 % fetal bovine serum (Invitrogen, Carlsbad, CA, USA) at 37 °C in a 5 % CO2 atmosphere.
siRNA transfection
SChLAP1 siRNA and negative control siRNA were purchased from Sigma-Aldrich. The sequence was 5′-CCAAUGAUGAGGAGCGGGA-3′ [13]. The cells were incubated with either SChLAP1 siRNA or negative control siRNA using Lipofectamine 2000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocols.
Real-time quantitative PCR
Total RNA was extracted from the tissue samples through using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the protocols. cDNA was synthesized from total RNA through using the M-MLV Reverse Transcriptase (Promega, USA) in a 25-μl volume. The primer sequences for SChLAP1 primers and GAPDH were described in the previous works [13]. qPCR was performed in a total reaction volume of 20 μl, which includes 1 μl of first-strand cDNA, 0.4 μl of forward primer, 0.4 μl of reverse primer, 10 μl of 2× All-in-One™ qPCR Mix (GeneCopoeia Inc, Rockville, MD, USA), 0.4 μl of 50× ROX reference dye and 7.8 μl of double-distilled water. The reactions were performed in triplicate by the ABI PRISM 7000 Fluorescent Quantitative PCR System (Applied Biosystems, Foster City, CA, USA). GAPDH was chosen as the internal control. Relative expression fold changes of SChLAP1 were calculated by using the comparative ΔC t method \(\left(\text{value of}\,2^{{- {\Delta }C_{t} \,({\text{SChLAP}}1 \hbox{-} {\text{GAPDH}})}}\right)\) [13]. These experiments were repeated three times.
Cell growth assay
Cell growth was detected using Cell Counting Kit-8, CCK-8 (Beyotime Institute of Biotechnology, Shanghai, China) according to instructions. Cells were grown in a 96-well plate for 24 h and cultured in normal medium. Then, cells were transfected with SChLAP1 siRNA or negative control siRNA. At 0, 24, 48 and 72 h after transfection, the absorbance in each well was measured at 490 nm with a microplate reader. These experiments were repeated three times.
Flow cytometry analysis of cell apoptosis
Cells were transfected with SChLAP1 siRNA or negative control siRNA and cultured in normal medium. At 48 h post-transfection, cells were harvested and resuspended in fixation fluid. Five microliters of AnnexinV-FITC and 2 μl propidium iodide were added to 500 μl cell suspension. Cell apoptosis was then determined by using flow cytometry analysis (BD, USA). These experiments were repeated three times.
Wound healing assay
Cell motility was determined by wound healing assay. At 24 h post-transfection, a wound field was created using a sterile 200 μl pipette tip in about 90 % confluent cells. The cells were incubated for 16 h at 37 °C, and then the migration of cells was monitored with a digital camera system. The cell migration distance (μm) was calculated by the software program HMIAS-2000. These experiments were repeated at least three times.
Statistical analysis
The SChLAP1 RNA expression changes between bladder cancer tissues and matched normal tissues were analyzed using paired samples t test. In the CCK-8 assay, flow cytometry analysis and wound healing assay, the differences between cells transfected with SChLAP1 siRNA and cells transfected with negative control siRNA were analyzed using ANOVA. All these statistical analyses were performed by using SPSS (version 17.0 SPSS Inc.). A p value of <0.05 was considered to be statistically significant.
Results
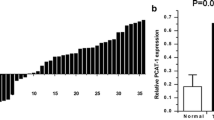
SChLAP1 was overexpressed in bladder cancer
The relative expression level of SChLAP1 was determined by using real-time qPCR in a total of 36 patients with bladder cancer. The SChLAP1 expression change between bladder cancer tissues and matched normal tissues is shown in Fig. 1. SChLAP1 was overexpressed in bladder cancer tissues compared to paired normal tissues (p = 0.002). These results indicated that SChLAP1 should function as an oncogenic gene in bladder cancer. Patients’ clinical parameters are listed in Table 1.
Silencing SChLAP1 inhibited cell growth
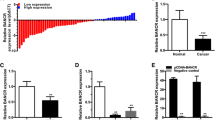
We further detected whether SChLAP1 promotes cell growth in bladder cancer. Bladder cancer T24 and 5637 cells were transfected with SChLAP1 siRNA or negative control siRNA. The inhibitory rate (SChLAP1 siRNA/negative control siRNA) was 85.42 ± 3.47 % in T24 cells and 86.12 ± 4.33 % in 5637 cells, respectively. Data were shown as mean ± SD. Then, the cell growth curves of bladder cells were determined by CCK-8 assay. Cell growth arrest was observed in both T24 cells (Fig. 2a) (p < 0.001) and 5637 cells (Fig. 2b) (p < 0.001) when transfected with SChLAP1 siRNA. These results demonstrated that SChLAP1 should increase cell growth in bladder cancer.
Silencing SChLAP1 inhibited cell growth. Cell proliferation was detected by CCK-8 assay. After transfection of SChLAP1 siRNA or negative control siRNA, OD values were measured. ANOVA was used for the comparison of curves of cell growth. a Cell growth arrest was observed in bladder cancer T24 cells (p < 0.01). b Cell growth arrest was observed in bladder cancer 5637 cells (p < 0.01). Data were shown as mean ± SE. Each experiment in both cell lines was performed in triplicate for three independent times
Silencing SChLAP1 induced apoptosis
After that, we asked the question of whether SChLAP1 can also suppress cell apoptosis in bladder cancer. Bladder cancer T24 and 5637 cells were transfected with SChLAP1 siRNA or negative control siRNA. At 48 h post-transfection, the cell apoptosis of both T24 (Fig. 3a) and 5637 cells (Fig. 3b) was determined by flow cytometry analysis. Induced cell apoptosis was observed in both T24 (p = 0.038) and 5637 cells (p = 0.014) transfected with SChLAP1 siRNA (Fig. 3c, d). These results confirmed that SChLAP1 inhibits cell apoptosis in bladder cancer.
Silencing SChLAP1 induced apoptosis. Forty-eight hours after transfection of SChLAP1 siRNA or negative control siRNA, the cell apoptosis changes were determined by flow cytometry analysis. a Representative images of flow cytometry analysis in T24 cells. b Representative images of flow cytometry analysis in 5637 cells. c Cell apoptosis induction was observed in bladder cancer T24 cells (p < 0.01). d Cell apoptosis induction was observed in bladder cancer 5637 cells (p < 0.01)
Silencing SChLAP1 inhibited cell migration
Finally, we determined whether SChLAP1 promotes cell migration in bladder cancer. Bladder cancer T24 and 5637 cells were transfected with SChLAP1 siRNA or negative control siRNA, and the cell migration changes of bladder cells were determined by wound healing assay. Cell migration arrest was observed in both T24 cells (Fig. 4a) (p = 0.022) and 5637 cells (Fig. 4b) (p = 0.014) as expected. These results confirmed that SChLAP1 increases cell migration in bladder cancer.
Discussion
As a noncoding gene, SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex [13]. SChLAP1 impairs SNF5-mediated gene expression regulation and genomic binding [13]. However, the relationship between SChLAP1 and bladder cancer is still unclear and mysterious.
This study provided novel insights into the function of SChLAP1 in bladder cancer. The first objective of this study was to test the hypothesis that the expression levels of SChLAP1 were up-regulated in most bladder cancer tissue samples. This hypothesis was confirmed by the qPCR results. We found that SChLAP1 was overexpressed in bladder cancer. A deregulated lncRNA should play a vital role in regulating cancer cell biological behavior. So the second objective of the study was to test the hypothesis that down-regulation of SChLAP1 inhibited cell proliferation or migration and promoted apoptosis. So we detected the cell growth, apoptosis and migration by silencing SChLAP1 in two bladder cancer cell lines. Cell growth arrest, increased apoptosis and suppressed migration were observed in bladder cells transfected with SChLAP1 siRNA. These findings suggested that SChLAP1 may play essential functions in the carcinogenesis of bladder cancer.
In summary, SChLAP1 plays a tumor promoter role in bladder cancer. More work will be needed to determine the molecular mechanisms of SChLAP1 as a candidate biomarker for bladder cancer in the clinic.
References
Egerod FL, Bartels A, Fristrup N, Borre M, Ørntoft TF, Oleksiewicz MB et al (2009) High frequency of tumor cells with nuclear Egr-1 protein expression in human bladder cancer is associated with disease progression. BMC Cancer 9:385
Rosser CJ, Urquidi V, Goodison S (2013) Urinary biomarkers of bladder cancer: an update and future perspectives. Biomark Med 7:779–790
Xue Y, Ma G, Zhang Z, Hua Q, Chu H, Tong N, Yuan L, Qin C, Yin C, Zhang Z, Wang M (2015) A novel antisense long noncoding RNA regulates the expression of MDC1 in bladder cancer. Oncotarget 6:484–493
Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F, Liu Y (2014) TGF-β-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin Cancer Res 20(6):1531–1541
Tan J, Qiu K, Li M, Liang Y (2015) Double-negative feedback loop between long non-coding RNA TUG1 and miR-145 promotes epithelial to mesenchymal transition and radioresistance in human bladder cancer cells. FEBS Lett 589(20 Pt B):3175–3181
Li HJ, Li X, Pang H, Pan JJ, Xie XJ, Chen W (2015) Long non-coding RNA UCA1 promotes glutamine metabolism by targeting miR-16 in human bladder cancer. Jpn J Clin Oncol 45(11):1055–1063
Yan TH, Lu SW, Huang YQ, Que GB, Chen JH, Chen YP, Zhang HB, Liang XL, Jiang JH (2014) Upregulation of the long noncoding RNA HOTAIR predicts recurrence in stage Ta/T1 bladder cancer. Tumour Biol 35(10):10249–10257
Batista PJ, Chang HY (2013) Long non-coding RNAs: cellular address codes in development and disease. Cell 152:1298–1307
Sahu A, Iyer MK, Prensner JR et al (2014) The role of long noncoding RNA SChLAP1 in prostate cancer. Cancer Res 74(19 Supplement):541
Prensner JR, Zhao S, Erho N et al (2014) RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. Lancet Oncol 15(13):1469–1480
Mehra R, Udager AM, Ahearn TU, Cao X, Feng FY, Loda M, Petimar JS, Kantoff P, Mucci LA, Chinnaiyan AM (2015) Overexpression of the long non-coding RNA SChLAP1 independently predicts lethal prostate cancer. Eur Urol. doi:10.1016/j.eururo.2015.12.003
Mehra R, Shi Y, Udager AM, Prensner JR, Sahu A, Iyer MK, Siddiqui J, Cao X, Wei J, Jiang H, Feng FY, Chinnaiyan AM (2014) A novel RNA in situ hybridization assay for the long noncoding RNA SChLAP1 predicts poor clinical outcome after radical prostatectomy in clinically localized prostate cancer. Neoplasia 16(12):1121–1127
Prensner JR, Iyer MK, Sahu A et al (2013) The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet 45(11):1392–1398
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Jianjun Zhang and Zhenfeng Shi have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Zhang, J., Shi, Z., Nan, Y. et al. Inhibiting malignant phenotypes of the bladder cancer cells by silencing long noncoding RNA SChLAP1. Int Urol Nephrol 48, 711–716 (2016). https://doi.org/10.1007/s11255-016-1230-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11255-016-1230-2