Abstract
Surra is a parasitic disease caused by Trypanosoma evansi and transmitted non-cyclically by biting flies. The disease significantly affects the health, productivity, and market value of camels thereby constituting a major constraint to food safety, security, and economy. This is the first study on the prevalence of surra in northwestern Nigeria, using a range of diagnostic tests along the parasitological-serological-molecular continuum hence, emphasizing it as a major enzootic risk for camels in Nigeria. In this cross-sectional study, 600 blood samples were collected from camels at major abattoirs in northwestern Nigeria and evaluated for the prevalence of T. evansi using parasitological (Giemsa staining), serological (CATT/T. evansi), and molecular (VSG-PCR and sequencing) methods. The overall prevalence of surra recorded in this study was 5.3%, 11.5%, and 22.5% using Giemsa-stained blood smears, CATT/T. evansi, and VSG-PCR respectively. However, higher prevalence rates at 6.0%, 13.7%, and 26.7% by Giemsa-stained blood smears, CATT/T. evansi, and VSG-PCR were recorded in Katsina State compared with results from Kano State. A significantly (p < 0.05) higher prevalence by VSG-PCR was observed when compared with both parasitological and serological methods used. Although age and body condition scores were associated (p < 0.05) with surra prevalence in sampled camels, no seasonal association (p > 0.05) was recorded. Sequencing of the VSG region of Trypanosoma spp. Further confirmed the presence of T. evansi as the aetiological agent of surra from the sampled camels. Findings from this study call for the implementation of adequate control measures aimed at reducing the impact of T. evansi infections on camel production in Nigeria.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Trypanosomosis in camels also referred to as “surra” is a vector-borne disease (VBD) that constitutes a major threat to farm animals in tropical and sub-tropical countries including Nigeria. The disease is caused by Trypanosoma evansi which belongs to the sub-genus Trypanozoon. Microscopically, the organism is long and slender with a prominent undulating membrane and long free flagellum (Getachew 2005). The disease is endemic in Africa, Asia, and South America and it is transmitted mechanically by the bites of hematophagous flies: Tabanids, Stomoxys, and Hippoboscids (Eyop and Matios 2013).
Trypanosomosis in camels is considered the most serious animal protozoan disease in African countries that depend on camels as an important source of food and income for millions of herders (Kamidi et al. 2017). The disease causes production losses, anaemia, weight loss, and abortion in a range of domestic species in Africa, Asia, and South America. Camel trypanosomosis is usually asymptomatic but can be fatal when not properly diagnosed, and treatment is initiated early in the course of the disease (Desquesnes et al. 2013).
Laboratory diagnosis of Surra is often recommended for the confirmation of infection (OIE 2012; Hassan-Kaddle et al. 2019). Although the standard trypanosome detection methods (STDMs) have been used over the years in the diagnosis of animal trypanosomosis, neither the parasitological nor the serological methods are sensitive and specific enough to differentiate between the various species of Trypanosoma in animals. Thus, various genetic, and molecular methods have been developed to overcome the limitation of STDMs (Barghash et al. 2016). To date, the molecular diagnostic techniques for the diagnosis of Trypanosoma infection offer better results. Furthermore, the increased sensitivity and specificity of the molecular techniques lies in the ability to detect all the stages of infection and the low levels of Trypanosoma spp. DNA in blood and tissue samples from animals. To this end, several primers have been developed for the amplification and sequencing of different target genes or regions such as ribosomal DNA, internal transcribed spacer region (ITS), kinetoplast DNA, and variable surface glycoprotein (VSG) genes of the parasites (El-Wathig et al. 2016; Tehseen et al. 2017).
A recent study of surra in northwestern Nigeria using the conventional parasitological methods reported a prevalence of 31.5% (Argungu et al. 2015). Therefore, to update the information on surra in this region, this study was conducted on camels slaughtered in the main abattoirs in Kano and Katsina cities, northwestern Nigeria. Specifically, this study aimed to determine the prevalence of trypanosomosis using parasitological, serological, and molecular methods and to assess the risk factors for surra in the study areas. Also, genetic characterization and the phylogenetic relationship of sequences in this study were compared to sequences in the GenBank.
Materials and methods
Ethical approval
All experimental protocols and animal work were approved by the Animal Use and Care Committee of the National Veterinary Research Institute (NVRI) Vom, Plateau State, Nigeria with the certification ID: NVRI AEC REF No. ACE/02/88/20.
Study area
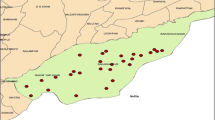
The study was conducted at the central abattoirs of Katsina (latitudes 11° 08ʹ N and 13° 22ʹ N and longitude 6° 52ʹ E and 9° 20ʹ E) and Kano (latitude 12° 40ʹ N and 10° 31ʹ N and longitude 7° 40ʹ E and 9° 30ʹ E) states located in the Northwestern region of Nigeria (Fig. 1). The climate of the two study areas is composed of two major seasons, the dry and wet seasons. The wet season starts in May and ends in September or early October, while the dry season begins in October and ends in April or early May. The mean annual rainfall is about 690 mm while the mean annual temperature ranges between a maximum of 43 °C and a minimum of 29 °C. The vegetation is mainly savanna, climatically defined as Sudan savanna, which is characterized by the presence of scattered trees and shrubs in open grassland (Wakawa et al. 2016). The choice of Katsina and Kano States in the Northwestern region was based on the presence of camel international markets in these two states and both states are located along the major trans-Sahara animal trade routes to Nigeria.
Sample size determination
The sample size was determined based on the prevalence of 27% (Enwezor and Anthony 2005) with a 95% confidence level and 5% precision as recommended by Thrusfield (2007). A total of 600 camels (Camelus dromedarius) were examined in this survey with 300 camels sampled from each of the states. One hundred and fifty camels were sampled during the dry and wet seasons in each of the states.
Sampling
This study was conducted on apparently healthy camels brought for slaughter in the main abattoirs in Kano and Katsina cities, northwestern Nigeria. In each of the cities, the major abattoir where a large number of camels was slaughtered for human consumption was chosen purposively. The camels slaughtered were sourced directly from the camel market or from farmers who rear the camels to fatten them before selling them to the meat sellers. On every sampling day, apparently healthy camels were selected for the study. The sampling period covered the dry and wet seasons. In total, 135 male and 165 female camels were sampled in Kano State, while 103 males and 197 females were sampled in Katsina State. The camels were examined before slaughter to determine their ages according to their dentition as described by Johnson (2003). Animals were categorized based on age 1–3 years, 4–6 years, 7–9 years, and 10–12 years. Furthermore, the camels were categorized based on the body conditions score as good, fair, or poor according to Salah et al. (2019).
Blood samples from camels were collected by jugular venipuncture for laboratory diagnosis. About 3 ml of blood from each camel was collected into plain tubes and then kept at room temperature (25 °C) until visible clot retraction was seen. The clotted samples were then centrifuged at × 300 g for 5 min, and the serum was aliquotted and stored at − 20 °C until serological analysis was performed. Four milliliters of blood was placed into a tube containing ethylene diamine tetraacetic acid (EDTA) for parasitological and molecular analysis. In the field, the blood samples were kept in a cold box packed with ice before transportation to the Parasitology laboratory of the National Veterinary Research Institute (NVRI), Vom, Plateau State, Nigeria where they were preserved at − 20 °C until further analysis.
Parasitological examination
For the parasitological diagnosis for the presence of Trypanosoma spp. from the blood samples collected, microscopic examination of Giemsa-stained thin smears was performed (OIE, 2012). Briefly, blood samples were processed for microscopic examination according to standard procedures (Soulsby and Mӧnnig 1982). Stained blood smears were examined under the microscope using the oil immersion objective for the detection of Trypanosoma spp. A minimum of 50 microscopic fields was examined before the result was determined.
Serological examination
Commercially available Card Agglutination Test (CATT/T. evansi) kits were purchased from the Laboratory of Serology, Institute of Tropical Medicine, Antwerp, Belgium, and used in this study. Serum samples were tested with Card Agglutination Test for Trypanosomosis (CATT/T. evansi) following the instruction of the manufacturer with slight modifications (Ibrahim et al. 2011). Briefly, one drop of test serum was diluted 1:4 in CATT-buffer. The mixture was then pipetted onto a plastic-coated test card. One drop of CATT reagent was added and the reaction mixture was spread out using a clean stirring rod. The reaction mixture was allowed to react on the card with manual rotation for 5 min. Positive reactions were interpreted based on the appearance of blue granular agglutinations visible to the naked eye. Positive and negative controls were included in each reaction run.
Molecular characterization
DNA was extracted from the whole blood collected from each of the sampled animals using a quick-DNA miniprep kit (Zymo Research) according to the manufacturer’s instructions. The eluted DNA was then stored at − 20 °C until PCR analysis. Published primers, TE-FOR-(5ʹ-TGCAGACGACCTGACGCTACT-3ʹ) and TE-REV-(5ʹ-CTCCTAGAAGCTTCGGTGTCCT-3ʹ) for the amplification of the 227-bp fragments of T. evansi, were used in this study (Wuyts et al. 1994). The PCR amplification of the samples was performed in a 25-µl reaction that contained 2.5 µl of genomic DNA extract, 0.5 µl of 20 µM primer (TE-FOR and TE-REV), 12.5 µl of one Taq® Quick-Load® 2 × Master Mix with Standard Buffer (New England Bio Labs), and the volume made up with 9.0 µl Nuclease Free Water (Promega®). Amplification was conducted on an Applied Biosystem®9700 PCR Machine. The reaction conditions were as follows: initial denaturation at 95 °C for 4 min followed by 30 cycles of denaturation at 95 °C for 1 min, annealing at 60 °C for 1 min, extension at 72 °C for 1 min and final extension at 72 °C for 10 min. The PCR products were visualized in 1.5% agarose gel stained with ethidium bromide.
DNA of T. evansi extracted from the blood of a camel positive by microscopic examination and distilled water were included in every PCR run as positive and negative controls, respectively. The gel was observed for the appropriate size DNA band under a UV trans-illuminator.
Positive amplicons were sent to a commercial sequencing company (Macrogen Europe, Netherlands) for sequencing in the forward direction. Sequences obtained were manually edited and compared with the sequences available in the GenBank database using the Basic Local Alignment Sequence Techniques (BLASTn) algorithm hosted by the National Centre for Biotechnology Information, Bethesda, MD, USA (www.blast.ncbi.nlm.nih.gov/blast.cgi). The sequences obtained in this study were deposited in the GenBank with the accession number (MZ394796 and MZ394795) for T. evansi from Katsina and MZ394797 for T. evansi from Kano State. Phylogenetic analysis was performed to compare the relationship between nucleotide sequences detected in this study with those in GenBank database. The evolutionary history was inferred using the maximum likelihood method based on the Kimura 2-parameter model. The bootstrap consensus tree was inferred from 1000 replications. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 14 nucleotide sequences. Codon positions included were 1st + 2nd. There were a total of 304 positions in the final dataset. No outgroup was included in the analysis since we only examined the relationship between the taxa. Evolutionary analysis was conducted in the Molecular Evolutionary Genetics Analysis package (MEGA 5).
Data analysis
Data generated during the study were entered into Microsoft Excel and analyzed using the R statistical software (R Core Team. 2013). The association between prevalence and risk factors was assessed using the chi-square test. The level of significance was set at p ≤ 0.05.
Results
The overall prevalence rates of T. evansi in the study area were 5.3%, 11.5%, and 21.5% by microscopy, serology, and PCR, respectively (Table 1). However, using the three methods explored in this study, a state-wise comparison indicated a higher prevalence of Surra in Katsina State compared with Kano State (Table 1). Female camels and those between 1 and 3 years of age were most affected with T. evansi compared with male and other age groups. In Katsina State camels with good body condition scores were not infected with T. evansi, but infection with T. evansi was recorded among camels with good body condition in Kano State (Table 1).
There was a significant difference (p < 0.05) between the T. evansi prevalence detected by the three diagnostic methods used in this study. PCR was the most sensitive followed by serology whereas microscopic detection was the least sensitive (Table 2). Furthermore, the agarose gel electrophoresis of the amplified PCR products from positive samples revealed a 227-bp fragment (Fig. 2). Regardless of the diagnostic method used, T. evansi prevalence was associated with body condition scores and the age of the animals. Furthermore, the sex of the camels was associated with the prevalence of T. evansi based on serology and PCR methods employed. However, there was no association between the prevalence of T. evansi in camels and the different seasons (Table 2).
The maximum likelihood phylogenetic tree generated a topology showing the T. evansi from Nigeria forming a cluster with high bootstrap with T. evansi from Iran (GenBank: MF188845.1) as well as T. evansi isolated from Dog in India (GenBank: MG600142.1). Other Trypanosoma spp. form distinct clades according to their sequence similarities (Fig. 3).
Discussion
Camels will continue to constitute an important part of the lives and livelihood of subsistent farmers in Nigeria, both as draught animals and a source of protein. However, diseases such surra causes a setback to camel productivity and wellbeing with a net socio-economic consequence to the farmers. In this study, low to high prevalence (5.3–21.5%) of T. evansi was detected in northwestern Nigeria depending on the diagnostic method used. The relatively low prevalence recorded in this study is consistent with earlier reports from some parts of Nigeria (Mbaya et al. 2010; Wakil et al. 2016) and other parts of the world (Dia et al. 1997; Tehseen et al. 2015; Olani et al. 2016; Bala et al. 2018; Hassan-Kadle et al. 2019). However, a higher prevalence of 31.5% trypanosomosis using conventional parasitological methods was reported in camels in this region of Nigeria (Argungu et al. 2015). The same authors reported a significantly higher prevalence of surra in female than male camels, similar to our findings by serology and PCR, but not by microscopy. Such differences can be attributed to the subjective interpretation of microscopic results.
The low prevalence recorded by the microscopic examination method could be due to its low sensitivity. However, the CATT/T. evansi serological assay detected higher cases of T. evansi infections from the sampled camels than by the microscopy. The disadvantage though is the inability of this test to differentiate between active infection and antibodies from treated animals. It has been reported that following treatment, antibodies from the treated animal remain in blood circulation up to nearly 4 weeks; thus, such animals will be detected as positive cases (Olaho-Mukani et al. 1996; Thammasart et al. 2001; Singh and Chaudhri 2002; Aregawi et al. 2015; Birhanu et al. 2015; Tehseen et al. 2015; Mohamoud 2017). Therefore, this fact necessitates getting a reliable history of the animal regarding recent anti-trypanosome treatment before sample collection to make CATT/T. evansi and all other antibody detecting tests are more reliable.
The PCR diagnosis gave a higher prevalence than the other two methods used in this study. This finding attests to the ability of this method to detect and amplify low levels of parasite DNA in blood circulation. Unlike the blood smear examination by microscopy where a high level of parasitemia, as well as the morphology of the parasite, is required to detect a positive sample, the PCR is reputed to be sensitive at detecting low parasitemia (Abdel-Rady 2006). However, some factors such as the presence of PCR inhibitors during DNA extraction have been reported to limit the sensitivity of PCR (Shyma et al. 2013). Generally, the prevalence recorded by PCR was significantly higher than the results from the other methods used in this study. This is in agreement with other studies (Birhanu et al. 2015; Ereqat et al. 2020). This further confirms the fact that PCR is an accurate, more sensitive, and specific method in the diagnosis of trypanosomes infection than the parasitological and serological methods. More so, it also overcomes the problem of non-specific reactions in the case of serological methods. The PCR can also detect low parasitemia is associated with chronic infections (Abdel-Rady 2006). The results from this study agree with the work of Nahla et al. (2011) who reported a higher prevalence (90.0%) of surra using molecular technique (PCR) than both serological (CATT/T. evansi) (47.6%) and the parasitological (3.7%) techniques. Conversely, Tehseen et al. (2015) reported prevalence rates of 0.7%, 47.7%, and 30.5% through parasitological, serological, and molecular techniques. This is not surprising because in serological techniques antibodies can remain in circulation for several months after treatment, thus given a false-positive result.
The phylogenetic analysis showed that the nucleotide sequences of T. evansi from Nigerian camels formed a monophyletic cluster with sequences in GenBank from camels, dogs, and horses from Iran, Iraq, India, and Malaysia. The clustering pattern observed in this study is similar to a recent report from Palestine (Ereqat et al. 2020). This is a strong indication of the monomorphic nature of T. evansi.
A sex-wise comparison indicated that female camels sampled in this study had a higher prevalence of surra than male camels. This might be attributed to stressor other sex-related physiological conditions including pregnancy and/or lactation which may reduce disease resistance in female camels and render them more susceptible to infections (Bhutto et al. 2009). There was no relation between surra prevalence and the season in this study. This finding is at variance with previous reports from different parts of the world (Lӧhr et al. 1985; Singh and Joshi 1991; Kashiwazaki et al. 1998; Jindal et al. 2005; Desquesnes et al. 2013). This could be attributed to vector density and differences in climatic conditions in the various parts of the works. The prevalence of T. evansi among camel herds is strongly dependent on the vector population, vegetation, and suitable breeding habitat for hematophagous flies (Mohammed 1999). However, our observation in this study suggests that most of the camels harbor chronic infection; hence, there was no seasonal difference in the prevalence related to vector abundance as earlier suggested by some researchers (Batra et al. 1994; Soodan et al. 1995). Furthermore, younger camels examined in this study were more predisposed to T. evansi infection than older camels. This is in agreement with the earlier reports in Nigeria (Mbaya et al. 2010; Kassa et al. 2011). Age susceptibility and lack of premunity have been suggested to account for the higher incidence of T. evansi in young camels (Soulsby and Mӧnnig 1982; Njiru et al. 2002).
In this study, camels in poor or fair body conditions had a higher prevalence of surra compared with those in good body condition. This finding is in agreement with the results reported by Eyop & Matios (2013) but contrary to the report of Idehen et al. (2018). The body condition score is related to the plain of nutrition of the animals, hence, their ability to mount resistance to infections including surra. Animals with poor body conditions are malnourished and therefore susceptible to disease conditions. There is often a relationship between the season and body condition score of camels due to feed scarcity especially in extensively managed animals. The extensive system of camel husbandry and management practiced by farmers in the study area is supported by the readily available pasture for camels during the rainy season compared to the dry season. There is a need for the provision of feed supplements to the camels, especially during the dry season. This will alleviate the effects of the food scarcity that is common during the dry season. Also, adequate veterinary care should be provided to the camels to alleviate the effects of different animal diseases including surra.
Conclusion
This study reports the prevalence of surra in northwestern Nigeria using three diagnostic methods. Based on our knowledge, this is the first report of T. evansi infection in camels using three diagnostic methods along the parasitological-serological-molecular continuum, especially the use of PCR and sequencing to confirm the diagnosis. Taken together, surra is prevalent in camels in northwestern Nigeria and constitutes a constraint to camel productivity in the area. Adequate control measures aimed at reducing the impact of trypanosomes on camel production in the study area is recommended.
Data availability
Not applicable.
Code availability
Not applicable.
References
Abdel-Rady, A. (2006). Comparison of card agglutination test and parasitological methods for the diagnosis of camel trypanosomosis in Egypt. In: Proceedings of the International Scientific Conference on Camels, 10–12 May, 2006. Qassim University, Buraidah, Kingdom of Saudi Arabia, 571–576.
Aregawi, W.G.,Kass, S.T., Tarekgn,K.D., Birhanu, W.T., Haile, S.T. & Kiflewahid, F.W. (2015). Parasitological and serological study of camel trypanosomosis (Surra) and associated risk factors in Gabi Rasu Zone, Afar, Ethiopia, Journal of Veterinary Medicine and Animal Health.7(6): 234-240
Argungu, S.Y., Bala, A.Y. & Liman, B. (2015). Pattern of Trypanosoma evansi infection among slaughtered camels (Camelus dromedaries) in Sokoto central abattoir. Journal of Zoological and Bioscience. 2(4), 1-7.
Bala, A.E., Abakar, A.D., Mohammed, M.S. & Abbas, M.A. (2018). Prevalence of Trypanosoma evansi in camels in four states of Great Butana, Sudan. Journal of Entomology and Zoology Studies, 3(3), 33-37.
Barghash, S.M., Darwish, A.M., & Abou-Elnaga, T.R. (2016). Molecular characterization and phylogenetic analysis of Trypanosoma evansi from local and imported camels in Egypt. Journal of Phylogenetic & Evolution Biology 4:169
Batra, U. K., Kumar, A. &Kulshreshtha, R. C. (1994). A study on surra in bovines in some parts of Haryana. Indian Veterinary Journal, 71, 971-974.
Bhutto, B., Gadahi, J. A., Shah, G., Dewani, P. &Arijo, A. G. (2009). Field investigation on the prevalence of trypanosomiasis in camels in relation to sex, breed and herd size. Pakistan Veterinary Journal, 30, 173-7.
Birhanu, H., Fikru,R., Said, M., Kidane, W., Gebrehiwot, T., Hagos, A., Alemu, T., Dawit, T., Berkvens, D., Goddeeris, M.B. & Buscher, P.(2015). Epidemiology of Trypanosoma evansi and Trypanosoma vivax in domestic animals from selected districts of Tigray and Afar regions, Northern Ethiopia. Parasites & Vectors.8:212
Desquesnes, M., Holzmuller, P., Lai, D.H., Dargantes, A., Lun, Z., &Jittaplapong, S. (2013). Trypanosoma evansi and Surra review and perspectives on origin, history, distribution, taxonomy, morphology, hosts, and pathogenic effects. Biomed Research International. 194176, 22
Dia, M. L., Diop, C. &Aminetou, M. (1997). Some factors affecting the prevalence of Trypanosoma evansi in camels in Mauritania. Veterinary Parasitology, 72(2), 111-120.
El-Wathig, M., Faye, B., Thevenon, S., Ravel, S., &Bossard, G. (2016). Epidemiological survey of camel trypanosomosis in Al-jouf, Saudi Arabia based on PCR and ELISA. Emirates Journal of food and Agriculture. 28:212-6
Enwezor,F.N.C. & Anthony, K. B.S. (2005). Camel trypanosomosis-a review. Veterinarski arhiv 75(5): 439-452
Ereqat, S., Naseredin, A., Al-Jawabreh, A., Al-Jawabreh, H., Al-Laham, N. & Abdeen, Z. (2020). Prevalence of Trypanosoma evansi in livestock in Palestine. Parasites Vectors 13:21 https://doi.org/10.1186/s13071-020-3894-9
Eyop, E. &Matios, L. (2013): Review on camel trypanosomosis (Surra) due to Trypanosoma evansi: Epidemiology and host response. Journal of Veterinary Medicine and Animal Health, 5(12), 334-345.
Getachew, A (2005). Trypanosomosis in Ethiopia. Review Article, Ethiopia Journal of Biological Science. 4(1):95
Hassan-Kadle, A.A., Ibrahim, M.A., Nyingilli, S.H., Yusuf, A.A., Vieira, J.W.S., & Vieira, C.F.R. (2019). Parasitological, serological and molecular survey of camel trypanosomosis in Somalia. Parasites & Vector, 12:598
Ibrahim, A.M., Ismail, A.A., Majid, A.A., Sidig, H.S., Osman, M.E., & Rahman, A, H. (2011). Prevalence of camel trypanosomosis and its effect on PCV as health indicator in the Sudan. University of Khartoum Journal of veterinary Medicine and Animal Production. 2:138-50
Idehen, C. O., Ishola, O. O., Adeyemi, I. G., Abongaby, G. C., Olaleye, O. O., Aluma, A. L., Opabunmi, R. O. &Obaloto, O. B. (2018).Prevalence of African trypanosomosis in cattle and sheep in Bassa Local Government Area of Plateau State, Nigeria. Sokoto Journal of Veterinary Sciences, 16(3), 11-17.
Jindal, N., Gupta, S. L., Batra, M. & Singh, R. (2005). A note on prevalence of surra in bovines in Haryana. Indian Veterinary Journal, 82, 1114-1115.
Johnson, R.F. (2003). The stockman’s handbook by Ensiminger, 2nd Ed. P539
Kamidi, C.M., Saarman, N.P., Dion, K., Mireji, P.O., Ouma, C., &Murilla, G. (2017). Multiple evolutionary origins of Trypanosoma evansi in Kenya. PLoS Neglected Tropical Disease. 11: e0005895
Kashiwazaki, Y., and Pholpark, M., Polsar, C. &Pholpark, S. (1998). Haemoparasite infection in newly introduced dairy cattle in Loei Province, Thailand: Trypanosoma evansi antigen levels by ELISA referring to abortion. Veterinary Parasitology, 80, 99-109.
Kassa, T., Tadesse, E. & Hassen, C. (2011). Prevalence of camel trypanosomosis and its vectors in Fentale district, South East Shoa zone, Ethiopia. Veterinarski Arhiv, 81,611-621.
Lӧhr, K. F., Pholpark, S., Srikijakarn, L., Thaboran, P., Bettermann, G. &Staak, C. (1985). Trypanosoma evansi infection in buffaloes in northeast Thailand. I. Field investigations. Tropical Animal Health and Production, 17, 121-125.
Mbaya, A. W., Ibrahim, U. I., &Apagu, S. T. (2010) Trypanosomosis of the dromedary camel (Camelus dromedaries) and its vector in the Tse-tse free arid zone of North-Eastern, Nigeria. Nigeria Veterinary Journal, 31(3), 154-163.
Mohammed, R. (1999). Camel Trypanosomosis: Prevalence and drug sensitivity test in Dire Dowa administrative council eastern Ethiopia. DVM Thesis FVM, AAU, Debre Zeit Ethiopia.
Mohamoud, A.H.I. (2017) Sero-prevalence study of camel trypanosomiasis in selected villages of Galkayo, Somalia. Open Journal of Veterinary Medicine, 7. 31
Nahla, O.M.A., Hamid, I.M.N., & Hamid, S.A. (2011).Molecular diagnosis of Trypanosoma evansi infection in Camelus dromedarius from eastern and western regions of the Sudan. Emirate Journal of food and Agriculture. 23(4): 320-329
Njiru, Z.K., Bett, O., Ole-Mapeny, I.M., Githiori, J.B., &Ndung, J.M. (2002) Trypanosomosis and helminthosis in camels; comparison of ranch and traditional camel management systems in Kenya. Journal of Camel Practice Research 55, 67-71.
OIE–World organization for animal health. Trypanosoma evansi infection (surra). Chapter 3.1.21. In: Manual of diagnostic tests and vaccines for terrestrial animals 2012. 7thed.Paris:OIE;2012. http://www.oie.int/fileadmin/Home/eng/Healthstandards/tahm/3.01.21TRYPANOSURRA.pdf. Assessed13 June 2021
Olaho-Mukani, W., Nyang’ao, J. M. N. &Ouma, J. O. (1996). Use of Suratex for field diagnosis of patent and non-patent Trypanosoma evansi infections in camels. British Veterinary Journal,152, 109-111.
Olani, A., Habtamu, Y., Wegayehu, T. (2016). Prevalence of camel trypanosomosis (surra) and associated risk factors in Borena zone, southern Ethiopia. Parasitology Research 115,1141–1147. https://doi.org/10.1007/s00436-015-4845-9
Salah, A. A. Robert, I D., & Mohamed A. S (2019). Prevalence and distribution of Trypanosoma evansi in camels in Somaliland Tropical Animal Health & Production 51 (8), 2371–2377: https://doi.org/10.1007/s11250-019-01947-6.
Shyma, K. P., Gupta, S. K., Singh, A., Chaudhri, S.S. & Gupta, J. P. (2013). Detection of Trypanosoma evansi in whole blood of domestic animals by DNA amplification method. Indian Journal of Animal Research, 47, 456-9.
Singh, A. &Chaudhri, S. S. (2002). Comparison of efficiency of parasitological methods with Ag-ELISA in Trypanosoma evansi infected crossbred calves. Indian Journal of Animal Science,72(2), 117-119.
Singh, B. & Joshi, S. J. (1991). Epidemiology, clinico-pathology and treatment of clinical Trypanosoma evansi infection in buffalo (Bubalus bubalis). Indian Veterinary Journal, 68, 975-979.
Soodan, J. S., Singh, K. B., Juyal, P. D. &Khahra, S. S. (1995). Incidence of Trypanosoma evansi infection in equines in Punjab state. Journal of Veterinary Parasitology, 9(2), 133-134.
Soulsby, E.J.L & Mӧnnig, H.O. (1982). Helminths, arthropods and protozoa parasites of domesticated Animals, 7th Edition. Bailliére Tindal, London, pp 232-233.
Tehseen, S., Jahan, N., Desquesnes, M., Shahzad, I., & Qamar, M.F. (2017). Field investigation of Trypanosoma evansi and comparative analysis of diagnostic tests in horses from Bahawalpur, Pakistan. Turkish Journal of Veterinary and Animal science.41:288–93
Tehseen, S., Johan, N., Qamar, M. F., Desquesnes, M., Shahzad, M. I., Deborggroev, S. & Buscher, P. (2015). Parasitological, serological and molecular survey of Trypanosoma evansi infection in dromedary camels from Cholistan Desert, Pakistan. Parasites& Vectors, 8, 1-11
Thammasart, S., Kanitpun, R., Saithasao, M. & Kashiwazaki, Y. (2001).Preliminary studies by ELISA on the antigen and antibody with Trypanosoma evansi in cattle. Tropical Animal Health Production, 33(3), 189-199.
Thrusfield, M. (2007). Veterinary Epidemiology, (3rd ed). London Blackwell Science Ltd.
Wakawa, I.D., Adams, L.I. and Bichi, A.M. (2016). Tree species composition within Kano state university of science and technology Wudil, Kano state, Nigeria. Journal of Research in Forestry, wildlife and environment. 8(2): 100-111
Wakil, Y., Lawal, J. R., Gazali, Y. A., Mustapha, F. B., Bello, A. M., Mshelia, E. S. &Ayomikun, A. M. (2016). Survey on prevalence of haemoparasites of trade camels (Camelus dromedaries) in Maiduguri; Nigeria. Journal of Veterinary Medicine and Animal Science, (2), 7-10.
Wuyts, N., Chodesajjawatee, N. &Panyim, S. (1994). A simplified and highly sensitive detection of Trypanosoma evansi by DNA amplification. Southeast Asian Journal of Tropical Medicine Public Health, 25, 266-271.
Acknowledgements
We acknowledge the Nigerian Institute for Trypanosomiasis Research (NITR) and National Veterinary Research Institute (NVRI) both in Vom, Plateau State, Nigeria, for helping out in the field and laboratory diagnosis respectively.
Author information
Authors and Affiliations
Contributions
MSA and DAD collected samples, carried out laboratory work, analyzed data, and prepared the manuscript. JAY and DGA were involved in conceiving the project, the study design, and reviewing the manuscript. KJ carried out the phylogenetic analysis and contributed to data analysis. RRC and OOO managed the technical aspect of the studies and finalizing the manuscript. TDA and PJG contributed to the molecular and parasitological analysis of the samples. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mamman, S.A., Dakul, D.A., Yohanna, J.A. et al. Parasitological, serological, and molecular survey of trypanosomosis (Surra) in camels slaughtered in northwestern Nigeria. Trop Anim Health Prod 53, 537 (2021). https://doi.org/10.1007/s11250-021-02891-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11250-021-02891-0