Abstract
The lipase E hormone-sensitive (LIPE) enzyme is one of the lipolytic enzymes, and it plays a key role in the regulation of adipose tissue deposition. This study was conducted to investigate the possible association between the LIPE gene variations and the main body weight measurements in Awassi sheep. A total of 160 of sexually mature Awassi rams (Ovis aries) that aged between 2 and 3 years were included in the present study. Genomic DNA was extracted and two specific PCR amplicons were designed to amplify two coding regions within the LIPE gene. Genotyping experiments were performed using polymerase chain reaction-single-strand conformational polymorphism (PCR-SSCP). Two different SSCP banding patterns were identified, CC and CD in exon 2, and AA and AT in exon 9. Five novel single-nucleotide polymorphisms (SNPs) were detected by sequencing, namely g.151C > A and g.198C > T in exon 2, and g.213G > C, g.226G > T, and g.232A > C in exon 9. Haplotype block analysis showed strong linkage disequilibrium values between the two SNPs in exon 2 and the three SNPs in exon 9. Association analysis of haplotypes with carcass traits demonstrated a significantly higher dressing percentage (P < 0.05) and lower fat tail weight (FTW) in CACT and GCGTAC haplotypes made these haplotypes more favorable for human consumption. The current research is the first one to report a tight association between the LIPE genetic polymorphism and the dressing percentage and FTW traits, suggesting a pivotal role played by these co-inherited SNPs in the metabolism of carcass traits in sheep.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lipolysis in adipose tissue plays a major role in energy homeostasis and body weight and is regulated, at least in part, by the multiple lipolytic genes (Nishizawa and Shimomura 2019; Kulyte et al. 2020). Lipolytic genes are shown to be related to several body weight measurements in a variety of animal species, and thus, they have the potential to provide an insight into growth traits in sheep (Carlsson et al. 2006). Lipolytic genes have recently demanded more attention because of their inevitable role in obesity and diseases (Luglio et al. 2015). One of these genes is the hormone-sensitive lipase (HSL), or lipase E hormone-sensitive (LIPE) gene. The ovine LIPE gene is a highly conserved gene, which consists of 10 exons and located on chromosome 14 (Jiang et al. 2014). Due to the versatile roles of this gene, its expression is not only confined in adipose tissues. Instead, LIPE gene is also highly expressed in other tissues, such as ovary, testis, heart, adrenal glands, skeletal, and muscle tissues (Qiao et al. 2007). The LIPE gene encodes HSL, a multifunctional enzyme that is involved in the control of hormone-stimulated lipolysis, free fatty acid biosynthesis, and mobilization (Zechner et al. 2012; Fang et al. 2017). It acts as a rate-limiting enzyme in the conversion of stored intracellular triacylglycerol to diacylglycerol and releases non-esterified fatty acids (Xue et al. 2015; Jocken et al. 2018). Furthermore, HSL enzymes play a key role in the determination of insulin resistance degree and tissue content of diacylglycerol (Hsiao et al. 2013). Two different forms of LIPE enzymes have been identified in sheep. Both forms exhibit considerable homology with LIPE amino acid sequences with several other mammalian species. This homology suggests a remarkable functional constraint over evolutionary time (Lampidonis et al. 2011). In dairy goats, LIPE gene variation may exert an essential role in altering the mobilization of medium-chain and unsaturated fatty acids and body composition traits (Zidi et al. 2010). It is well established that the genetic polymorphism of the LIPE gene has been associated with multiple carcass traits in several species. Goszczynski et al. (2014) have reported high polymorphism in the LIPE gene and confirmed a significant association of this gene variation with the fatty acid composition in cattle. Similarly, it has been found that single-nucleotide polymorphisms (SNPs) of the LIPE gene have been associated with the carcass and meat quality traits in cattle (Fang et al. 2014). Significant associations between the variation in the LIPE gene and subcutaneous fat deposition, including an increased body mass index, waist circumference, and obesity in humans, have been observed (Carlsson et al. 2006). Given the essential role the LIPE gene plays in fat metabolism, it is suggested that the nucleic acid substitution within this gene may cause alterations in the function of its encoded LIPE enzyme and may result in varied phenotypic traits in the studied population. Unfortunately, only limited numbers of studies have investigated the association of this gene with the primary productive traits of sheep (Yang et al. 2014). Keeping in mind the above aspects, the purpose of this study was aimed to investigate the possibility of the association between polymorphisms of the LIPE gene with several production traits in the Iraqi native Awassi sheep. To the best of our knowledge, the present manuscript is the first one to describe the association of the LIPE gene polymorphisms with several fat depositions and composition traits in sheep.
Materials and methods
Sheep population and carcass growth trait measurements
The study was conducted according to the regulations of the international recommendations for the care and use of animals (Federation of Animal Science Societies 2010) and animal experimentation was approved by Al-Qasim Green University (Approval No. 12.10.15). A total of 160 of sexually mature Awassi rams (Ovis aries) aged between 2 and 3 years were included in the present study. The referred animals were the progeny of the different males and females and they were randomly selected from three flocks in the middle Euphrates regions of Iraq. Any relationship between the parents of the selected animals was not recorded. In each studied flock, 10–12 rams were randomly allocated to mate with about 20–25 ewes per ram, with male identification recorded. All the selected animals were confirmed to be healthy in terms of the absence of any noticeable clinical symptoms. Animals were kept on natural pasture during spring and autumn, while in winter, animals were kept indoors and fed about 2.5% of their live body weight daily, comprising a mixture of barely (59%), bran (40%), and salt (1%) concentrates. From the period of October 2017 to June 2018, several phenotypic measurements were taken from the Awassi sheep from the main abattoirs in the referred regions, such as live body weight (BW), back fat thickness (BFT), abdominal fat (AF), fat tail weight (FTW), and carcass weight, which were recorded at the specified slaughterhouses as described by Rehman et al. (2013). Dressing percentage of each animal was calculated according to the following formula (Warriss 2000):
Genomic DNA extraction
Genomic DNA was extracted from the whole blood by a universal salting-out technique (Al-Shuhaib 2017). The integrity of the extracted DNA was evaluated by direct electrophoresis on 0.8% agarose gel, while the purity of the extracted DNA was assessed by a nanodrop spectrophotometric method (BioDrop μLITE, BioDrop, UK).
Polymerase chain reaction
Two pairs of specific primers were designed by NCBI primer BLAST software to amplify the biggest two coding exons within the LIPE gene, including the LIPE-exon-2 and LIPE-exon-9 fragments, of 342 bp and 278 bp, respectively (Fig. 1a). Further details of the designed primers are described in Table 1. The PCR reaction was performed using the AccuPower PCR PreMix (Bioneer, South Korea). Each 20 μl of PCR premix contained 250 μM of dNTPs, 10 mM of Tris-HCl (pH 9.0), 30 mM of KCl, 1 U of Top DNA polymerase, and 1.5 mM of MgCl2. The PCR reaction mixture was completed with 10 pmol of each primer and 10–30 ng of genomic DNA. The PCR program was initial denaturation at 94 °C for 5 min, followed by 30 cycles of denaturation (94 °C for 30 s), annealing (61.8 °C for LIPE-exon-2 and 60.4 °C for LIPE-exon-9 for 30 s), elongation (72 °C for 30 s), and a final extension (72 °C for 5 min). The PCR products were verified by electrophoresis on a 1.5% (w/v) agarose gel, and photos were taken using a photodocumentation unit (ChemiDoc, Bio-Rad, USA). It was confirmed that all electrophoretic PCR bands were specific before being submitted to the next genotyping step.
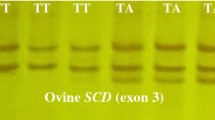
Genomic organization of the LIPE gene and its PCR-SSCP-sequencing strategy. (A) PCR design of two PCR specific primer pairs for the amplification of 342 bp and 278 bp in exon 2 and exon 9, respectively. (B) Post-PCR genotyping using SSCP technique, in which both exons showed two different genotypes. (C) Sequencing reaction interpretation of the detected genotypes, in which two SNPs were detected in the exon 2 and three SNPs were identified in exon 9. (D) Characterization of the observed SNPs, in which one silent p.L83= and one missense p.T67K SNPs were observed in the exon 2, and three missense p.A638P, p.G642V, and p.H644P SNPs were observed in the exon 9. The cyan color refers to the presence of a particular SNP in both genotypes, while the pink color refers to the detection of a particular SNP in one specific genotype. The positions of primers and the nomenclature of the observed SNPs are listed according to NCBI Reference Sequence: NC_019471.2 following the nomenclature described in varnomen.hgvs.org/
Single-strand conformation polymorphism
The initial denaturation of the PCR amplicons, as well as SSCP protocol, was performed according to Al-Shuhaib et al. protocol (Al-Shuhaib et al. 2018). The samples were loaded into 8% SSCP gels (acrylamide:bis-acrylamide = 37.5:1). The running conditions were set at 200 V, 100 mA for 4 h at a constant temperature of 20 °C using mini-wide gels (216 × 110) mm with gel thickness 1.0 mm (JY-CZ-B, Junyi-Dongfang Electrophoresis Equipment). The DNA fragments in the gels were visualized by silver nitrate as described by Byun et al. (2009).
Sequencing of PCR amplicons
Each observed PCR-SSCP electrophoretic pattern was commercially sequenced from both directions according to the instruction manuals recommended by sequencing laboratories (Macrogen Inc. Geumcheon, Seoul, South Korea). The reference databases of the two LIPE fragments were retrieved from the NCBI website (https://www.ncbi.nlm.nih.gov). Subsequently, the sequences of the observed genotypes were edited, aligned, and analyzed by BioEdit Sequence Alignment Editor Software Version 7.1 (DNASTAR, Madison, WI, USA). The electropherogram variations were then visualized by SnapGene Viewer Ver. 4.0.4, (http://www.snapgene.com). The novelty of the observed SNPs was confirmed by the dbSNP database (https://www.ncbi.nlm.nih.gov/projects/SNP/). Further confirmations of the observed SNPs were obtained from the ensemble genome browser 95 (https://asia.ensembl.org/index.html).
Statistical analysis
The allele and genotype frequencies of both studied loci of LIPE gene were estimated by direct counting, and the differences of the observed and expected frequencies of exon 2 and exon 9 genotypes were estimated using chi-square test to verify if the targeted ovine population was in Hardy-Weinberg equilibrium. Genetic indices of Awassi sheep, including observed heterozygosity (Ho) and expected heterozygosity (He), were performed by PopGen32 software, v. 1.31 (Yeh et al. 1999). Pairwise linkage disequilibrium (LD) between SNPs was calculated using SHEsis software (She and He 2006). All statistical analyses were performed using SPSS v23.0 (IBM, NY, USA). General linear mixed-effects models (GLMMs) were used to evaluate the effect of genotype on phenotypic traits. The model used to test the SNP genotype effect was fitted as follows:
where Yijk is the phenotypic traits observed, μ is the overall mean for each trait, Gi is the fixed effect associated with the ith SNP genotype or haplotype, αj is the random effect of jth sire, and eijk is the random error for ijk. Preliminary statistical analyses indicated that age, season, and nutrition were not found to affect carcass characteristics and thus they were not included in the model.
Results
Polymorphism of the LIPE gene
Both amplified loci of the LIPE gene were observed alongside with two SSCP banding patterns. Exon 2 showed two genotypes, one with two single-strand (ss) DNA bands, CC genotype, and the other one with three ssDNA bands, CD genotype. Similarly, exon 9 showed two genotypes, one with three ssDNA bands, AA genotype, and the other one with four ssDNA bands, AT genotype, within the same gel conditions (Fig. 1b). The nucleic acid substitutions of the detected genotypes were analyzed by comparing DNA sequencing electropherograms of both observed genotypes in both sequences of exon 2 and exon 9, respectively (Fig. 1c).
LIPE gene sequence interpretation
No previously published data concerning the LIPE gene were deposited under Awassi breed in the NCBI GenBank database. Therefore, the exon 2 and exon 9 DNA sequences obtained in this study, and those previously reported for other highly related ovine sequences (DQ647326.1 and NM_001128154.1 for exon 2, and DQ647326.1, NM_001128154.1, KC585035.1, KC585036.1 for exon 9), were compared to identify polymorphic sites. Based on alignments and annotations conducted by Blastn, dbSNP, ensemble, and DNA STAR tools, the novelty of all the five detected SNPs was confirmed in the investigated Awassi population. Two transition SNPs were detected in exon 2: c.49839273C > A, with a missense effect of p.T67K that was observed in both genotypes, and c.49839320C > T, with a silent effect of p.L83= that was observed only in the CD genotype. However, both transition/transversion forms were detected in exon 9, in which three SNPs, c.49847654G > C, c.49847667G > T, and c.49847673A > C, with three respective missense effects of p.A638P, p.G642V, and p.H644P were observed (Table 2). While p.G642 and p.H644P were detected in both AA and AT genotypes, p.A638P was detected only in the AT genotype (Fig. 1d).
The genetic diversity of the observed variations and association of polymorphisms with carcass traits in Awassi breed
Frequencies of individual alleles and genotypes in the analyzed populations of Awassi sheep are presented in Table 3. The χ2 test showed that the polymorphism of the LIPE gene at both loci did not deviate from the Hardy-Weinberg equilibrium (P < 0.05). The observed heterozygosity was lower than its expected value. This entailed a low level of genetic diversity. Pairwise linkage disequilibrium (LD) between five SNPs was calculated in the Awassi breed. Two SNPs (g.151C > A and g.198C > T) in exon 2 and three SNPs (g.213G > C, g.226G > T, and g.232A > C) in exon 9 were in high LD (r2 > 0.9), signifying a closely and strongly linked between pairs of SNPs. Concerning exon 2 variation, differences between combined genotypes were statistically significant just for FTW and dressing percentage (P < 0.05), in which individuals with CACT haplotype showed lower FTW and higher dressing percentage than those with CCCC haplotype. Similarly, variation in the exon 9 appeared to exhibit significant effects on FTW and dressing percentage. The individuals with GCGTAC haplotype showed a significant association with lower FTW and higher dressing percentage (P < 0.05) than those with GGGGAA haplotypes (Table 4). Thus, sheep with CACT and GCGTCA haplotypes had lower FTW and higher dressing percentages and could be used in the improvement of meat quality in Awassi breed. Haplotype analysis of five SNPs revealed a statistically significant association with carcass traits, indicating that these variants of the LIPE loci could be helpful genetic markers in sheep breeding.
Discussion
LIPE gene encodes the HSL enzyme, an intracellular neutral lipase, which hydrolyzes a variety of esters and participated in the mobilization of free fatty acids to the body. Since the LIPE gene is directly involved in regulating lipolysis, alterations in its sequences are expected to modify the fatty acid composition of tissues. For this reason, mutations in LIPE gene may affect the chemical composition of meat and alter meat quality traits (Goszczynski et al. 2014). This study describes a tight association between the LIPE gene variation and carcass measurements in sheep. This association was carried out based on the PCR-SSCP genotyping of the LIPE gene in 160 Awassi rams. The polymorphisms of exon 2 and exon 9 were associated with dressing percentage and FTW measurements in the investigated Awassi breed. Regarding the exon 2, only one SNP was detected with a silent effect, p.L83=. Although a silent SNP may change the protein conformation and translation kinetics (Komar 2007; Hunt et al. 2009), the currently observed silent SNP had obvious association with the studied carcass measurements. Concerning exon 9, three SNPs with three different missense effects were observed, p.A638P, p.G642V, and p.H644P. However, the observed association in the sheep between the exon 2 and exon 9 genetic variations of the LIPE gene and the carcass traits is quite remarkable since no previous study has demonstrated such association (Yang et al. 2014). However, our results are in line with several reports that demonstrated the linkage of LIPE variation with several carcass traits in cattle and pigs (Lei et al. 2005; Fang et al. 2014; Goszczynski et al. 2014; Xue et al. 2015). While it is likely that the association between the LIPE genetic polymorphisms with economic traits is may be informative to improve the quality of meat in sheep (Goszczynski et al. 2014), likewise, involving haplotype analysis can also improve the power of association studies with carcass traits in sheep (An et al. 2017). Based on haplotype analysis, sheep with the CACT and GCGTAC haplotypes had lower FTW and higher dressing percentages and are thus more favored for human consumption than those with the CCCC and GGGGAA haplotypes. However, consumers prefer leaner meat, since the increased FTW burdens breeders in terms of reproduction, growth, and carcass marketing conditions. Moreover, the presence of more tail fat in the body lowers the quality of meat and requires more energy than the production of lean tissue. Besides, consumers are not usually preferring fat dispersed throughout the tail over the meat fibers (Gokdal et al. 2003). Consequently, increased FTW reduces the commercial value of the carcass (Bingol et al. 2006). Therefore, it can be stated that the higher proportion of the fat in the carcass lowers its desirability to the consumers (Bingol et al. 2006; Skapetas et al. 2006). In agreement with our findings, a tendency for genotypes with lower FTW was found to have a higher percentage of fat in carcass cuts (Ünal et al. 2006). Though lipolysis is controlled by the expression of many lipolytic genes that may exhibit multifactorial regulation of this critical metabolic activity (Fan et al. 2019), this study detected a tight correlation between the polymorphisms of LIPE gene with FTW and dressing percentage. Accordingly, meat quality seems to be under the detected genetic polymorphism of the LIPE gene. Therefore, it is maybe rational to consider the present haplotype analysis as a valuable tool for assessing the degree of LIPE gene association with carcass traits.
Conclusion
Haplotype analysis of five novel SNPs observed in the LIPE gene demonstrated a tight linkage to some carcass and lipid traits in sheep. Since it exhibited significant association (P < 0.05) with higher dressing and lower FTW values in the Awassi breed. Therefore, the LIPE gene variation is an interesting marker for the assessment of carcass traits, and it can be recommended for future marker-assisted selection programs.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Al-Shuhaib, M.B.S., 2017. A universal, rapid, and inexpensive method for genomic DNA isolation from the whole blood of mammals and birds. Journal of Genetics, 96, 171–176
Al-Shuhaib, M.B.S., Al-Kafajy, F.R., Badi, M.A., AbdulAzeez, S., Marimuthu, K., Al-Juhaishi, H.A.I., Borgio, J.F., 2018. Highly deleterious variations in COX1, CYTB, SCG5, FK2, PRL and PGF genes are the potential adaptation of the immigrated African ostrich population. Computers in biology and medicine, 100, 17–26
An, Q., Zhou, H., Hu, J., Luo, Y., Hickford, J.G.H., 2017. Haplotypes of the ovine Adiponectin gene and their association with growth and carcass traits in New Zealand Romney lambs. Genes, 8(6), 160
Bingol, M., Aygün, T., Gökdal, Ö., Yılmaz, A., 2006. The effects of docking on fattening performance and carcass characteristics in fat-tailed Norduz male lambs. Small Ruminant Research, 64, 101–106
Byun, S.O., Fang, Q., Zhou, H., Hickford, J.G.H., 2009. An effective method for silver-staining DNA in large numbers of polyacrylamide gels. Analytical Biochemistry, 385, 174–175
Carlsson, E., Johansson, L.E., Strom, K., Hoffstedt, J., Groop, L., Holm, C., 2006. The hormone-sensitive lipase C-60G promoter polymorphism is associated with increased waist circumference in normal-weight subjects. International Journal of Obesity, 30, 1442
Fan, H., Hou, Y., Sahana, G., Gao, H., Zhu, C., Du, L., Zhao, F., Wang, L., 2019. A Transcriptomic Study of the Tail Fat Deposition in Two Types of Hulun Buir Sheep According to Tail Size and Sex. Animals, 9(9), 655
Fang, X.B., Zhang, L.P., Yu, X.Z., Li, J.Y., Lu, C.Y., Zhao, Z.H., Yang, R.J., 2014. Association of HSL gene E1-c.276C>T and E8-c.51C>T mutation with economical traits of Chinese Simmental cattle. Molecular Biology Reports, 41, 105–112
Fang, X., Zhao, Z., Jiang, P., Yu, H., Xiao, H., Yang, R., 2017. Identification of the bovine HSL gene expression profiles and its association with fatty acid composition and fat deposition traits. Meat Science, 131, 107–118
Federation of Animal Science Societies, 2010. Guide for the Care and Use of Agricultural Animals in Research and Teaching. Champaign, IL 61822, 3rd edition
Gokdal, O., Aygun, T., Bingol, M., Karakus, F., 2003. The effects of docking on performance and carcass characteristics of male Karaka lambs. South African Journal of Animal Science, 33(3), 185–192
Goszczynski, D.E., Mazzucco, J.P., Ripoli, M.V., Villarreal, E.L., Rogberg-Muñoz, A., Mezzadra, C.A., Melucci, L.M., Giovambattista, G., 2014. Characterization of the bovine gene LIPE and possible influence on fatty acid composition of meat. Meta Gene, 2, 746–760
Hsiao, P.J., Chen, Z.C., Hung, W.W., Yang, Y.H.C., Lee, M.Y., Huang, J.F., Kuo, K.K., 2013. Risk interaction of obesity, insulin resistance and hormone-sensitive lipase promoter polymorphisms (LIPE-60 C>G) in the development of fatty liver. BMC Medical Genetics, 14(1), 54
Hunt, R., Sauna, Z.E., Ambudkar, S.V., Gottesman, M.M., Kimchi-Sarfaty, C., 2009. Silent (synonymous) SNPs: should we care about them? Methods in Molecular Biology, 578, 23–39
Jiang, Y., Xie, M., Chen, W., Talbot, R., Maddox, J. F., Faraut, T., Wu, C., Muzny, D.M., Li, Y., Zhang, W., Stanton, J-A., Brauning, R., Barris, W.C., Hourlier, T., Aken, B.L., Searle, S.M.J., Adelson, D.L., Bian, C., Cam, G.R., Chen, Y., Cheng, S., DeSilva, U., Dixen, K., Dong, Y., Fan, G., Franklin, I.R., Fu, S., Fuentes-Utrilla, P., Guan, R., Highland, M.A., Holder, M.E., Huang, G., Ingham, A.B., Jhangiani, S.N., Kalra, D., Kovar, C.L., Lee, S.L., Liu, W., Liu, X., Lu, C., Lv, T., Mathew, T., McWilliam, S., Menzies, M., Pan, S., Robelin, D., Servin, B., Townley, D., Wang, W., Wei, B., White, S.N., Yang, X., Ye, C., Yue, Y., Zeng, P., Zhou, Q., Hansen, J.B., Kristensen, K., Gibbs, R.A., Flicek, P., Warkup, C.C., Jones, H.E., Oddy, V.H., Nicholas, F.W., McEwan, J.C., Kijas, J.W., Wang, J., Worley, K.C., Archibald, A.L., Cockett, N., Xu, X., Wang, W., Dalrymple, B.P., 2014. The sheep genome illuminates biology of the rumen and lipid metabolism. Science, 344, 1168–1173
Jocken, J.W.E., González Hernández, M.A., Hoebers, N.T.H., van der Beek, C.M., Essers, Y.P.G., Blaak, E.E., Canfora, E.E., 2018. Short-Chain Fatty Acids Differentially Affect Intracellular Lipolysis in a Human White Adipocyte Model. Frontiers in Endocrinology, 8, 372
Komar, A.A., 2007. SNPs, silent but not invisible. Science, 315, 466–467
Kulyte, A., Lundbäck, V., Lindgren, C.M., Luna, J., Lotta, L.A., Langenberg, C., Arner, P., Strawbridge, R.J., Dahlman, I., 2020. Genome-wide association study of adipocyte lipolysis in the GENetics of adipocyte lipolysis (GENiAL) cohort. Molecular Metabolism, 34, 85–96
Lampidonis, A.D., Rogdakis, E., Voutsinas, G.E., Stravopodis, D.J., 2011. The resurgence of Hormone-Sensitive Lipase (HSL) in mammalian lipolysis. Gene, 477, 1–11
Lei, M.G., Wu, Z.F., Deng, C.Y., Dai, L.H., Zhang, Z.B., Xiong, Y.Z., 2005. Sequence and polymorphism analysis of porcine hormone-sensitive lipase gene 5′-UTR and exon I. Acta Genetica Sinica, 32, 354–359
Luglio, H.F., Sulistyoningrum, D.C., Susilowati, R., 2015. The role of genes involved in lipolysis on weight loss program in overweight and obese individuals. Journal of Clinical Biochemistry and Nutrition, 57, 91–97
Nishizawa, H., Shimomura, I., 2019. Fat cell lipolysis and future weight gain. Journal of Diabetes Investigation, 10(2), 221
Qiao, Y., Huang, Z., Li, Q., Liu, Z., Hao, C., Shi, G., Dai, R., Xie, Z., 2007. Developmental changes of the FAS and LIPE mRNA expression and their effects on the content of intramuscular fat in Kazak and Xinjiang sheep. Journal of Genetics and Genomics, 34(10), 909–917
Rehman, A., Rehman, S., Akhtar, S., Jan, I., Qureshi, M.S., Rahman, S.U., 2013. Determination of meat yield and dressing percentage of Turki, Afghan Arabi and Baluchi sheep breeds in Afghanistan. Sarhad Journal of Agriculture, 29(1), 113–117
She, Y.Y., He, L., 2006. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Research, 16, 851–851
Skapetas, B., Sinapis, E., Hatziminaouglou, J., Karalazos, A., Katanos, J., 2006. Effect of age at slaughter on carcass characteristics and carcass composition in lambs of mountain Greek breeds. Czech Journal of Animal Science, 51(7), 311
Ünal, N., Akçapınar, H., Aytac, M., Atasoy, F., 2006. Fattening performance and carcass traits in crossbred ram lambs. Medycyna Weterynaryjna, 62, 401–404
Warriss, P.D., 2000. Meat Science. CABI Publish. New York, USA, 1st edition
Xue, W., Wang, W., Jin, B., Zhang, X., Xu, X., 2015. Association of the ADRB3, FABP3, LIPE, and LPL gene polymorphisms with pig intramuscular fat content and fatty acid composition. Czech Journal of Animal Science, 60, 60–66
Yang, G., Forrest, R., Zhou, H., Hickford, J., 2014.Variation in the ovine hormone-sensitive lipase gene (HSL) and its association with growth and carcass traits in New Zealand Suffolk sheep. Molecular Biology Reports, 41, 2463–2469
Yeh, F.C., Yang, R., Boyle, T., 1999. POPGENE: Version 1.31. Microsoft window-based freeware for population genetic analysis, University of Alberta. Edmonton
Zechner, R., Zimmermann, R., Eichmann, T.O., Kohlwein, S.D., Haemmerle, G., Lass, A., Madeo, F., 2012. FAT SIGNALS–lipases and lipolysis in lipid metabolism and signaling. Cell Metabolism, 15, 279–291
Zidi, A., Fernandez-Cabanas, V.M., Carrizosa, J., Jordana, J., Urrutia, B., Polvillo, O., Gonzalez-Redondo, P., Gallardo, D., Amills, M., Serradilla, J.M., 2010. Genetic variation at the goat hormone-sensitive lipase (LIPE) gene and its association with milk yield and composition. Journal of Dairy Research, 77, 190–198
Acknowledgments
The authors are grateful to the staff of Barakat Abu al Fadhl Al-Abbas (AS) Station for raising sheep and to Al-Khafeel Co. (Karbala, Iraq) for their facilities that provided the Awassi sheep population. This research did not receive any specific funding. The authors are grateful to the staff of Paper True for proofreading and editing of the manuscript.
Funding
The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Al-Thuwaini, T.M., Al-Shuhaib, M.B.S., Lepretre, F. et al. Co-inherited novel SNPs of the LIPE gene associated with increased carcass dressing and decreased fat-tail weight in Awassi breed. Trop Anim Health Prod 52, 3631–3638 (2020). https://doi.org/10.1007/s11250-020-02400-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11250-020-02400-9