Abstract
Genome editing using CRISPR/Cas9 has been highlighted as a powerful tool for crop improvement. Nevertheless, its efficiency can be improved, especially for crops with a complex genome, such as soybean. In this work, using the CRISPR/Cas9 technology we evaluated two CRISPR systems, a one-component vs. a two-component strategy. In a simplified system, the single transcriptional unit (STU), SpCas9 and sgRNA are driven by only one promoter, and in the conventional system, the two-component transcriptional unit (TCTU), SpCas9, is under the control of a pol II promoter and the sgRNAs are under the control of a pol III promoter. A multiplex system with three targets was designed targeting two different genes, GmIPK1 and GmIPK2, coding for enzymes from the phytic acid synthesis pathway. Both systems were tested using the hairy root soybean methodology. Results showed gene-specific edition. For the GmIPK1 gene, edition was observed in both configurations, with a deletion of 1 to 749 base pairs; however, the TCTU showed higher indel frequencies. For GmIPK2 major exclusions were observed in both systems, but the editing efficiency was low for STU. Both systems (STU or TCTU) have been shown to be capable of promoting effective gene editing in soybean. The TCTU configuration proved to be preferable, since it was more efficient. The STU system was less efficient, but the size of the CRISPR/Cas cassette was smaller.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Over many years, several methodologies have been developed for plant genome editing, such as the use of meganucleases (Adli 2018), zinc-finger nucleases (ZFNs) (Davies et al. 2017) and TALENs (transcription activator-like effector nucleases) (Du et al. 2016). However, these strategies have shortcomings that limit effective use of these technologies, such as low editing efficiency, complicated vector assembly and off-target mutations (Gaj et al. 2013; Adli 2018). In this context, CRISPR/Cas (Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR associated protein) has emerged and become prevalent because of its simplicity when recognizing the site to be modified and flexibility of use (Pellagatti et al. 2015; Wolter et al. 2019).
CRISPR/Cas was originally part of the prokaryotic immune system (Wiedenheft et al. 2012). This system is represented by the presence of a Cas endonuclease with two catalytic domains responsible for the cleavage of the double-strand DNA: HNH domain and RuvC nuclease and a single guide RNA complex (sgRNA), which is the fusion of a mature crRNA (CRISPR-derived RNA) and a tracrRNA (trans-activating RNA), creating a functional structure for activation of the endonuclease and recognition of the target sequence (Shan et al. 2013; Barrangou 2015). A Protospacer Adjacent Motifs (PAM) downstream to target sequence determines the anchorage location and the site of the double strand break (DSB) on the DNA strands (Jinek et al. 2012).
Despite major advances in conventional plant breeding, the development of improved plant varieties is proceeding less quickly than necessary, given the increased demand for food caused by the rapid global population growth (Gao 2018; Ahmar et al 2020). Germplasm accesses sometimes have no information available or absence of natural variations to be used for the development of events with desirable characteristics (Marathe et al 2018). For some crops, such as soybean, which have a complex genome (Schmutz et al. 2010), precise and efficient strategies for gene function analysis and crop improvement are of great interest, mainly for breeding programs. The CRISPR/Cas system has already been used to edit genes in soybean (Chilcoat et al. 2017; Cai et al. 2018; Bao et al. 2019; Cheng et al. 2019). However, due to the relatively long time required to obtain a transgenic soybean plant, it is important to have a practical system to validate genome-editing constructs before generating genetically modified plants. Although there are several in silico prediction tools for the design/choice of sgRNA, rapid in vivo experimental studies might be more reliable in determining the best target sequences (Lee et al. 2017; Zhang et al. 2016).
To find the appropriate system to edit the soybean genome, we evaluated the efficiency of a single transcriptional unit (STU) strategy for the CRISPR/Cas9 system compared to a two-component transcriptional unit (TCTU) using the hairy root-based expression system, evaluating a multiplex sgRNAs. We chose two genes (GmIPK1 and GmIPK2) coding for enzymes from the phytate synthesis pathway (Sparvoli and Cominelli 2015) as a model.
Materials and methods
Selection of CRISPR/Cas target sites
The soybean genomic sequences for two inositol-pentakisphosphate 2-kinase genes (Fig. 1), Glyma.14G072200 and Glyma.12G240900 (GmIPK1 and GmIPK2, respectively), were obtained from Phytozome (https://phytozome-next.jgi.doe.gov, Goodstein et al. 2012). The soybean gene models were based on the Wm82.a2.v1 genome assembly (Schmutz et al. 2010). These genes were chosen based on the fact that they are potential candidates for decreased phytic acid content in seeds. GmIPK1 (Glyma.14G072200) gene is 4158 nucleotides in length distributed in seven exons. Additionally, it has two paralogs, namely Glyma.04G030000 and Glyma.06G03010. Although the gene on chromosome 14 shares similarity at the transcript level with the rest (85.9% and 85% similarity, respectively), they are different at the DNA level. We focused on the GmIPK1 gene on chromosome 14 because it has the highest expression levels in immature soybean seeds (Yuan et al. 2012), while Glyma.12G240900 (GmIPK2) is a much shorter gene (1630 nucleotides) with a single exon.
Scheme showing the targeted sequences for IPK1 gene (a) and IPK2 gene (b) and location of the primers. Blue arrow: exons; light orange oblong: introns; gray oblong: 5′ and 3′UTR (untranslated region). L: Location of target. The scheme shows all primer positions used in this work. The sequence primers are in the supplementary file. (Color figure online)
All possible Streptococcus pyogenes Cas9 (SpCas9) target sites within the obtained sequences were identified with Geneious Prime 2019.2.3 (https://www.geneious.com). Potential off-target effects were calculated according to the method previously developed by Hsu et al. (2013). Predicted on-target activity was estimated with the online software GPP sgRNA Designer (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design) using Azimuth 2.0 (Doench et al 2016, Sanson et al. 2018). Three target sites were selected for each gene based on their genomic locations, potential off-target score and predicted on target activity (Table 1).
Vector construction
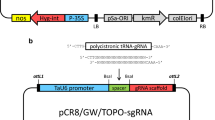
The binary CRISPR/Cas9 vector p201-EGFP-C9 was assembled to test the STU and TCTU systems (Fig. 2). The vector was constructed using the p201-EGFP-C9, which contains the EGFP (Enhanced Green Fluorescent Protein) (Chiu et al. 1996) reporter gene under the control of the CsVMV promoter (from cassava vein mosaic virus) (Verdaguer et al. 1996), the nos gene terminator (An et al. 1985) and the coding sequence for SpCas9 (Mali et al. 2013), which was obtained as Addgene plasmid #41815 and is under control of the GmUbi3 promoter (Chiera et al. 2007) and the PsRbcs terminator (Schardl et al. 1987). The sgRNAs of the TCTU configurations are under the control of the MtU6 promoter from Medicago truncatula (Kim et al. 2013). In the STU system (Wang et al. 2018), each sgRNA target sequence was combined with the optimized scaffold (Dang et al. 2015) and separated by a unique sequence (UNS), and these were used as linkers to facilitate cloning (Torella et al. 2014).
Schematic diagram of two CRISPR/Cas9 expression systems: dual promoter system (TCTU) and single transcriptional unit (STU) system. Hairy root vector CRISPR/Cas9 is used to edit the GmIPK1 and GmIPK2 genes with the two systems: TCTU (a) and STU (b). The Cas 9 endonuclease from Streptococcus pyogenes (SpCas9) is under the control of the ubiquitin-3 promoter from glycine max (orange). The cassette of the EGFP reporter gene is under the control of the CsVMV (cassava vein mosaic virus) promoter (light green) and T-nos terminator (nopaline synthase). In the TCTU system each sgRNA is driven by the U6 promoter from Medicago truncatula separated for a UNS sequence. In the STU system the sgRNAs are only separated for the UNS sequence
The CRISPR/Cas9 TCTU configuration cassettes were assembled and cloned into the p201-EGFP-C9 digested with SpeI, generating the final 15,454 bp vector (MtU6-sgRNA(thrice)-GmUbi-SpCAs9-RbcsT) (Fig. 2a). For assembly of the STU vectors, the cassettes were cloned into the p201-EGFP-C9 digested with AvrII, generating the final 14,396 bp vectors (GmUbi-SpCAs9-sgRNA(thrice) -RbcsT) (Fig. 2b).
Soybean hairy root transformation
The Glycine max cultivar Jack was used in this study. This variety is the best for somatic embryogenesis and plant regeneration (Raza et al. 2020). Soybean hairy root transformation was performed as previously described by Jacobs et al. (2015) with no selection pressure. EGFP fluorescence was used to detect transgenic roots 20 days after transformation. Each positive root was considered as a single event and used for downstream analysis. Additionally, PCR was used to detect changes. A 1322-bp amplicon was expected for the GmIPK1 gene using the primers IPK1-97F: ACACAATTCCTTTCCCACCA and IPK1-1399R: AGCAGAGGCTAGATCCTTGA. For the GmIPK2 gene, a 1053-bp amplicon was expected using primers IPK2-80F: TTGCATTGCTTTGTGTAAGG and IPK2-1113R: CTGCGACACTAATTCAAGCA.
Detection of CRISPR/Cas9-mediated gene editing
Genomic DNA was isolated from GFP-positive roots (approximately 2 mm from the root tip) using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA). Gene editing patterns were determined by amplicon sequencing using specific primers tailed with partial TruSeq Illumina adapter sequences (Supplementary Table 1). PCR products from the same sample were pooled and individually barcoded using indexed universal iTru/iNext primers. Indexed samples were pooled in equimolar amounts and sequenced on an Illumina MiSeq using 150-bp paired-end (2 × 150 PE) mode at the Georgia Genomics and Bioinformatics Core, University of Georgia. Amplicon sequencing data were analyzed using AGEseq software (Xue and Tsai 2015).
Results
A single transcriptional unit (STU) system was compared to the two-component transcriptional (TCTU) system in soybean using hairy root transformation. We evaluated two strategies to express multiple sgRNAs for the genes GmIPK1 and GmIPK2 that code for two different phytic acid biosynthetic enzymes: GmIPK2 codes for an inositol-polyphosphate multikinase, mainly involved in the production of inositol 1,3,4,5,6-pentakisphosphate, while GmIPK1 codes for an inositol-pentakisphosphate 2-kinase, which phosphorylates inositol 1,3,4,5,6-pentakisphosphate in position 2, releasing phytic acid (Sparvoli and Cominelli, 2015). Genomic sequences of both genes were obtaining from the G. max genome (assembly Wm82.a2.v1, Schmutz et al. 2010). A total of 46 SpCas9 target sites were identified for GmIPK1, 22 of them in the coding sequence (CDS), from which 3 target sites were chosen. Targets were located in exon 1, exon 2 and exon 3, respectively, and were named GmIPK1-target1, GmIPK1-target2 and GmIPK1-target3. Their potential off-target score ranged from 1.87 to 7.11% (all below the 10% threshold) with predicted on-target activity ranging from 45.12 to 67.65%. All three targets are in sense orientation. Of the 65 target sequences identified from the GmIPK2 gene, 47 targeted the CDS. The three selected guides for this gene were all chosen in the anti-sense orientation and separated by at least 75 bp. Their potential off-target score ranged from 3.27 to 7.96% and predicted on-target activity between 57.96 and 66.46%. Selected targets for GmIPK2 were named GmIPK2-target1, GmIPK2-target2 and GmIPK2-target3. Details on selected targets are summarized on Table 1.
Four different p201-EGFP-C9 plasmids, IPK1-TCTU, IPK1-STU, IPK2-TCTU and IPK2-STU, were used for soybean hairy root transformation. For each construct, ten cotyledons from germinating seeds were transformed (Fig. 3a), and transgenic hairy roots were confirmed by visual inspection of EGFP florescence (Fig. 3b). PCR analysis was used for a primary evaluation to detect changes before Sanger sequencing; 1322- and 1053-bp amplicons were expected for the GmIPK1 and GmIPK2 genes, respectively (Fig. 3c). Of a total of 15 individual EGFP-positive roots collected from each transformation, genomic DNA was isolated, and targeted amplicon sequencing libraries were prepared to determine CRISPR/Cas9-mediated gene editing.
a Hairy-root process and detection of fragments Preparation of the cotyledons, showing the cut for inoculation (outlined in red); b roots grown in MS medium under EGFP flashlight; c detection of the fragment of the GmIPK1 gene by PCR on agarose gel under UV light, expected fragment of 1233 bp. Lines 1–6: IPK1-STU, lines 7–11: IPK1-TCTU, +: plant control. d Detection of the fragment of the GmIPK2 gene by PCR on agarose gel under UV light, expected fragment of 1053 bp. Lines 1–5: IPK1-STU, lines 7–10: IPK1-TCTU, +: plant control. M: 1-kb ladder LIBPBio
Illumina MiSeq paired-end sequencing technology yielded a total of 376,180 paired-end clean reads from GmIPK1 and GmIPK2 amplicon-seq libraries. Subsequent analysis showed that 180,971 (48.11%) and 195,209 (51.89%) paired-end reads were obtained from GmIPK1 and GmIPK2, respectively. CRISPR/Cas-induced gene modification patterns were analyzed using AGEseq software (Supplementary Table 2 and Supplementary Table 3). The STU system showed very low editing rates for the GmIPK1 gene, with wild-type sequences detected on 99.1% of the reads. Six of 15 samples had no edits, while 5 only had < 2% editing at 2 of the 3 sites. Editing occurred mainly in four different events. GmIPK1-STU-#04 reads with a 1-bp deletion were found in all three sites at a low but detectable level (< 0.3%). The sequence between GmIPK1-Target 1 and GmIPK1-Target 3 (751 bp) was deleted in 156 of the reads (3.99% and 5.40%) (Fig. 4). GmIPK1-STU-#06 only displayed modifications for GmIPK1-Target 3, with a 5-bp deletion detected in 19% of the reads. Modifications were identified in all three sites of the GmIPK1-STU-#08 event (Fig. 4). A 2-bp deletion was detected in GmIPK1-Target 1 with a frequency > 87%. At the same time, a 4-bp deletion sequence was observed in 18.52% of the reads of GmIPK1-Target 3. Finally, a reduced percentage of the reads mapped to GmIPK1-Target 2 harbored modifications, with nearly 4% of the reads showing a 3-bp deletion and another 3% showing a single base insertion. For GmIPK1-STU-#09, a 749-bp deletion comprising the sequence between GmIPK1-Target 1 and GmIPK1-Target 3 was observed.
Comparison of the single transcriptional unit (STU) and two-component transcriptional unit (TCTU) CRISPR/Cas9 gene editing systems in soybean hairy roots for the GmIPK1 gene. a Schematic illustration of the GmIPK1 gene (Glyma.14G072200). All exons are represented by solid green arrows, and introns are represented as lines. Both the 5′ and 3′ untranslated regions (UTR) are depicted as gray boxes. GmIPK1-Target 1, GmIPK1-Target 2 and GmIPK1-Target 3 are represented by blue, pink and yellow arrows, respectively. b CRISPR/Cas9 induced mutations by the STU and TCTU systems. Wild-type sequences are in green, deletions are shown as dashes, and SNPs are shown in black. The targeted sequences are highlighted in blue, pink and yellow, and the PAM is underlined in red. Percentages next to the sequences indicate the number of reads mapped over the total number of reads sequenced for a given target within each sample. ND: Not detected. (Color figure online)
Higher indel frequencies were observed when the TCTU system was tested on the GmIPK1 gene (73.20% reads carried a mutation) (Fig. 4). Different sgRNA sequences led to a wide collection of mutations, ranging from small deletions (1 to 12 bp) or insertions (only 1 bp) to large deletions that removed the sequences between two sgRNAs. Only two events (GmIPK1-TCTU-#02 and GmIPK1-TCTU-#14) harbored no modifications. The most commonly found mutations for both STU and TCTU are as follows: for IPK1-Target 3–751 bp, −4 bp and −1 bp; for GmIPK1-Target 2 −1 bp (STU), −3 and −7 bp only in TCTU; for IPK1-Target 1–751 bp, −3 bp and −4 bp (only for the TCTU). As previously detected in IPK1-STU samples, deletions of about 751 bp (comprising the sequence between GmIPK1-Target 1 and GmIPK1-Target 3) are frequently present in IPK1-TCTU samples (8/15). Additional gene segment deletions between GmIPK1-Target 1 and GmIPK1-Target 2 (GmIPK1-TCTU-#04 and GmIPK1-TCTU-#06), GmIPK1-Target 2 and GmIPK1-Target 3 (GmIPK1-TCTU-#05) were detected as well. Average editing frequencies of 89.30%, 69.13% and 79.39% were estimated for GmIPK1-Target 1, GmIPK1-Target 2 and GmIPK1-Target 3, respectively. However, some single events such as GmIPK1-TCTU-#09, GmIPK1-TCTU-#12 and GmIPK1-TCTU-#15 show editing levels > 95% in all three sites.
For the GmIPK2 gene (Fig. 5), the STU system delivered overall 96.38% of wild-type sequencing reads. GmIPK2-Target 1 and GmIPK2-Target 3 showed similar editing rates (12.71% and 13.76%, respectively). In contrast, GmIPK2-Target 2 presented an almost undetectable editing rate of 0.04%. The most identified modification was a deletion of about 215 bp between the GmIPK2-Target 1 and GmIPK2-Target 3 target sites. This modification was present in all 15 events evaluated using the STU system with frequencies up to 54.08%/33.71% (event GmIPK2-STU-#11). No plants were identified without CRISPR/Cas9-mediated edits. Smaller deletions (1 or 2 bp) were also common modifications identified on IPK2-STU events. Again, the TCTU system induced a much higher number of modifications than the STU system (79.33% and 96.38%), but the differences between the STU and TCTU systems were less pronounced for GmIPK2 than for GmIPK1. Despite all events showing some kind of edition, GmIPK2-Target 2 had the lowest editing rate with only a 0.67% rate (10 out of the 15 events showed only wild-type reads). The most frequent edit was the excision of the fragment between GmIPK2-Target 1 and GmIPK2-Target 3. Another large deletion, i.e., between GmIPK2-Target 1 and GmIPK2-Target 2, was only detected in event GmIPK2-TCTU-#05. Events GmIPK2-TCTU-#08 and GmIPK2-TCTU-#04 presented a higher editing rate, ranging between from 60.85% and 41.57%. Only IPK2-TCTU-#04 presented any editing on GmIPK2-Target 2. GmIPK1-TCTU-#02 library had some issues and was sequenced at low depth coverage, resulting in no sequencing information available on the GmIPK1-target 2.
Comparison of the single transcriptional unit (STU) and two-component transcriptional unit (TCTU) CRISPR/Cas9 gene editing systems in soybean hairy roots for the GmIPK2 gene. a Schematic representation of the GmIPK2 gene (Glyma.12G240900). All exons are represented by solid green arrows, and introns are shown as lines. The 5′ and 3′ untranslated regions (UTR) are shown as gray boxes. GmIPK2-Target 1, GmIPK2-Target 2 and GmIPK2-Target 3 are represented by blue, pink and yellow arrows, respectively. b CRISPR/Cas9 induced mutations in both STU and TCTU systems. Wild-type sequences are in green, deletions are shown as dashes, and SNPs are shown in black. The targeted sequences are highlighted blue, pink and yellow, and the PAM is highlighted and underlined in red. Percentages next to sequences indicate the number of reads mapped over the total number of reads sequenced for a given target within each sample. ND: Not detected. (Color figure online)
Discussion
Although CRISPR technology has been revolutionizing gene edition since its discovery, new advances have made this system even more efficient (Fiaz et al 2019). Traditionally, the CRISPR/Cas9 system uses a combination of two types of promoters, one from the class II Pol promoter that regulates Cas endonuclease expression and other from the class III that regulates the sgRNA (Tang et al. 2019). Some limitations have been become apparent after it was developed because of the poor characterization of pol III promoters in some organisms (Sun et al. 2015) and uncoordinated activity of Cas9 and sgRNA expression, given they are being driven by different promoters (Tang et al. 2019), or repetitive use of the sequence that might cause variation of expression levels and transgene silencing (Ma et al. 2015). The CRISPR/Cas9 system can be simplified even further without compromising its efficiency. A more compact system such as the STU design would enhance flexibility and simplify gRNA construction. In this context, we evaluated the efficiency of editing two soybean genes by using CRISPR/Cas9 with the sgRNA in two different configurations, i.e., the conventional (TCTU) and simplified (STU) strategies. It was possible to reduce the CRISPR/Cas9 expression cassette from 7050 bp in the TCTU system to 5990 bp in the STU system, a reduction of > 1000 bp. Given that each sgRNA has 120 bp, the remaining 1000 bp is equivalent to approximately eight new sgRNAs in a simplified design, which means eight new targets for other genes in a multiplex system.
In this work, when we compare the systems, the TCTU and STU gave gene-specific results, but both were able to create indels from DSBs. These included deletions ranging from 1 to 10 bp for IPK2. For IPK1, the deletion size varied from 1 to 12 bp. They also included 1-bp insertions that were detected for both genes. In addition, we obtained a 752-bp deletion in GmIPK1 gene when the fragment between GmIPK1-Target 1 and -Target 3 was deleted. For the GmIPK2 gene, the largest deletion was 217 bp, which is the distance between GmIPK2-Target 1 and -Target 3. Overall, the major difference is that the TCTU system consistently gave higher editing frequencies for both genes. This difference in frequency may simply indicate that the TCTU is the superior system. The STU system employed here was based on reports by Wang et al. (2018) and Mikami et al. (2017) wherein Cas9 multiplexed gRNAs were shown to achieve editing efficiencies from 50 to 94%. An advantage of relying on the plant endogenous processing machinery to cleave the RNA is that it simplifies gRNA construct assembly. Other approaches have used the Cys4 RNA cleaving system (Cermak et al. 2017), exogenous ribozymes (Gao et al. 2015; Tang et al. 2016) and the polycistronic tRNA-gRNA gene system (Xie et al. 2015) to release individual gRNAs, and these approaches may circumvent the low frequencies obtained here with the STU system. A similar simplified transcriptional unit CRISPR system (STU) was tested in rice and showed editing rate efficiencies between 29 and 38%, and the efficiency remained the same when compared to the traditional system. Point mutations were observed at the cleavage site or at most two bases ahead (Tang et al. 2016). With additional optimization, the mutation rate was close to 50% across six targets (Tang et al. 2019). Different endonucleases were tested with the STU configuration (FnCpf1, LbCpf1 or Cas9) using a multiplex system, and the efficiency was > 50% (Wang et al. 2018).
The successful activity from the CRISPR/Cas9 complex activity is associated with target sequence characteristics (Doench et al. 2014), but we are not aware of reports describing preferences based on target position within a multiplex system. Notably, the entire fragment between targets 1 and 3 was more frequently removed for the GmIPK2 gene (smaller gene) as all events from both STU and TCTU had this deletion in frequencies up to 60.8% for event 4 from the TCTU system. For the GmIPK1 gene, the entire segment between targets 1 and 3 (approximately 715 bp long, depending on the cut) was lost in 4 of 15 events with the STU system and 8 of 15 events for the TCTU system. Edits for target 2 of the GmIPK2 gene were rare. The GmIPK2 is the smaller gene, and the proximity of the target sites (101 bp between target 1 and 2, 76 bp between target 3 and 2) might have led to interference with the Cas endonucleases.
In diploid cells, three zygosities are possible, monoallelic, heterozygous diallelic (more commonly referred to as biallelic) and homozygous diallelic (Luttgeharm et al. 2017). Knowing the nature of the mutations helps understand the efficiency of each system. In our hands, biallelic editing is rare for the STU system, although event 8 for GmIPK1 had an 87.5% biallelic editing frequency for target 1, consisting of a 2-bp deletion. For GmIPK1 with the TCTU system, all three targets were biallelic for events 4 and 9, while events 6 and 8 were monoallelic for targets 1 and 3. Event 10 was also biallelic for targets 1 and 3, but monoallelic for target 2. Diallelic edits were predominantly heterozygous. None of the targets in the GmIPK2 gene resulted in diallelic edits with either system.
In conclusion, the TCTU and STU systems were effective in editing soybean genes coding for enzymes from the phytic acid synthesis pathway, though the higher editing frequencies obtained with the TCTU system make this the preferred technology. This technology will be the foundation for efficiently editing genes in the soybean genome as well as production of low phytic acid genotypes.
Availability of data and materials
All raw data using in this manuscript will be made available upon request.
References
Adli M (2018) The CRISPR tool kit for genome editing and beyond. Nat Commun 9:1911
Ahmar S, Gill RA, Jung KH, Faheem A, Qasim MU, et al. (2020). Conventional and molecular techniques from simple breeding to speed breeding in crop plants: recent advances and future outlook. Int J Mol Sci. 2020 21(7):2590
An G, Watson BD, Stachel S, Gordon MP, Nester EW (1985) New cloning vehicles for transformation of higher plants. EMBO J 4:277–284
Bao A, Chen H, Chen L, Chen S, Hao Q et al (2019) CRISPR/Cas9-mediated targeted mutagenesis of GmSPL9 genes alters plant architecture in soybean. BMC Plant Biol 19(1):131
Barrangou R (2015) Diversity of CRISPR-Cas immune systems and molecular machines. Genome Biol 16:247
Cai Y, Chen L, Sun S, Wu C, Yao W, et al. (2018) CRISPR/Cas9-Mediated deletion of large genomic fragments in soybean. Int J Mol Sci 19.
Cermak T, Curtin SJ, Gil-Humanes J, Cegan R, Kono TJY et al (2017) A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell 29:1196–1217
Cheng Q, Dong L, Su T, Li T, Gan Z et al (2019) CRISPR/Cas9-mediated targeted mutagenesis of GmLHY genes alters plant height and internode length in soybean. BMC Plant Biol 19:562
Chiera JM, Bouchard RA, Dorsey SL, Park EH, Buenrostro-Nava MT, Ling PP, Finer JJ (2007) Isolation of two highly active soybean (Glycine max (L.) Merr.) promoters and their characterization using a new automated image collection and analysis system. Plant Cell Rep 26:1501–1509
Chilcoat D, Liu Z-B, Sander J (2017) Use of CRISPR/Cas9 for Crop Improvement in Maize and Soybean. Prog Mol Biol Transl Sci 149:27–46
Chiu W-L, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J (1996) Engineered GFP as a vital reporter in plants. Curr Biol 6:325–330
Dang Y, Jia G, Choi J, Ma H, Anaya E et al (2015) Optimizing sgRNA structure to improve CRISPR-Cas9 knockout efficiency. Genome Biol 16:280
Davies JP, Kumar S, Sastry-Dent L (2017) Use of Zinc-Finger Nucleases for Crop Improvement. Progress in Molecular Biology and Transl Sci 149:47–63
Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M et al (2014) Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol 32:1262–1267
Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Root DE (2016) Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34(2):184–191
Du H, Zeng X, Zhao M, Cui X, Wang Q et al (2016) Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. J Biotechnol 217:90–97
Fiaz S, Ahmad S, Riaz A, Noor MA, Wang X et al (2019) Applications of the CRISPR/Cas9 System for Rice Grain Quality Improvement: Perspectives and Opportunities. Int J Mol Sci 20(4):888
Gaj T, Gersbach CA, Barbas CF 3rd (2013) ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31:397–405
Gao Y, Zhang Y, Zhang D, Dai X, Estelle M, Zhao Y (2015) Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. Proc Natl Acad Sci U S A 112(7):2275–2280
Gao C (2018) The future of CRISPR technologies in agriculture. Nat Rev Mol Cell Biol 19:275–276
Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic acids research, 40(Database issue), D1178–D1186.
Hsu PD, Scott DA, Weinstein J, Ran FA, Konermann S et al (2013) DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31(9):827–832
Jacobs TB, LaFayette PR, Schmitz RJ, Parrott WA (2015) Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol 15:16
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA et al (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821
Kim GB, Nam YW (2013) Isolation and characterization of Medicago truncatula U6 promoters for the construction of small hairpin RNA-mediated gene silencing vectors. Plant Mol Biol Report 31(3):581–593
Lee CM, Zhu H, Davis TH, Deshmukh H (2017) Bao G (2017) Design and Validation of CRISPR/Cas9 Systems for Targeted Gene Modification in Induced Pluripotent Stem Cells. Methods Mol Biol 1498:3–21
Luttgeharm KD, Wong K-S, Siembieda S (2017) Heteroduplex cleavage assay for screening of probable zygosities resulting from CRISPR mutations in diploid single cell lines. Biotechniques 62:268–274
Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B et al (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8:1274–1284
Mali P, Yang LH, Esvelt KM, Aach J, Guell M, DiCarlo JE et al (2013) RNA-Guided human genome engineering via Cas9. Science 339(6121):823–826
Marathe A, Krishnan V, Mahajan MM, Thimmegowda V, Dahuja A, et al, (2018). A. Characterization and molecular modeling of Inositol 1,3,4 tris phosphate 5/6 kinase-2 from Glycine max (L) Merr.: comprehending its evolutionary conservancy at functional level. 3 Biotech. 8 (1):50.
McCarty NS, Graham AE, Studená L, Ledesma-Amaro R (2020) Multiplexed CRISPR technologies for gene editing and transcriptional regulation. Nat Commun 11(1):1281
Mikami M, Toki S, Endo M (2017) In Planta Processing of the SpCas9-gRNA Complex. Plant Cell Physiol 58:1857–1867
Pellagatti A, Dolatshad H, Valletta S, Boultwood J (2015) Application of CRISPR/Cas9 genome editing to the study and treatment of disease. Arch Toxicol 89:1023–1034
Punjabi M, Bharadvaja N, Sachdev A, Krishnan V (2018) Molecular characterization, modeling, and docking analysis of late phytic acid biosynthesis pathway gene, inositol polyphosphate 6-/3-/5-kinase, a potential candidate for developing low phytate crops.8 (8): 344
Raza G, Singh MB, Bhalla PL (2020) Somatic Embryogenesis and Plant Regeneration from Commercial Soybean Cultivars. Plants 9(1):38
Sanson KR, Hanna RE, Hegde M, Donovan KF, Strand C, et al (2018) Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat Commun 9:5416
Schardl CL, Byrd AD, Benzion G, Altschuler MA, Hildebrand DA et al (1987) Design and construction of a versatile system for the expression of foreign genes in plants. Gene 61:1–11
Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T et al (2010) Genome sequence of the palaeopolyploid soybean. Nature 463:178–183
Shan Q, Wang Y, Li J, Zhang Y, Chen K et al (2013) Targeted genome modification of crop plants using a CRISPR-Cas system: Nat Biotechnol 31(8):686–688
Sparvoli F, Cominelli E (2015) Seed Biofortification and Phytic Acid Reduction: A Conflict of Interest for the Plant? Plants 4:728–755
Sun X, Hu Z, Chen R, Jiang Q, Song G, et al. (2015) Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci Rep 5.
Tang X, Zheng X, Qi Y, Zhang D, Cheng Y, Tang A, Voytas DF et al (2016) A single transcript CRISPR-Cas9 system for efficient genome editing in plants. Mol Plant 9:1088–1091
Tang X, Ren Q, Yang L, Bao Y, Zhong Z et al (2019) Single transcript unit CRISPR 2.0 systems for robust Cas9 and Cas12a mediated plant genome editing. Plant Biotechnol J 17:1431–1445
Torella JP, Lienert F, Boehm CR, Chen JH, Way JC et al (2014) Unique nucleotide sequence-guided assembly of repetitive DNA parts for synthetic biology applications. Nat Protoc 9:2075–2089
Verdaguer B, de Kochko A, Beachy RN et al (1996) Isolation and expression in transgenic tobacco and rice plants, of the cassava vein mosaic virus (CVMV) promoter. Plant Mol Biol 31:1129–1139. https://doi.org/10.1007/BF00040830
Wang M, Mao Y, Lu Y, Wang Z, Tao X, et al. (2018) Multiplex gene editing in rice with simplified CRISPR-Cpf1 and CRISPR-Cas9 systems: J Integr Plant Biol. 2018;60(8):626–631.
Wiedenheft B, Sternberg SH, Doudna JA (2012) RNA-guided genetic silencing systems in bacteria and archaea. Nature 482:331–338
Wolter F, Schindele P, Puchta H (2019) Plant breeding at the speed of light: the power of CRISPR/Cas to generate directed genetic diversity at multiple sites. BMC Plant Biol 19:176
Xie K, Mikenberg B, Yang Y (2014) Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci USA 112(11):3570–3575
Xue LJ, Tsai CJ (2015) AGEseq: Analysis of Genome Editing by Sequencing. Mol Plant 8:1428–1430
Yuan FJ, Zhu DH, Tan YY, Dong DK, Fu XJ et al (2012) Identification and characterization of the soybean IPK1 ortholog of a low phytic acid mutant reveals an exon-excluding splice-site mutation. Theor Appl Genet 125(7):1413–1423
Zhang Z, Mao Y, Ha S, Liu W, Botella JR et al (2016) A multiplex CRISPR/Cas9 platform for fast and efficient editing of multiple genes in Arabidopsis. Plant Cell Rep 35:1519–2153
Acknowledgements
This study was supported in part by Empresa Brasileira de Pesquisa Agropecuária (Embrapa), with funds allocated to the Georgia Agricultural Experiment Stations (Athens, GA, USA) and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brazil). J. Carrijo was supported by a fellowship from Cordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Brazil).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by JC, EB and NT. The first draft of the manuscript was written by JC and GRV, and all authors commented on and revised previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Consent to participate and publication
All authors consented to their participation and the publication of this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Carrijo, J., Illa-Berenguer, E., LaFayette, P. et al. Two efficient CRISPR/Cas9 systems for gene editing in soybean. Transgenic Res 30, 239–249 (2021). https://doi.org/10.1007/s11248-021-00246-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-021-00246-x