Abstract
Genetically modified (GM) pigs hold great promises for pig genetic improvement, human health and life science. When GM pigs are produced, selectable marker genes (SMGs) are usually introduced into their genomes for host cell or animal recognition. However, the SMGs that remain in GM pigs might have multiple side effects. To avoid the possible side effects caused by the SMGs, they should be removed from the genome of GM pigs before their commercialization. The Cre recombinase is commonly used to delete the LoxP sites-flanked SMGs from the genome of GM animals. Although SMG-free GM pigs have been generated by Cre-mediated recombination, more efficient and cost-effective approaches are essential for the commercialization of SMG-free GM pigs. In this article we describe the production of a recombinant Cre protein containing a cell-penetrating and a nuclear localization signal peptide in one construct. This engineered Cre enzyme can efficiently excise the LoxP-flanked SMGs in cultured fibroblasts isolated from a transgenic pig, which then can be used as nuclear donor cells to generate live SMG-free GM pigs harboring a desired transgene by somatic cell nuclear transfer. This study describes an efficient and far-less costly method for production of SMG-free GM pigs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Genetically modified (GM) pigs are valuable for swine breeding, human biomedicine and life sciences (Hammer et al. 1985; Montag et al. 2018; Tong et al. 2011; Whyte and Prather 2011; Zhang et al. 2018). During the production of GM pigs, selectable marker genes (SMGs) such as enhanced green fluorescence protein (EGFP) and neomycin resistance (Neo) genes are usually introduced into the genome of GM pigs. These integrated SMGs are useful for identification of GM pigs, or the selection of the GM somatic cells for the production of GM pigs by somatic cell nuclear transfer (SCNT) (Lai et al. 2002; Naruse et al. 2005; Park et al. 2001; Wu et al. 2013). However, the presence of SMGs in GM animals might cause the following problems: (1) the expression of the SMGs or the regulatory elements of the SMGs might interfere with the expression pattern of adjacent integrated foreign genes or endogenous genes, and thus result in undesired phenotypes; (2) the proteins encoded by some SMGs, such as the neomycin resistance gene, may confer host GM animals resistance to some drugs and thus undermine their therapeutic effects when they are used to treat infections in host GM animals; (3) the presence of SMGs in GM animals will restrict the next round of genetic manipulation in the same host, because given that the number of currently available SMGs is very small, it is hard to find a new SMG that is not carrying by the host for the next round of genetic modification. To prevent the above mentioned problems, SMGs should be removed from the genome of GM pigs.
The Cre/LoxP system is commonly employed to delete the SMGs from the genome of GM animals (McLellan et al. 2017; Sauer and Henderson 1988; Sunaga et al. 1997). Here, the Cre enzyme catalyzes the recombination of the SMG-flanking LoxP sites, resulting in removal of the SMGs from the DNA fragment between the two LoxP sites (Hoess and Abremski 1984; Hoess et al. 1982).
Recently, SMG-free GM pigs have been produced by SCNT via transfection of Cre mRNA into somatic cells harboring LoxP-flanked SMGs (Bi et al. 2016), or by injection of Cre mRNA into fertilization-derived zygotes carrying LoxP-flanked SMGs (Whitworth et al. 2018). However, due to its low stability, Cre mRNA may be degraded after being delivered into cells or zygotes. Furthermore, the injection of Cre DNA, mRNA or protein into zygotes to remove SMGs may result in mosaic animals with incomplete SMG deletion as Cre-catalyzed SMG excision at times occurs only in partial blastomeres after cleavage of injected embryos (Sunaga et al. 1997; Araki et al. 1995; Kaartinen and Nagy 2001). Therefore, a more suitable approach is to use Cre proteins, with their longer half-life, to remove SMGs from somatic cells, and consequently using them as nuclear donors for the production of SMG-free GM animals through SCNT (Kang et al. 2018). However, the efficiency of this approach still needs to be further improved to achieve efficient production of GM pigs.
Hence, we engineered a Cre protein carrying a cell-penetrating peptide called transactivator of transcription (TAT) (Vives et al. 1997), and a nuclear localization signal (NLS) peptide (Kalderon et al. 1984). We then used this recombinant Cre enzyme to efficiently remove the SMGs from the genome of ear fibroblasts isolated from a GM pig carrying an anti-porcine circovirus type 2 (PCV2) short hairpin RNA (shRNA) transgene. These SMG-free fibroblasts were then used to successfully generate healthy SMG-free GM pigs by SCNT.
Materials and methods
Plasmid construction
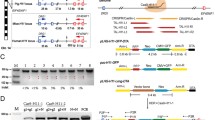
A DNA fragment carrying a designed anti-PCV2 shRNA expression cassette and a LoxP-flanked SMG (Neo-2A-EGFP) expression cassette was synthesized and inserted into the piggyBac plasmid pCyL50 (Wang et al. 2008) to generate the plasmid pCyL50-shRNA (see the plasmid map in Fig. 1a). The piggyBac transposase expression plasmid mPB (Cadinanos and Bradley 2007) was a kind gift from The Wellcome Trust Sanger Institute (Cambridgeshire, UK). The HTNCre recombinase expression plasmid pTriEx-HTNC (Plasmid #13763) was purchased from Addgene (Watertown, MA, USA).
Production of GM founder pigs by cytoplasmic injection combined with piggyBac transposition-mediated gene transfer. a Map of the anti-PCV2 shRNA expression plasmid. pB 3′TR: pB transposon 3′ terminal repeat element. H1: H1 promoter. Anti-PCV2 shRNA: anti-PCV2 hairpin siRNA. LoxP: a DNA sequence specifically recognized by Cre recombinase. CMV: cytomegalovirus promoter. Neo: neomycin-resistant gene. 2A: 2A peptide. EGFP: enhanced green fluorescent protein gene. pB 5′TR: pB transposon 5′ terminal repeat element. HindIII: the restriction enzyme used for Southern blot and inverse PCR analysis. Probe: the probe used for Southern blot analysis. P1, P15, P2, P4, P13, P14, P16 and P3 are the primers used for analysis in this study (also see Table 1). b EGFP expression in produced GM founder piglets. Representative pictures of piglets and their organs showing that green fluorescence was clearly observed on GM piglet and their organs, but not visualized on wild-type (WT) piglets and their organs under blue light. (Color figure online)
Cytoplasmic injection
Cytoplasmic injection of the in vivo fertilization-derived pig zygotes was performed as we previously described (Li et al. 2014). Briefly, zygotes of Duroc breed were flushed out from the oviducts of artificially inseminated sows. The pCyL50-shRNA (10 ng/µL) and mPB (40 ng/µL) plasmids were diluted in 10 mM Tris-HCl (pH 7.6) and 0.25 mM EDTA (pH 8.0) and backfilled into a bevel-tipped injection capillary pipette with a 10-µm internal diameter. One-cell stage embryos were immobilized to a holding pipette by suction, while the injection capillary pipette was manually pushed though the zona pellucida and plasma membrane. Approximately 10 pL of plasmid solution was injected into the embryo cytoplasm using a pressure-controlled Eppendorf transjector 5246 (Eppendorf, Hamburg, Germany). Injected embryos were cultured in PZM3 medium (Yoshioka et al. 2003) at 39 °C with 5% CO2, 7% O2, 88% N2 and 100% humidity for 20 h, and then transferred into the oviducts of estrous-synchronized recipient sows.
EGFP expression analysis
EGFP expression on GM pigs and their tissues was visualized by a ‘living organisms’ fluorescent protein observation system consisting of a blue light lamp with maximum excitation at 488 nm and goggles with light filters (Model: FBL, BLS Ltd., Hungary) and photographs of wild-type (WT) and GM pigs were taken consecutively. The EGFP expression in GM cells was observed by inverted microscope ECLIPSE Ti-U (Nikon, Japan).
Real-time quantitative PCR analysis
Total RNA was extracted from ear tissue of each GM founder pigs using the MirVana miRNA Isolation Kit (Thermo Fisher Scientific, USA). Primer sets P5 and P6 (for primer and probe sequences see Table 1) were used to synthesize cDNA of shRNA by TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher). Primer sets P7 + P8, P10 + P11 and P9 + P12 were used to amplify the cDNA of shRNA transgene, the internal control miR16 gene and probe by TaqMan Universal PCR Master Mix II (no UNG) and SYBR® Select Master Mix (Thermo Fisher). The relative transgene expression level was calculated by the 2−ΔΔCt method based on the threshold cycle (Ct) values. Sanger sequencing of the qPCR products was used to validate transgene sequences.
Southern blot analysis
Ten µg genomic DNA isolated from each of 6 founder GM pigs and 1 WT pig was restriction digested with Hind III, purified by ethanol precipitation and then separated by electrophoresis in a 1% agarose gel. The digested DNA was then transferred to a nylon membrane (Amersham Biosciences, UK) by the capillary transfer method. The membrane was prehybridized overnight at 42 °C with prehybridization buffer and then hybridized with an 980 bp EGFP gene probe amplified by primer sets P13 + P14 (for primer sequences see Table 1) and labeled with digoxigenin by using a PCR DIG Probe Synthesis Kit (Roche, USA). Hybridization and washing were performed with DIG-High Prime DNA Labeling and Detection Starter Kit II (Roche). After hybridization, the membrane was incubated for 30 min in blocking solution and subsequently incubated for another 30 min in Anti-Digoxigenin-AP antibody solution (Roche). The membrane was then incubated for 15–20 min at 15–25 °C with 1 mL of CSPD ready-to-use, and the Southern photograph was captured with the EC3 imaging system (UAP, USA).
Inverse PCR analysis
Genomic DNA extracted from one of the GM pigs (GM3) was digested with restriction enzyme Hind III (Thermo Fisher). The digestion product was purified by a DNA purification column (Qiagen, China). The purified DNA was eluted with 50 µL of ddH2O. After adjustment with ddH2O and T4 ligase buffer to a final required volume prior to ligase enzyme addition, T4 ligase was added to a final concentration of 10 U/µL in a 1000 µL final ligation mixture. The ligation reaction was allowed to proceed 12–16 h by incubation at 16 °C and the ligated circularized DNA was purified via a Qiagen DNA purification column after elution from the column with 50 µL of ddH2O. One µL of the ligation product was used as a template for the PCR reaction with primer sets: P1 + P2, or P3 + P4 (for primer location see Fig. 1a, for primer sequences see Table 1). The resulting PCR products were cloned by ligation into a T-A vector (Life Technologies, USA) and sequenced for correctness. The obtained sequences were compared to the transgene vector sequence and the Sus scrofa genomic DNA sequence database (Build Sscrofa10.2) by NCBI BLAST (https://blast.ncbi.nlm.nih.gov) to identify the integration sites of the PB transposon.
Production of HTNCre recombinase
In order to produce His-tagged HTNCre protein, the HTNCre recombinase expression plasmid pTriEx-HTNC was transformed into Escherichia coli BL21 (DE3). BL21 bacteria were inoculated at 1:50 into 100 mL Luria–Bertani (LB) medium containing 50 mg/mL Ampicillin and grown at 37 °C until the OD600 value reach 1.0. Expression of plasmid-encoded protein was induced by addition of IPTG at a final concentration of 0.1 mM, followed by incubation overnight at 16 °C. Cells were harvested by centrifugation and lysed by sonification. The supernatant of the lysate was purified by SP-FF column and Superdex 200 column (Invitrogen, China). The His-tagged HTNCre protein was eluted from the column with PBS, 10% glycerol, pH7.4 buffer solution, and analyzed by SDS-PAGE.
HTNCre recombinase transduction
Primary fibroblast cells were isolated from the ear tissue of GM3 founder pig and cultured in 6-well plates with Dulbecco’s Modified Eagle Medium (DMEM, ThermoFisher) supplemented with 10% Fetal Bovine Serum (FBS, Thermo Fisher) and 2% antibiotics (penicillin–streptomycin, Thermo Fisher) at 37.5 °C under 5% CO2. Once the cultured fibroblasts reached 80% confluence, they were washed twice with DPBS before addition of Opti-MEM® I medium (Thermo Fisher) containing 200 µg/mL of purified HTNCre recombinase. After incubation at 37.5 °C under 5% CO2 for 3 h, the cells were washed with DPBS two times and cultured for another 24 h. Subsequently, the cells were trypsinized and reseeded on 10 cm dishes with a density of about 200 cells/dish in DMEM containing 10% FBS without selection drugs. After about 10 days of culture, well-separated colonies were isolated by cloning cylinders and transferred to 24-well plates. At sub-confluence, about one third of cells of each colony were transferred to 6-well plates first and then to 6 cm dishes.
Flow cytometric analysis
Following the procedures described above, GM3 founder pig-derived fibroblasts were incubated with Opti-MEM® I medium (Thermo Fisher) containing different concentrations of purified HTNCre or TAT-Cre (a gift from Qiuyan Li, see Kang et al. 2018) for 3 h, then were trypsinized and analyzed by the Plus personal flow cytometer Accuri™ C6 (BD, USA) according to the instructions of the manufacturer.
PCR analysis
Genomic DNA was isolated from GM ear fibroblast or tissues using Tissue DNA Extraction Kit (Omega, USA). To assess the recombination efficiency of the HTNCre recombinase, a 980 bp fragment of the EGFP gene, a 3211 bp fragment covering the marker gene expression cassette between two LoxP sites or a 530 bp residual fragment after the deletion of marker gene were amplified by PCR using primer sets P13 + P14, P15 + P16, respectively (for primer sequences see Table 1, for primer locations on the plasmids see Figs. 1a, 4d) by using KOD-FX PCR Kit (TOYOBO, Japan). PCR was performed as follows: 94 °C for 2 min, followed by 35 cycles of 98 °C for 10 s, 60 °C for 30 s, 68 °C for 3 min, and then 72 °C for 10 min. The PCR products were sequenced to confirm their identity.
Somatic cell nuclear transfer
The SMG-free GM fibroblasts were used as donor cells to produce SMG-free GM piglets by SCNT. The SCNT procedures were carried out as we described previously (Li et al. 2013).
Results
Production and identification of GM founder pigs
A piggyBac donor plasmid carrying an anti-PCV2 shRNA expression cassette and a SMG expression cassette was constructed (Fig. 1a). In this plasmid, the LoxP-flanked SMG expression cassette consists of a human cytomegalovirus (CMV) promoter, and a Neo gene linked with an EGFP gene by a 2A peptide (Fig. 1a). The piggyBac transposon plasmid was injected with a piggyBac transposase expression plasmid into the cytoplasm of in vivo fertilization-derived one-cell zygotes to produce anti-PCV2 GM pigs. A total of 97 injected embryos were transferred into 6 recipient sows, 4 of them farrowed and delivered 23 live piglets, 6 of which expressed EGFP on their bodies (Table 2 and Fig. 1b). No abnormal behavior or phenotype was observed on these 6 EGFP-positive GM founder pigs. One GM founder pig was dissected at the age of one month to observe EGFP expression on its internal organs. EGFP expression was observed on most of its internal organs (Fig. 1b), but not on the liver and spleen (data not shown). We are not sure if this GM founder pig is mosaic, because some studies showed that the EGFP transgene expression was not observed on some tissues of transgenic animals due to silence of the EGFP transgene in the corresponding tissues, instead of mosaic integration of the EGFP transgene (Vasey et al. 2009; Zeng et al. 2016).
Transgene expression and integration in GM founder pigs
The qPCR results demonstrated that the 6 GM founder pigs expressed different levels of anti-PCR2 siRNA in their ear tissues (Fig. 2a). These 6 GM founder pigs carried different copy number of transgenes, ranging from 1 to 4 copies per individual, as indicated by the Southern blot analysis (Fig. 2b). To our surprise, the anti-PCV2 siRNA expression level seems had no positive correlation with the transgene copy number in 6 GM founder pigs (Fig. 2a, b). This suggested that the transgene integration site, other than the copy number of the integrated transgene, may also affect transgene expression level in host GM animals.
Analysis of the transgene in the genome of GM founder pigs. a Real-time quantitative PCR analysis of the relative expression of anti-PCV2 siRNA in the ear tissue of different GM founder (F0) pigs. The experiment was performed in triplicate and the data was presented as mean ± SEM. b Southern blot analysis of transgene integration pattern in six GM and one WT pigs. The copy number of transgene in individual GM founder pigs varies from 1 to 4. Founders GM3 and GM4 carried only one copy of the transgene. c Inverse PCR analysis of the transgene integration site in GM3 founder pig. “Query” represents the inverse PCR-identified genomic sequences flanking the inserted pB transposon in GM3. “Sbjct” represents the genomic sequences found by blasting the “Query sequences” against the pig (sus scrofa) genome. The “Sbjct sequences” locate on chromosome 10 fully match the inverse PCR-identified genomic sequences flanking the inserted pB transposon. The sequencing results of the inverse PCR amplification product shows the 5′ terminal sequences of the inserted pB transposon and its 5′ flanking genomic sequences, and the 3′ terminal sequences of the inserted pB transposon and its 3′ flanking genomic sequences. TTAA is the target insertion site for piggyBac transposon
Inverse PCR was used to identify the integration site of the single copy of transgene in the genome of GM3 founder pig, which was located at an intergenic region of chromosome 10 (Fig. 2c).
The GM3 founder pig was selected to isolate fibroblasts from its ear tissue for subsequent experiments as this GM founder pig was expressing a relative high level of anti-PCV2 siRNA and carrying only one copy of transgene in its genome.
Production of recombinant Cre (HTNCre) recombinase
Soluble cell-permeant HTNCre protein was expressed and purified from the supernatant fraction of the cleared lysate of E. coli BL21 (DE3) cells transformed with the HTNCre recombinase expression plasmid pTriEx-HTNC. This recombinant enzyme contained, besides the Cre recombinase, a trans-activator of transcription for transduction (TAT) of the HTNCre enzyme which enhances cell transduction, and a nuclear localization signal peptide for translocation of the HTNCre enzyme into the cell nucleus (Fig. 3a). The purity and molecular weight of the purified HTNCre recombinase, as calculated via SDS-PAGE, was 95% and 42.76 kDa, respectively (Fig. 3b).
Production of recombinant Cre recombinase. a Map of the prokaryotic plasmid pTriEx-HTNCre for expression of cell-permeable and nuclear-localised His-TAT-NLS-Cre (HTNCre) recombinase. H6: 6 × His tag, boxed with yellow, used for purification of HTNCre protein. TAT: trans-activator of transcription from human immunodeficiency virus (HIV), boxed with blue, used for transduction of the HTNCre enzyme into cells. NLS: nuclear localisation signal from simian virus 40 (SV40), boxed with green, used for translocation of the HTNCre enzyme into the cell nucleus. Cre: recombinase gene from bacteriophage P1. b Expression and purification of HTNCre from bacteria as analyzed by SDS-PAGE. MW: molecular weight marker. 1–3 and 5: crude lysate of induced bacteria transfected with the pTriEx-HTNCre plasmid. 4: crude lysate of non-induced bacteria transfected with the pTriEx-HTNCre. 6: supernatant of induced bacteria lysate. 7: precipitation of induced bacteria lysate. 8: purified product. (Color figure online)
The efficiency of HTNCre-mediated SMG deletion in pig fibroblasts
The flow cytometric analysis indicated that treatment of 100, 200 and 300 µg/mL of HTNCre protein to GM3 founder pig fibroblasts resulted in successful SMG deletion in 86.4%, 95.2% and 93.3% treated cells (Fig. 4). However, transduction of 200 µg/mL of a regular Cre enzyme, TAT-Cre, into GM pig fibroblasts only removed SMG from the genome of 45.8% cells (Fig. 4). These results suggested that HTNCre was highly efficient in deleting LoxP-flanked SMGs from the genome of GM pig cells. In addition, we observed that the pig fibroblasts treated with 100–300 µg/mL of HTNCre enzyme showed no significant decrease in cell viability, compared to non-treated control pig fibroblasts (data not shown). This suggested that HTNCre has no significant cytotoxic effect on pig fibroblast when using at the concentrations of 100–300 µg/mL.
Flow cytometric analysis of the efficiency of HTNCre-mediated SMG deletion in GM3 founder pig-derived fibroblasts. The percentage in rectangle V2-L and V2-R defines the ratio of EGFP-negative and EGFP-positive fibroblasts, respectively. TAT-Cre is a control recombinase that does not carry the nuclear location signal peptide
Selection and identification of SMG-free GM pig fibroblasts
EGFP-negative cell colonies were obtained after treatment with the HTNCre enzyme of the GM3 founder pig-derived EGFP-positive fibroblasts (Fig. 5a). The PCR and the sequencing results indicated that the HTNCre-catalyzed recombination successfully removed the SMG fragment from the genome of all 7 tested EGFP-negative cell colonies (Fig. 5b–d).
Selection and Identification of SMG-free GM3 founder pig-derived fibroblasts. a The green fluorescence phenotype of fibroblast cell colonies with (lower panel) or without (upper panel) successful deletion of SMG. The scale bar indicates 500 μm. b Schematic of HTNCre-mediated deletion of SMG between two LoxP sites. The PCR products amplified from undeleted and deleted SMG, by the primer set of P15 and P16, should be 3211 bp and 530 bp, respectively. c PCR analysis of SMG deletion in seven fibroblast cell colonies. MW: molecular weight. Negative: negative control; W, water, used as blank control; WT, wild-type pig. Positive: positive control; P, the anti-PCV2 shRNA expression plasmid; −Cre, the cells without HTNCre treatment; Mix, mixture of SMG deleted and undeleted cell colonies. C1–C7: seven individual fibroblast cell colonies generated following HTNCre treatment. d The sequencing result of the 530 bp PCR product amplified from one representative SMG-free cell colony
Production of SMG-free GM pigs
The SMG-free GM3 founder pig-derived fibroblasts were used as donor cells to produce SMG-free GM pigs by SCNT. A total of 1044 cloned embryos were generated and transferred into the oviducts of four recipient sows, two of which became pregnant. One sow aborted its single fetus at 4 months of gestation, the other pregnant sow farrowed one healthy piglet (Table 3).
We observed no green fluorescence from the live-born SMG-free GM piglet (Fig. 6a). PCR analysis results confirmed that neither the aborted cloned fetus nor the live-born cloned piglet carried the SMG in their genomes (Fig. 6b).
PCR analysis of the SCNT-produced SMG-free GM piglets and fetuses. a Analysis of the EGFP expression in SMF-free GM piglet. Green fluorescence was not visualized on SMG-free GM piglet, but was clearly observed on a control EGFP-positive transgenic piglet (an offspring of the GM3 founder pig) under blue light. b PCR analysis of the EGFP gene and SMG between the two LoxP sites in the genome of a WT piglet, a control EGFP-positive transgenic piglet (an offspring of the GM3 founder pig), a SMG-free GM fetus and a SMG-free GM piglet. (Color figure online)
Discussion
Here, we have demonstrated that 200 µg/mL of HTNCre-mediated SMG excision resulted in 95.2% of all fibroblast to be SMG free. We further showed that these cells were capable nuclear donors for SCNT, resulting in SMG-free GM pigs. The SMG deletion efficiency in pig fibroblasts treated with 200 µg/mL (equal to 4.6 µM) of HTNCre was significantly higher than those obtained with 0.5–4 µM of HNCre (His tag-NLS-Cre) or TAT-Cre (TAT-Cre) treatment in pig, goat and monkey fibroblasts (Kang et al. 2018; Lan et al. 2013; Peitz et al. 2002; Xu et al. 2008). This SMG excision efficiency was also higher than that reported following treatment with 0.5–4 µM of R9-Cre and CPP5-Cre, two other forms of recombinant Cre recombinases (Kang, et al. 2018).
We believe that the high efficiency of HTNCre-mediated SMG deletion may be due to the fusion of the TAT and the NLS peptides to the Cre protein. TAT has been frequently used as a cell-penetrating peptide or a protein transduction domain. In that capacity, it was used to deliver a large variety of molecules, including nucleic acids, peptides and proteins, into mammalian cells without causing apparent toxicity (Bechara and Sagan 2013; Brasseur and Divita 2010; Mae and Langel 2006; Milletti 2012; Wadia and Dowdy 2005). Specifically, the TAT peptide increases the transduction of fusion proteins into cells by the lipid raft-dependent micropinocytosis (Wadia et al. 2004). The NLS peptide on the other hand not only enhances the penetration of fusion proteins into cells but also facilitates the translocation of fusion proteins to the cell nucleus (Gu et al. 1993; Lin et al. 2004). The fusion of this NLS peptide to the Cre enzyme has been shown to increase the efficiency of Cre enzyme-mediated recombination (Peitz et al. 2002; Jo et al. 2001). In our study we were able to show that the fusing both, the TAT and NLS peptides to the Cre protein enhanced the SMG deletion efficiency in HTNCre-treated cells.
Currently, LoxP-flanked SMGs can be deleted from the genome of GM pigs by Cre recombinase in three ways: (1) by crossing the GM pigs with transgenic pigs expressing Cre recombinase in their germ cells to obtain SMG-free GM pigs from the resulting offspring (Song et al. 2016); (2) by injecting Cre protein, Cre mRNA, or Cre expression plasmid into zygotes from GM pigs, then transfer them into recipient sows, and finally identify SMG-free GM pigs from the injected embryos-derived piglets (Whitworth et al. 2018); ((3) by transfecting Cre protein, Cre mRNA, or Cre expression plasmid into GM somatic cells, then select SMG-free GM somatic cells to generate SMG-free GM pigs by SCNT (Bi et al. 2016; Kang et al. 2018). The first method mentioned above requires generation and maintenance of a Cre-expressing GM pig line, which is costly. It also takes a long time to obtain SMG-free GM pigs because the need to wait until the GM pigs reach sexual maturity to perform mating. In addition, the SMG-free GM pigs generated by this method will inherit half of their genome from the Cre-expressing transgenic pigs, therefore harboring the Cre-gene, an obstacle for commercial use of such GM pigs. With the second method described above the GM pigs also needs to reach sexual maturity to produce zygotes for injection. Furthermore, the SMG-free GM pigs generated with this approach may be mosaic as described above. Using the third method can remove the SMGs from the donor somatic cells for producing the primary (F0) generation of GM pigs or from the donor somatic cells isolated from the newborn F0 generation GM pigs, and thus takes a short time to obtain SMG-free GM pigs compared with the first and the second methods. The third approach represents the most expedient and risk-averse procedure of all three methods described, in particular when used in combination the HTNCre fusion protein described herein.
In this study, we selected a transgenic founder pig, GM3, as a nuclear donor to generate SMG-free anti-PCV2 GM pig. This GM3 founder pig expressed a high level of anti-PCV2 siRNA from a single transgene copy. Mating this heterozygous GM3 founders with WT pigs, will result in half offspring carrying a single copy of transgene at the same genomic location as the parental founder, which presumably should produce similar levels of transgene expression. In contrast, GM founders carrying multiple copies of independently integrated transgene, such as the GM1 and GM5 founder pigs shown in Fig. 2b, will produce GM offspring with varying degrees of transgene expression levels due to segregation of the multiple copies of transgene after their transmission from the same GM founder to its GM progenies (Zeng et al. 2016, 2017).
In summary, an engineered Cre fusion enzyme containing a TAT peptide and an NLS peptide, namely HTNCre, can efficiently delete the LoxP-flanked SMGs from the genome of pig somatic cells, which can subsequently be used as donor cells to generate healthy SMG-free GM pigs by SCNT.
References
Araki K, Araki M, Miyazaki J, Vassalli P (1995) Site-specific recombination of a transgene in fertilized eggs by transient expression of Cre recombinase. Proc Natl Acad Sci USA 92(1):160–164
Bechara C, Sagan S (2013) Cell-penetrating peptides: 20 years later, where do we stand? FEBS Lett 587(12):1693–1702
Bi Y, Hua Z, Liu X, Hua W, Ren H, Xiao H, Zhang L, Li L, Wang Z, Laible G, Wang Y, Dong F, Zheng X (2016) Isozygous and selectable marker-free MSTN knockout cloned pigs generated by the combined use of CRISPR/Cas9 and Cre/LoxP. Sci Rep 6:31729
Brasseur R, Divita G (2010) Happy birthday cell penetrating peptides: already 20 years. Biochim Biophys Acta 1798(12):2177–2181
Cadinanos J, Bradley A (2007) Generation of an inducible and optimized piggyBac transposon system. Nucl Acids Res 35(12):e87
Gu H, Zou YR, Rajewsky K (1993) Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell 73(6):1155–1164
Hammer RE, Pursel VG, Rexroad CJ, Wall RJ, Bolt DJ, Ebert KM, Palmiter RD, Brinster RL (1985) Production of transgenic rabbits, sheep and pigs by microinjection. Nature 315(6021):680–683
Hoess RH, Abremski K (1984) Interaction of the bacteriophage P1 recombinase Cre with the recombining site loxP. Proc Natl Acad Sci USA 81(4):1026–1029
Hoess RH, Ziese M, Sternberg N (1982) P1 site-specific recombination: nucleotide sequence of the recombining sites. Proc Natl Acad Sci USA 79(11):3398–3402
Jo D, Nashabi A, Doxsee C, Lin Q, Unutmaz D, Chen J, Ruley HE (2001) Epigenetic regulation of gene structure and function with a cell-permeable Cre recombinase. Nat Biotechnol 19(10):929–933
Kaartinen V, Nagy A (2001) Removal of the floxed neo gene from a conditional knockout allele by the adenoviral Cre recombinase in vivo. Genesis 31(3):126–129
Kalderon D, Roberts BL, Richardson WD, Smith AE (1984) A short amino acid sequence able to specify nuclear location. Cell 39(3 Pt 2):499–509
Kang Q, Sun Z, Zou Z, Wang M, Li Q, Hu X, Li N (2018) Cell-penetrating peptide-driven Cre recombination in porcine primary cells and generation of marker-free pigs. PLoS ONE 13(1):e190690
Lai L, Park KW, Cheong HT, Kuhholzer B, Samuel M, Bonk A, Im GS, Rieke A, Day BN, Murphy CN, Carter DB, Prather RS (2002) Transgenic pig expressing the enhanced green fluorescent protein produced by nuclear transfer using colchicine-treated fibroblasts as donor cells. Mol Reprod Dev 62(3):300–306
Lan C, Ren L, Wu M, Liu S, Liu G, Xu X, Chen J, Ma H, Cheng G (2013) Deletion of marker gene in transgenic goat by Cre/LoxP system. Sheng Wu Gong Cheng Xue Bao 29(12):1847–1854
Li Z, Shi J, Liu D, Zhou R, Zeng H, Zhou X, Mai R, Zeng S, Luo L, Yu W, Zhang S, Wu Z (2013) Effects of donor fibroblast cell type and transferred cloned embryo number on the efficiency of pig cloning. Cell Reprogram 15(1):35–42
Li Z, Zeng F, Meng F, Xu Z, Zhang X, Huang X, Tang F, Gao W, Shi J, He X, Liu D, Wang C, Urschitz J, Moisyadi S, Wu Z (2014) Generation of transgenic pigs by cytoplasmic injection of piggyBac transposase-based pmGENIE-3 plasmids. Biol Reprod 90(5):93
Lin Q, Jo D, Gebre-Amlak KD, Ruley HE (2004) Enhanced cell-permeant Cre protein for site-specific recombination in cultured cells. BMC Biotechnol 4:25
Mae M, Langel U (2006) Cell-penetrating peptides as vectors for peptide, protein and oligonucleotide delivery. Curr Opin Pharmacol 6(5):509–514
McLellan MA, Rosenthal NA, Pinto AR (2017) Cre-loxP-mediated recombination: general principles and experimental considerations. Curr Protoc Mouse Biol 7(1):1–12
Milletti F (2012) Cell-penetrating peptides: classes, origin, and current landscape. Drug Discov Today 17(15–16):850–860
Montag J, Petersen B, Flogel AK, Becker E, Lucas-Hahn A, Cost GJ, Muhlfeld C, Kraft T, Niemann H, Brenner B (2018) Successful knock-in of Hypertrophic Cardiomyopathy-mutation R723G into the MYH7 gene mimics HCM pathology in pigs. Sci Rep 8(1):4786
Naruse K, Ishikawa H, Kawano HO, Ueda H, Kurome M, Miyazaki K, Endo M, Sawasaki T, Nagashima H, Makuuchi M (2005) Production of a transgenic pig expressing human albumin and enhanced green fluorescent protein. J Reprod Dev 51(4):539–546
Park KW, Cheong HT, Lai L, Im GS, Kuhholzer B, Bonk A, Samuel M, Rieke A, Day BN, Murphy CN, Carter DB, Prather RS (2001) Production of nuclear transfer-derived swine that express the enhanced green fluorescent protein. Anim Biotechnol 12(2):173–181
Peitz M, Pfannkuche K, Rajewsky K, Edenhofer F (2002) Ability of the hydrophobic FGF and basic TAT peptides to promote cellular uptake of recombinant Cre recombinase: a tool for efficient genetic engineering of mammalian genomes. Proc Natl Acad Sci USA 99(7):4489–4494
Sauer B, Henderson N (1988) Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci USA 85(14):5166–5170
Song Y, Lai L, Li L, Huang Y, Wang A, Tang X, Pang D, Li Z, Ouyang H (2016) Germ cell-specific expression of Cre recombinase using the VASA promoter in the pig. FEBS Open Bio 6(1):50–55
Sunaga S, Maki K, Komagata Y, Ikuta K, Miyazaki JI (1997) Efficient removal of loxP-flanked DNA sequences in a gene-targeted locus by transient expression of Cre recombinase in fertilized eggs. Mol Reprod Dev 46(2):109–113
Tong J, Wei H, Liu X, Hu W, Bi M, Wang Y, Li Q, Li N (2011) Production of recombinant human lysozyme in the milk of transgenic pigs. Transgenic Res 20(2):417–419
Vasey DB, Lillico SG, Sang HM, King TJ, Whitelaw CB (2009) CMV enhancer-promoter is preferentially active in exocrine cells in vivo. Transgenic Res 18(2):309–314
Vives E, Brodin P, Lebleu B (1997) A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J Biol Chem 272(25):16010–16017
Wadia JS, Dowdy SF (2005) Transmembrane delivery of protein and peptide drugs by TAT-mediated transduction in the treatment of cancer. Adv Drug Deliv Rev 57(4):579–596
Wadia JS, Stan RV, Dowdy SF (2004) Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat Med 10(3):310–315
Wang W, Lin C, Lu D, Ning Z, Cox T, Melvin D, Wang X, Bradley A, Liu P (2008) Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc Natl Acad Sci USA 105(27):9290–9295
Whitworth KM, Cecil R, Benne JA, Redel BK, Spate LD, Samuel MS, Prather RS, Wells KD (2018) Zygote injection of RNA encoding Cre recombinase results in efficient removal of LoxP flanked neomycin cassettes in pigs. Transgenic Res 27(2):167–178
Whyte JJ, Prather RS (2011) Genetic modifications of pigs for medicine and agriculture. Mol Reprod Dev 78(10–11):879–891
Wu Z, Xu Z, Zou X, Zeng F, Shi J, Liu D, Urschitz J, Moisyadi S, Li Z (2013) Pig transgenesis by piggyBac transposition in combination with somatic cell nuclear transfer. Transgenic Res 22(6):1107–1118
Xu Y, Liu S, Yu G, Chen J, Chen J, Xu X, Wu Y, Zhang A, Dowdy SF, Cheng G (2008) Excision of selectable genes from transgenic goat cells by a protein transducible TAT-Cre recombinase. Gene 419(1–2):70–74
Yoshioka K, Suzuki C, Itoh S, Kikuchi K, Iwamura S, Rodriguez-Martinez H (2003) Production of piglets derived from in vitro-produced blastocysts fertilized and cultured in chemically defined media: effects of theophylline, adenosine, and cysteine during in vitro fertilization. Biol Reprod 69(6):2092–2099
Zeng F, Li Z, Cai G, Gao W, Jiang G, Liu D, Urschitz J, Moisyadi S, Wu Z (2016) Characterization of growth and reproduction performance, transgene integration, expression, and transmission patterns in transgenic pigs produced by piggyBac transposition-mediated gene transfer. Anim Biotechnol 27(4):245–255
Zeng F, Li Z, Zhu Q, Dong R, Zhao C, Li G, Li G, Gao W, Jiang G, Zheng E, Cai G, Moisyadi S, Urschitz J, Yang H, Liu D, Wu Z (2017) Production of functional human nerve growth factor from the saliva of transgenic mice by using salivary glands as bioreactors. Sci Rep 7:41270
Zhang X, Li Z, Yang H, Liu D, Cai G, Li G, Mo J, Wang D, Zhong C, Wang H, Sun Y, Shi J, Zheng E, Meng F, Zhang M, He X, Zhou R, Zhang J, Huang M, Zhang R, Li N, Fan M, Yang J, Wu Z (2018) Novel transgenic pigs with enhanced growth and reduced environmental impact. Elife 7:e34286
Acknowledgements
This study was supported by a grant from a National Science and Technology Major Project of China (Grant Number: 2016ZX08006002), and two grants from the Department of Science and Technology of Guangdong Province, China (Grant Numbers: 2018B020203002 and 2018ML1101).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Huang, X., Zou, X., Xu, Z. et al. Efficient deletion of LoxP-flanked selectable marker genes from the genome of transgenic pigs by an engineered Cre recombinase. Transgenic Res 29, 307–319 (2020). https://doi.org/10.1007/s11248-020-00200-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-020-00200-3