Abstract
A chimeric Bacillus thuringiensis toxin (Bt) gene, cry2AX1was cloned in a bi-selectable marker free binary vector construct. The cry2AX1 gene, driven by the Chrysanthemum rbcS1 promoter, was introduced into JK1044R, the restorer line (Oryza sativa L. ssp. Indica) of a notified commercially grown rice hybrid in India, by Agrobacterium-mediated transformation. Its effect against two major lepidopteran insect pests viz., yellow stem borer (YSB) Scirpophaga incertulas, rice leaf folder (RLF) Cnaphalocrocis medinalis and one minor insect pest, oriental army worm (OAW) Mythimna separata was demonstrated through bioassays of transgenic rice plants under laboratory and greenhouse conditions. The rbcS1 promoter with chloroplast signal peptide was used to avoid Cry2AX1 protein expression in rice seed endosperm tissue. A total of 37 independent transformants were generated, of which after preliminary molecular characterization and YSB bioassay screening, five events were selected for their protein expression and bioefficacy against all three rice insect. One elite transgenic rice line, BtE15, was identified with Cry2AX1 expression ranging from 0.68 to 1.34 µg g−1 leaf fresh weight and with 80–92 % levels of resistance against rice pests at the vegetative and reproductive stages. Increase in Cry2AX1 protein concentration was also observed with crop maturity. The Cry2AX1protein concentration in the de-husked seeds was negligible (as low as 2.7–3.6 ng g−1). These results indicate the potential application of cry2AX1 gene in rice for protection against YSB, RLF and OAW.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice is a staple food for nearly half of the world’s population. Among the biotic constraints, insect pests have been identified as the major factor causing damage to rice yield (Waddington et al. 2010). Stem borers are major pests of rice worldwide and about fifty species from two orders, lepidoptera and diptera are reported to affect rice and among these, yellow stem borer (YSB) Scirpophaga incertulas present mainly in Asia is responsible for steady annual yield loss of 5–10 % and with occasional localized outbreaks of up to 60 % (Pathak and Khan 1994; Bandong and Litsinger 2005). Other lepidopterans such as rice leaf folder (RLF) Cnaphalocrocis medinalis, rice case worm Nymphulade punctalis, oriental army worm (OAW) Mythimna separata, and swarming caterpillar Spodoptera mauritia are also known to cause substantial economic loss to rice crop. In India, average yield losses in recent years due to rice stem borers and leaf folders accounts for 30 and 10 per cent, respectively (Krishnaiah and Varma 2012). Selvaraj et al. (2012) showed that the joint incidence of YSB and RLF causes more damage than when these insects attack separately. Though OAW is considered a minor pest, it causes severe loss to rice yield in endemic years (Mishra et al. 2013).
Chemical pesticides are predominantly used to control rice insect pests. However, increasingly, application of chemical pesticides is becoming less effective in controlling YSB, RLF and OAW due to their feeding nature, wherein either they live inside the stem or inside a folded leaf nests or in the soil avoiding direct contact with the applied pesticides. Moreover, indiscriminate use of pesticides in rice is becoming a major concern for rice ecosystem affecting the beneficial insects (Waddington et al. 2010). The other means of controlling insect pests such by deploying native resistance genes are not very promising since the cultivated rice germplasm has a very low level of resistance to lepidopteran pests, particularly to stem borers (Khan et al. 1991; Visalakshmi et al. 2014). This clearly indicates the need to develop other approaches to control rice lepidopteran insects.
Application of insecticidal Bt cell suspension spray is a well-known traditional method to control lepidopteran insect pests of several important crops (Nester et al. 2002). Many different insecticidal cry genes encoding the insecticidal proteins have been identified in various Bt strains. Transgenic expression of many of these Cry proteins demonstrated their potential as a viable technology to control many different insect pests (Kumar et al. 2008). Transgenic rice expressing proteins from the cry1A gene family were shown to be effective against a variety of lepidopteran rice insects. Transgenic cry1Ab rice was effective against YSB (Wu et al. 1997; Datta et al. 1998), and against rice striped stem borer (SSB) Chilo suppressalis (Ghareyazie et al. 1997). Transgenic cry1Ac rice was shown to be effective against YSB (Nayak et al. 1997; Khanna and Raina 2002) and also against RLF, Melanitis leda ismene (Rice green caterpillar) and Pelopidas mathias (Rice skipper) (Kim et al. 2009). Transgenic rice expressing cry1Ab and cry1Ac genes separately were found to be effective against SSB, YSB and RLF (Wang et al. 2010). Transgenic rice plants expressing fusion proteins of Cry1Ac and Cry1I (Yang et al. 2014) and that of Cry1Ab and Cry1Ac (Cheng et al. 1998) were reported to be effective against RLF and SSB. Transgenic rice expressing cry1C* gene provided resistance against YSB, SSB and RLF (Tang et al. 2006; Ye et al. 2009). Field trial with rice lines expressing cry1Ac and cry2A genes separately exhibited high level of resistance to YSB, RLF and rice skipper (Bashir et al. 2004). Transgenic rice with codon optimized cry2A was effective against YSB and RLF (Chen et al. 2005) in laboratory and field studies. Transgenic rice lines expressing three classes of Bt proteins (CrylAc, Cry2A, and Cry9C) separately were found to be effective against two rice borers YSB and SSB in laboratory and field studies (Chen et al. 2008). However none of these transgenic rice plants were studied for its effectiveness against a minor but increasingly important insect, OAW in addition to YSB and RLF.
Cry2A family of proteins was demonstrated to have a broad-spectrum toxicity against lepidopterans (Mandal et al. 2007) as well as against dipteran insects (Widner and Whiteley 1989). Generally, Cry2A protoxins are about half the size (65–70 kDa) of Cry1A protoxins (130–140 kDa) and therefore should be expressed at higher levels in transgenics. Recently, a chimeric cry2AX1 gene was artificially created by fusing cry2Aa and cry2Ac genes. It was reported to be toxic against Helicoverpa armigera and Spodoptera litura in transgenic tobacco (Udayasuriyan 2012; Jayaprakash et al. 2014) and tomato (Balakrishnan et al. 2012) and against RLF in transgenic rice (Manikandan et al. 2014a). We reasoned that given the reported broad spectrum toxicity of Cry2A proteins against various insects, Cry2AX1 may also provide broad protection against lepidopteran pests in transgenic rice.
Constitutive promoters are used most frequently in transformation studies. However, constitutive expression of transgenes generally attracted criticisms for their potential to develop target resistance against target insects and more environmental spread of the transgenic protein. Qiu et al. (2010) addressed this issue by expressing hybrid Cry1Ab/Ac Bt protein driven by the maize PEPC promoter. The transgene expressed only in the leaf and stems of rice and provided resistance against RLF. Another tissue specific promoter, rbcS, has also been utilized in rice for driving expression of transgenic cry1C* (Ye et al. 2009), cry1Ac (Kim et al. 2009) and cry1Ab (Qi et al. 2013) genes. Qi et al. 2013, demonstrated that in addition to tissue specific expression, when the transgenic protein was targeted to chloroplasts and resulted into better protein stability and expression (Qi et al. 2013).
Here, we report the development of a transgenic Indica rice line expressing a synthetic cry2AX1 gene. The cry2AX1 gene is driven by a green tissue-specific promoter (rbcS1) and targeted to the chloroplast to drive high expression. The Cry2AX1 protein is expressed in green tissues including leaf sheaths but with negligible expression in the de-hulled mature grains. The results indicate that cry2AX1 gene is effective against three different categories of lepidopteran pests; stem borer, leaf folder, army worm.
Materials and methods
Construction of a binary vector carrying the cry2AX1 gene
The chimeric cry2AX1 gene (Udayasuriyan et al. 2010) consists of a total 633 amino acids. The N-terminal of consisting of residues, 1–585 are from of Cry2Aa protein and C-terminal residues 586–633 corresponds to 576–623 of Cry2Ac isolated from indigenous strains of Bacillus thuringiensis. The cry2AX1 gene was codon optimized to improve expression in crop plants (NCBI accession number GQ332539.1). The 1.936 kb cry2AX1 gene was cloned into the NcoI and BamHI sites of the ImpactVector 1.4 (Krens et al. 2005) downstream to Chrysanthemum rbcS1 promoter along with chloroplast target signal peptide sequence and upstream of rbcs terminator. The cry2AX1 gene cassette located between attR sites in the ImpactVector 1.4 were then cloned into destination vector pMF1GFP-des (18.6 kb) by gateway recombination (Gateway LR Clonase® protocol, Invitrogen, life technologies, USA) protocol. The resulting binary vector is named as pJK2AX1 and has the nptII as plant selection marker gene. It also contains a codA based negative selection gene and a recombinase (recA) gene, which will be needed in the future to remove the marker cassettes from transgenic plants. The pJK2AX1 was mobilized into EHA105, a virulent Agrobacterium tumefaciens strain, by electroporation using a Multiporator (Eppendorf, Germany). Vector integrity and orientation was verified by HindIII restriction digestion, gene specific PCR and Sanger sequencing of T-DNA region.
Agrobacterium based transformation of JK1044R
JK1044R, the restorer parent of the commercial notified rice hybrid JKRH401 (Indian PPVFR code IET 18181) was used for transformation. Calli generated from scutellum of mature seeds were transformed using established Agrobacterium transformation, selection and regeneration protocol especially standardized for JK1044R restorer parent line, as described in Chakraborty et al. (2016). Transformation procedure with major modification of Hiei et al. (1994), using N6 basal media (Chu et al. 1975) with 2-4,D (2.0 mg L−1), NAA (0.5 mg L−1) and Kinetin (0.5 mg L−1) for embryogenic callus formation, whereas MS basal medium (Murashige and Skoog 1962) with Kinetin (2.5 mg L−1) and BAP (1.0 mg L−1) for plant selection and regeneration. A combination of G418 (30 mg L−1) and Paramomycin (70 mg L−1) was incorporated in antibiotic selection medium. The regenerated plantlets were sub-cultured onto half MS medium for rooting. Rooted plantlets were transplanted in pots filled with sterile clay-loam soil mixture and maintained in a containment greenhouse at 26 ± 2 °C with 16/8 h light/dark cycles until maturity. The panicles were bagged before floret opening and the mature seeds were harvested.
T1 and T2 generation advancement
The T1 and T2 seeds were germinated on Whatman No. 1 filter papers wetted with 1× Yoshida’s nutrient solution (Yoshida et al. 1976) supplemented with G418 (80 mg L−1) (Wang et al. 2007). Four days after germination, the seedlings were transferred onto 200 mL plastic cups containing 1:1 vermiculite and coco-peat mix (Keltech Energies Ltd. Bengaluru, India). The seedlings were maintained on 1× Yoshida’s nutrient solution. The bleached seedlings were discarded and the green seedlings were transplanted in pots in a containment greenhouse at 26 ± 2 °C with 16/8 h light/dark cycles until maturity.
Polymerase chain reaction analyses
Genomic DNA was isolated from leaf tissues by CTAB method (Dellaporta et al. 1983). The following PCR primers were used for detection of cry2AX1 gene: forward primer 5′-GCCTGAGCGAAAAGAGGAAG-3′, and reverse primer 5′-ACGTTGTTGCTCATGATCCT-3′. PCR reactions were carried out in 25 μL reaction volume in a thermocycler (EP Gradient, Eppendorf). PCR products were visualized and documented on a gel imaging system (molecular imager—XR system, Bio-Rad Laboratories, USA).
Qualitative and quantitative ELISA
Cry2A quali-plate and quanti-plate (Envirologix, USA) ELISA kits were used for qualitative and quantitative detection of Cry2AX1 in fresh tissues respectively. Fresh leaves, leaf-sheathes tissue or seeds were homogenized and used for detection of Cry2AX1 protein as per the manufacturer’s protocol. The OD was measured at 450 nm using an ELISA reader (Multiskan EX reader, MTX Lab Systems, Inc., USA).
Southern blot
Genomic DNA was isolated from leaf tissues by CTAB method (Dellaporta et al. 1983). About 10 µg of genomic DNA was digested with XhoI (Cat. No. R0146, NEB, UK) and was blotted onto a nylon membrane (Hybond N+, GE-Amersham, UK) using 20X SSC as transfer buffer. Thus a 200 bp PCR product amplified from the coding region of cry2AX1 gene downstream of XhoI site was used as a probe. The probe was radio-labeled with α32P-labeled dCTP (>3200 Ci/mol, Board of Radiation and Isotope Technology, Jonaki, India) using the random primer labeling kit (DecaLabel DNA labeling kit, Thermo Scientific, USA) as per the manufacturer’s instructions. Hybridization was performed in a rotary hybridization oven (Thermo Scientific), for 16 h at 62 °C. Membrane washing and signal detection were carried out by following Sambrook and Russell (2001).
YSB bioassays
YSB moths collected from rice fields around Hyderabad, India were brought to laboratory and allowed to lay eggs on rice leaves. Egg masses were collected and incubated at room temperature for hatching and freshly hatched neonate larvae were used for bioassays. For in vitro bioassay, about 5 cm long pseudo-stems were cut from rice plants at vegetative stage and were placed on a Petri plate lined with moist Whatman No. 1 filter paper. The cut ends of the pseudo-stem pieces were sealed with parafilms; five neonate larvae were released on each pseudo-stem and then incubated in the dark at room temperature. After 5 days the pseudo-stem pieces were dissected open and the number of live and dead larvae was counted. In some cases, a few larvae couldn’t be recovered since they might have been dead and degraded inside the pseudo-stem and were counted as dead.
In planta bioassays on whole plant were conducted inside a greenhouse following the protocols of Khanna and Raina (2002). Three neonates per tiller were released onto the active growing tillers or pre-booting panicles and cumulative deadhearts or whiteheads damage was estimated 15 days later for both transgenic and non-transgenic control.
Rice leaf folder bioassay
RLF moths were collected from commercial rice fields and released on 60-day old rice plants. It was then covered with Mylar tubes and 10 % honey solution was provided as food for the adults (Waldbauer and Marciano 1997). After 7–8 days, second instar larvae were collected. For in vivo bioassay, two larvae were released per tiller and kept within Mylar tubes in greenhouse. Damage scores were recorded 5 days after release of larva. Leaf damages were scored on the scale of 0–5 based on following criteria: 0, no leaf folding and no leaf scraping; 1, leaf folding but no leaf scraping; 2, leaf folding with less than 5 % leaf scraping; 3, leaf folding with about 10 % leaf scraping; 4, leaf folding with about 50 % scraping; and 5, leaf folding with more than 50 % leaf scraping. The experiments were repeated thrice. A different assay for insect mortality was also developed. In this assay, the mortality of insects were assessed by releasing two second-instar larvae on 5 cm long freshly harvested young leaves harvested from test entries. The detached leaves along with larvae were placed in Petri plates lined with moist Whatman No. 1 filter papers maintained at room temperature in the dark. Insect mortality data was recorded 5 days after releasing larvae.
OAW bioassay
Larvae of OAW were collected from commercial sorghum and maize fields around Hyderabad, India and cultured on artificial diet (Hattori and Atsusawa 1980). Pupa were collected and maintained on sterile vermiculite for about 10 days. Emerged moths were placed inside jars containing parafilms for egg laying. Five freshly hatched neonate larvae were released on each 5 cm long leaf blades detached from 60-day old rice plants. The leaf blades and the larvae were kept on Petri plates lined with moist Whatman No. 1 filter papers at room temperature in darkness for 5 days and insect mortality was recorded. Percent leaf area damage caused by OAW was scored on a scale of 0 to 5 based on following criteria: 0, no visible leaf damage; 1, less than 5 % leaf damage; 2, about 10 % leaf damage; 3, about 20 % leaf damage; 4, about 50 % leaf damage and 5, more than 50 % leaf damage. Growth of larvae and presence of feces were also recorded visually.
Data analyses
Percent mortality in in vitro pseudo-stem bioassays for YSB and leaf bioassays for RLF and OAW were calculated by dividing the total of dead and unrecovered larvae by the total number of larvae released. All percentage insect mortality and percentage plant damage data were arcsine transformed. Transformed mortality data and the quantitative protein expression data were analyzed using standard ANOVA, single factor (excel; microsoft) procedure. Differences between the treatment means were compared using the Fisher’s least significant difference (LSD) test and all differences were judged to be significant at p < 0.05.
Results
Development of transgenic rice plants and selection of good events
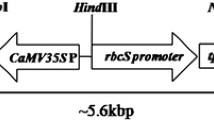
Agrobacterium tumefaciens EHA105 carrying pJK2AX1 (“Materials and methods” section and Fig. 1) was used for transformation of JK1044R rice line. A total of 66 independent putative transgenic plants were obtained and were grown to maturity. Out of these, 37 were found to carry the cry2AX1 gene (Supplementary Fig. 1) as can be detected by PCR assays. Seeds were harvested from these 37 putative events.
Schematic diagram of the T-DNA region of the binary vector pJK2AX1. Panel at the bottom describes each component of the construct; 1 GFP cassette as reporter marker; 2 codA + nptII cassette for positive and negative selection marker; 3 recombinase cassette for recombination marker; 4 insecticidal Cry2AX1 gene cassette for cry protein expression. Details of the abbreviations are as follows: LB left border, RS recombinase sites from Zygosaccharomyces rouxii; rbcSP, Rbcs1 promoter Chrysanthemum morifolium; E-GFP, enhanced green fluorescent protein gene; rbcST, RbcS1 terminator C. morifolium; 35SP, CaMV35S promoter; codA + nptII, fusion of codA gene for negative selection for fluoro-cytosine and nptII gene for positive selection for kanamycin; nosT, nopaline synthase terminator; recLBD, fusion of recombinase R-gene and ligand binding domain of the rat glucocorticoid receptor; TP, chloroplast target signal peptide; cry2AX1, cry2AX1 gene; RB, right border. The bold line represents 200 bp PCR fragment of cry2Ax1 gene used for preparation of radio-labelled probe for Southern blot hybridization analysis in the present study
In the T1 generation, five events showed albinism and stunted growth and were excluded. T1progenies from the rest of the 32 events were subjected to ELISA and 18 events were found to be positive. These positive events were subjected to YSB pseudo-stem in vitro bioassay (data not provided). The bioassay identified only five events, viz., BtE15, BtE52, BtE62, BtE66 and BtE68 exhibiting more than 70 % YSB larval mortality. These top five events were selected and ten positive T1 plants from each selected event were advanced to the T2 generation.
The homozygous T2 families were identified by germinating 100 seeds from each plant on G-418. Seedlings that didn’t carry the nptII gene showed bleaching effect. Out of 50 T2 families derived from five independent events, only 18 showed no bleaching effect, indicating no segregation of cry2AX1 in T2 generation and were considered as homozygous. The selected homozygous T2 families were subjected to further molecular characterization, such as PCR, ELISA and Southern blotting. Southern blotting confirmed that integration of cry2AX1 gene in events BtE15, BtE52, BtE62 and BtE68 carry only single copy of cry2AX1, whereas event BtE66 carry two copies of cry2AX1 gene (Supplementary Fig. 2).
Protein expression in green tissues and endosperm
The Cry2AX1 protein expression level was determined at two different growth stages, vegetative and reproductive in two different green tissues; leaf and leaf-sheath. The expression of Cry2AX1 protein of the top five transgenic events was ascertained by quantitative ELISA (Table 1). Protein expression ranged from 0.4 to 0.8 µg g−1 fresh tissue among the selected events at vegetative stage in both the green tissues. It was observed that the expression of Cry2AX1 was higher at the reproductive stage compared to the vegetative stage across all the tested events. Protein expression reached up 0.9 to 1.3 µg g−1 at reproductive stage in both the green tissues. Among the five events Cry2AX1 expression level in BtE66, BtE62 and BtE68 was found to be less than that of in BtE52 and BtE15 in both the stages. BtE15 showed significantly higher level of Cry2AX1 expression (0.68 µg g−1 and 1.18 µg g−1 in leaf and leaf sheath respectively) compared to the rest of the events. The expression of Cry2AX1 in the endosperm was found to be extremely low (2.7–3.6 ng g−1) when compared to green tissues such as leaf and leaf sheath (0.39–1.34 µg g−1) across all the selected five events.
Resistance to YSB in in vitro and in-planta bioassays
In order to ascertain the resistance of the transgenic cry2AX1 events against YSB, the T2 families of top five selected events were bio-assayed in vitro (Table 2; Fig. 2a, b). The mortality rate of neonate larva was in the rage to 64–88 % at vegetative stage and 72–92 % at reproductive stage while the controls showed only 6–8 % at both the stages. The larva that survived the assay were usually retarded in growth and died subsequently even when maintained on fresh control stem tissues (data not shown). This proved that the transgenic events are having good effect on YSB neonates. To confirm these results in another way, the same T2 families were also assayed for YSB resistance by in planta bioassay in the greenhouse (Table 2). Tiller damage (deadhearts) at vegetative stage was 1.4 % in event BtE15 and to a maximum of 6.0 % in event BtE68, whereas, the damage of control plants was 21.4 %. In planta bioassay at the reproductive stage revealed that the whitehead damage was as low as 0 % in event BtE15 and to a maximum of 0.6 % in BtE68, whereas the control plants showed about 17.9 % whiteheads damage. This proved the effectiveness of Cry2AX1 protein against YSB under laboratory condition and the event BtE15 performed better than the rest of the events.
a In-vitro YSB pseudo-stem bioassay of event BtE15 after dissected open (after 5 days) showing dead degenerating body of the larvae and b non-transgenic control with feeding damage along with larval growth. In-vitro leaf bioassay using 2nd instar RLF larva; c slight leaf scraping on event BtE15 compare to d non-transgenic control where severe leaf scraping was observed 5 days after release. In-vitro leaf bioassay using 1st instar OAW larvae; e showing mortality with no leaf damage on event BtE15 compare to f non-transgenic control which showed significant leaf area damages with larval growth and insect excreta accumulation
Resistance to RLF and OAW
The selected Cryy2AX1 events were also tested against RLF and OAW other lepidopteran pest of rice. Both detached leaf and in planta bioassays for RLF was performed. The detached leaf bioassay demonstrated RLF mortality up to 80 % in transgenic event BtE15, whereas, it was found to be not more than 5 % in control plants (Table 3). The live larvae recovered from transgenic plant treatments showed growth retardation, indicating starvation, which was further confirmed by correlating it with leaf damage and presence of excreta at the end of bioassay (Fig. 2c, d). In planta bioassay showed an average damage score of 0.9–1.3 in transgenic events, compared to 3.9 in non-transformed control plants (Table 3). Although the leaf folding was observed in most of the treatments the scraping on the folded transgenic leaves was found to be very limited compared to non-transgenic controls.
Similar results were consistently observed in OAW bioassay also. As shown in Table 3, transgenic events showed high mortality ranging from 72 to 92 % in selected events with almost nil to less than 5 % visible damage of the leaf area (score 0.2–1.2). Much less larval excreta were noticed in transgenic events when compared to non-transformed control plants (Fig. 2e, f). The larva that survived feeding on transgenic leaves were severely retarded, on the contrary larva fed with the leaves of the non-transgenic control grew normally.
Discussion
The current article discusses the development of cry2AX1 transgenic rice restorer line. We decided to transform an Indica and the parent of commercially successful hybrid rice in India. The logic was to reduce any potential linkage drag coming from non-elite backgrounds and also to retain the agronomic value of the final commercial product while saving time in backcross breeding. Regenerated transgenic cry2AX1 rice events were verified for stable gene integration.
Our strategy was to express the Cry2AX1 protein in green tissues in appropriate abundance, while restricting its expression in non-green tissue, such as endosperm. For this purpose, rbcS1 promoter from Chrysanthemum morifolium a dicot plant was utilized. In the present study we demonstrated negligible expression (2.7–3.6 ng g−1) of the Cry2AX1 protein in de-husked seeds while having adequate expression in the green tissues. Kim et al. (2009), reported cry1Ac protein expression in rice using a rice-rbcS promoter and a chloroplast target peptide from rice, wherein, negligible expression of Bt protein was noticed in the seeds. Similarly, Ye et al. (2009) reported, when Cry1C* gene was driven by the rice rbcS promoter, protein concentrations ranged from 0.71 to 3.13 µg g−1 leaf fresh weight and expression level in endosperm was found to be extremely low (2.6 ng/gm). But they also observed a declining trend of the Cry1C* protein concentration in the period between the vegetative and reproductive growth stages. Our work confirms the previous studies that rbcS promoter expresses negligibly in the rice seed endosperm.
We also wanted to express the transgenic protein at a very high level in the green tissues. Use of rbcS promoter to drive high expression was reported in literature. The Chrysanthemum rbcS1 was found to express beta-glucuronidase (gusA) 7–8 fold higher than the commonly used cauliflower mosaic virus (CaMV) 35S promoter when used in tobacco (Outchkourov et al. 2003), in the same study high expression levels (3–7 % of total soluble protein) were achieved when Chrysanthemum rbcS1 promoter was used in potato and tomato. Zheng et al. (2005) expressed Cry1Ca in transgenic shallots under the control of a Chrysanthemum rbcS1 promoter, and the transgenic plants showed high resistance to beet armyworm. Functionality of another dicot tomato rbcS promoter in monocot was previously demonstrated in rice (Kyozuka et al. 1993). However, we not only wanted to have high expression of the transgene but also wanted to avoid potential toxicity of the transgenic protein in the cytoplasm while maintaining a stable protein expression throughout the growth stages. So, a construct (Krens et al. 2005) was utilized the rbcS1 promoter and also fused therbcS1 chloroplast target peptide with the Cry2AX1 peptide so as to translocate the protein into chloroplast. Translocation of transgenic protein into chloroplast stroma shall provide higher stability and accumulation of Cry protein in chloroplast. It is well known that transgenic proteins get degraded much rapidly in free cytosol, compared to their presence in chloroplasts (Pillay et al. 2014). This phenomenon of increase of transgenic Bt protein levels because of translocation and accumulation into the chloroplast with the help of rbcS:tp sequence was also reported in Arabidopsis (Wong et al. 1992) and rice (Jang et al. 2002; Kim et al. 2009).
In the present study, protein concentration in green tissues (fresh leaf and leaf sheath) ranged between 0.39 and 1.34 µg g−1 (Table 1) which was found to be effective against YSB, RLF and OAW. It can be hypothesized that this level of Cry2AX1 protein may be tolerated to support normal plant phenotype. A wide variety of transgenic Bt protein concentration has been reported to be effective against YSB. This probably indicates that the required expression level of different Bt proteins are different. Tu et al. (2000) reported expression of 20.0 µg g−1 cry1Ab/Ac in transgenic CMS line (T51-1) of Minghui 63 which controlled 100 % of YSB in laboratory condition and 91.4–95.7 % in fields. Chen et al. (2005) reported Cry2A* protein concentration in four homozygous transgenic lines ranging from 9.65 to 12.11 µg g−1 of fresh leaf weight. YSB mortality in bioassays of these Cry2A* plants was found to be 100 %. These same events recorded deadheart symptom of 5.36–7.48 % in field which is approximately one-third of the control, with whiteheads ranging from 0.0 to 0.5 %. Zhao et al. (2004) reported a much lower expression 1.2 µg g−1 of CpTI + cry1Ac in B-line of Minghui 86 hybrid. And when the same event was tested in field condition, they reported 99.37 % control against YSB (Li et al. 2005a). Yang et al. (2014) reported a range of Bt protein (Cry1Ac/Cry1I) from 0.67 μg g−1 to 1.5 µg g−1 in green tissues to be effective against RLF and striped stem borer, C. suppressalis in transgenic rice. The protein expression level of the five selected Cry2AX1 transgenic lines was in line with some of the previous experiments dealing with different Cry proteins.
There was no information regarding the adequate Cry2AX1 protein concentration needed for high YSB mortality. The critical concentration of Cry2AX1 protein in transgenic plants for best resistance to YSB was primarily established by subjecting the transgenic events through YSB pseudo-stem bioassay. Three stunted events (BtE17, BtE27 and BtE36) were rejected though having high level of Cry2AX1 protein expression as well as high bioefficacy. Over-expression of transgenes may lead to severe negative effects and abnormal morphology leading to stunting and/or leaf chlorophyll bleaching in transgenic rice (Ku et al. 1999; Takeuchi et al. 2000; Tsuchida et al. 2001 and Taniguchi et al. 2008). Ten out of a total 18 T1 events screened by ELISA and for insect mortality resulted in low expression and low mortality (data not provided). Variability of Cry2AX1 protein concentration in five best selected events in the target tissues didn’t appear to be too high and was usually within a range (Table 1). And we presume that this will be the critical level of protein concentration. However, future experiments with purified proteins and development of protocol for growing YSB on artificial media may help address this issue critically.
Higher but range bound, accumulation of Cry2AX1 protein in target tissue would also result in enhanced bioefficacy against target pests. This hypothesis is in agreement with result of the current study, as the expression of Cry2AX1 protein was found to be higher in green tissues at later stages in all the tested transgenic events (Table 1). This data was also further supported by bioassays, wherein plants at reproductive stage were found to be more effective in resisting YSB compared to the plants at vegetative stage. This kind of accumulative expression might protect the transgenic Bt rice plants from YSB throughout the life cycle of the crop and especially more at the later stages when the pest pressure is usually higher. As reported earlier (Muralidharan and Pasalu 2006) damage at reproductive stage (whitehead) causes more yield reduction than damage at the vegetative stage (deadhearts). Kim et al. (2009) also demonstrated that targeting of Cry1Ac protein to chloroplasts using rbcS:tp system leads to high level of accumulation in the green tissues resulted in protection throughout the growth phase in field condition. However, Ye et al. (2009) observed a declining trend of the Cry1C* protein concentration in the period between the vegetative and reproductive growth stages. This may be because of the fact that Ye et al. (2009) didn’t use the chloroplast transit peptide in their construct. The enhanced protection displayed by the five Cry2AX1 events at the later growth stages will be valuable for future product development and needs to be validated under field condition.
In the present study, mortality of neonate YSB larva in cut-stem bioassay of the selected five events was found to be ranging from 64 to 88 %. However in-planta bioassay recorded deadheart damage of 0.0–3.9 % and whitehead damage of 0.0–0.7 %. In-planta bioassay results correlated more closely with the protein expression results. This confirms in-planta bioassays as more reliable than cut-stem bioassays (Khanna and Raina 2002).
In the current study, transgenic Cry2AX1 rice was effective in controlling RLF, and displayed range mortality (57.5–80.0 %) of 2nd instar RLF larva accompanied by feeding inhibition. However, though the leaf feeding was minimal (<5 %) in transgenic events, leaf folding was found to be common across all the events. This may be due to the fact that larvae usually fold the leaves first and then start feeding (Pathak and Khan 1994). Occurrence of folded leaves was also observed by Song et al. (2014) in transgenic Bt (Cry1Ab) rice bioassays against RLF, though the feeding area was less than 5 %, as opposed to 30 % in control. Li et al. (2005b) observed mortality of the third, fourth and fifth-instar RLF larvae of 81, 78 and 68 % respectively, after 72 h of feeding on transgenic Cry1Ab Bt rice, indicating that younger RLF larvae are more sensitive to Bt rice than older ones. Manikandan et al. (2014a, b) recently reported effectiveness of Cry2AX1 protein against RLF. The recombinant Cry2AX1 protein expressed in B. thuringiensis (13.33 ng µl−1) and E. coli cells (20 ng µl−1) reported mortality of RLF to be 86.7 and 56.7 % respectively (Manikandan et al. 2014a). Later, when the same Cry2AX1 protein was expressed in transgenic rice plants at ranges from 3.3 to 16.0 ng g−1 of fresh leaf tissue, RLF larval mortality was observed up to 40 % and significant reduction in leaf feeding (Manikandan et al. 2014b). This report is providing further evidence that Cry2AX1 is not only effective against RLF but also has a higher mortality rate of 77 % compared to 40 % reported by Manikandan et al. (2014b). This could be because of the gene construct used in our study was different compared to Manikandan et al. (2014b). Thus their results are not directly comparable to the present study.
Cry2AX1 transgenic rice was tested against OAW and we successfully demonstrated the potential usefulness of cry2AX1 transgenic rice in controlling OAW (Table 3). Kim et al. (2009), reported 20 % mortality of Armyworms under laboratory conditions when insects were fed with transgenic Cry1Ac. Yun et al. (2004) tested transgenic Cry1Ac in maize against OAW and found the gene to be effective. Other purified Cry proteins such as Cry1Ac (Jiang et al. 2010), Cry1Ab (Wang et al. 2013) and Cry1Ab/Cry9Aa fusion protein (Jianhua et al. 2011) were found to be effective against OAW in in vitro protein feeding assays.
Based on in-planta and in vitro insect bioassay studies conducted in the current study it is evident that the selected cry2AX1 transgenic rice events were significantly resistant against three different types of lepidopteran insect pests of rice i.e. YSB, RLF and OAW. All the five selected events displayed normal phenotype and provided resistance even at the late reproductive stage when the insect pressure is usually higher. This proves that Cry2AX1 is one of the most effective Bt gene against lepidopteran insects pests in rice. Among the five events, BtE15 had higher level of insect bioefficacy, and thus will have higher resistance to lepidopteran insect pests under field conditions. We plan to induce recombination process for marker free event generation and subsequently conduct field evaluation (pending approval from regulatory authorities in India).
References
Balakrishnan N, Ruturajrajan B, Naveenkumar A, Sozhavendan AE, Nandeesha P, Illakkiyapriya A, Balasubramani V, Kumar KK, Sudhakar D, Udayasuriyan V (2012) Genetic transformation of tomato with cry2AX1 gene for resistance to fruit borer. In: Proceeding of 3rd world congress on biotechnology. Hyderabad international convention centre (HICC), Hyderabad, 13–15 Sept 2012
Bandong JP, Litsinger JA (2005) Rice crop stage susceptibility to the rice yellow stemborer Scirpophaga incertulas (Walker) (Lepidoptera: Pyralidae). Int J Pest Manag 51:37–43
Bashir K, Husnain T, Fatima T, Latif Z, Mehdi SA, Riazuddin S (2004) Field evaluation and risk assessment of transgenic indica basmati rice. Mol Breed 13:301–312
Chakraborty M, Reddy PS, Narasu ML, Krishna G, Rana D (2016) Agrobacterium-mediated genetic transformation of commercially elite rice restorer line using nptII gene as a plant selectable marker. Physiol Mol Biol Plants, 1–10 (doi:10.1007/s12298-015-0334-y)
Chen H, Tang W, Xu CG, Li XH, Lin YJ, Zhang QF (2005) Transgenic indica rice plants harboring a synthetic cry2A* gene of Bacillus thuringiensis exhibit enhanced resistance against lepidopteran rice pests. Theor Appl Genet 111(7):1330–1337
Chen H, Mang G, Zhang QF, Lin YJ (2008) Effect of transgenic Bacillus thuringiensis rice lines on mortality and feeding behavior of rice stem borers (Lepidoptera: Crambidae). J Econ Entomol 101(1):182–189
Cheng X, Sardana R, Kaplan H, Altosaar I (1998) Agrobacterium-transformed rice plants expressing synthetic cry1Ab and cry1Ac genes are highly toxic to striped stem borer and yellow stem borer. Proc Natl Acad Sci USA 95:2767–2772
Chu CC, Wang CC, Sun CS, Msu C, Yin KC, Chu CY, Bi FY (1975) Establishment of an efficient medium for anther cultures of rice through comparative experiments on nitrogen sources. Sci Sin 18:659–668
Datta K, Vasquez Z, Tu J, Torrizo L, Alam MF, Oliva N, Abrigo E, Khush GS, Datta SK (1998) Constitutive and tissue-specific differential expression of the cryI(A)b gene in transgenic rice plants conferring resistance to rice insect pests. Theor Appl Genet 97(2):20–30
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA mini preparation: version II. Plant Mol Biol Rep 1:19–21
Ghareyazie B, Alinia F, Menguito CA, Rubia LG, Palma JM, Liwanag EA, Cohen MB, Khush GS, Bennett J (1997) Enhanced resistance to two stem borers in an aromatic rice containing a synthetic cry1A(b) gene. Mol Breed 3:401–414
Hattori M, Atsusawa S (1980) Mass rearing of the cabbage armyworm, Mamestra brassicae Linné and the common armyworm, Mythimna separata Walker (Lepidoptera, Noctuidae) on a simple artificial diet. Jpn J Appl Entomol Zool 24:36–38
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282
Jang IC, Lee KH, Nahm BH, Kim JK (2002) Chloroplast targeting signal of a rice rbcS gene enhances transgene expression. Mol Breed 9(2):81–91
Jayaprakash SP, Nandeesha P, Sozhavendan AE, Naveenkumar A, Illakiyapriya A, Balakrishnan N, Balasubramani V, Singh PK, Sudhakar D, Balasubramanian P, Udayasuriyan V (2014) Expression of a novel synthetic cry gene of Bacillus thuringiensis in transgenic tobacco confers resistance to Helicoverpa armigera and Spodoptera litura. J Pure Appl Microbiol 8(6):4687–4692
Jiang S-J, Luo L-Z, Hu Y, Zhang L (2010) Effects of Cry1Ac protein on growth and development, reproduction and flight potential of the oriental armyworm, Mythimna separata (Lepidoptera: Noctuidae). Acta Entomol Sin 53(12):1360–1366
Jianhua G, Zhang Y, Zhao Q, Lin C, Xu X, Zhicheng S (2011) Transgenicrice expressing a fusion protein of Cry1Ab and Cry9Aa confers resistance to a broad spectrum of lepidopteran pests. Crop Sci 51(6):2535–2543
Khan ZR, Litsinger JA, Barrion AT, Villanenva FFD, Fernandez NJ, Taylo LD (1991) World bibliography of rice stem borers. IRRI, Manila, pp 1794–1990 (Published by ICIPE)
Khanna HK, Raina SK (2002) Elite indica transgenic rice plants expressing modified cryIA(c) endotoxin of Bacillus thuringiensis show enhanced resistance to yellow stem borer (Scirpophaga incertulas). Transgenic Res 11(4):411–423
Kim EH, Suh SC, Park BS, Shin KS, Kweon SJ, Han EJ, Park SH, Kim YS, Kim JK (2009) Chloroplast-targeted expression of synthetic cry1Ac in transgenic rice as an alternative strategy for increased pest protection. Planta 230(2):397–405
Krens FA, Pelgrom KTB, Schaart JG, Den Nijs TPM, Rouwendal GJA (2005) Clean vector technology for marker-free transgenic crops. In: Tuberosa R, Phillips RL, Gale M (eds) Proceedings of the international congress of the wake of the double helix: from the green revolution to the gene revolution, pp 509–515
Krishnaiah K, Varma NRG (2012) Changing insect pest scenario in the rice ecosystem—a national perspective. Directorate of Rice Research Rajendranagar, Hyderabad, p 28
Ku MS, Agarie S, Nomura M, Fukayama H, Tsuchida H, Ono K, Hirose S, Toki S, Miyao M, Matsuoka M (1999) High-level expression of maize phosphoenol pyruvate carboxylase in transgenic rice plants. Nat Biotechnol 17(1):76–80
Kumar S, Chandra A, Pandey KC (2008) Bacillus thuringiensis (Bt) transgenic crop: an environment friendly insect-pest management strategy. J Environ Biol 29(5):641–653
Kyozuka J, McElroy D, Hayakawa T, Xie Y, Wu R, Shimamoto K (1993) Light-regulated and cell-specific expression of tomato rbcS-gusA and rice rbcS-gusA fusion genes in transgenic rice. Plant Physiol 102(3):991–1000
Li F-F, Ye G-Y, Chen X-X, Peng Y-F (2005a) Effects of transgenic Bt rice on the food consumption, growth and survival of Cnaphalocrocis medinalis (Guenée) larvae. Rice Sci 12(3):202–206
Li YR, Hu YQ, Zheng Y, Hu XB, Zhang MJ, Li BJ (2005b) Field evaluation of the resistance of transgenic rice expressing CpTI or CpTI+Bt to lepidopterous pests. J Fujian Agric For Univ Nat Sci Ed 34(2):181–184 (in Chinese)
Mandal CC, Gayen S, Basu A, Ghosh KS, Dasgupta S, Maiti MK, Sen SK (2007) Prediction-based protein engineering of domain I of Cry2A entomocidal toxin of Bacillus thuringiensis for the enhancement of toxicity against lepidopteran insects. Protein Eng Des Sel 20(12):599–606
Manikandan R, Sathish S, Balakrishnan N, Balasubramani V, Sudhakar D, Udayasuriyan V (2014a) Agrobacterium mediated transformation of indica rice with synthetic cry2AX1 gene for resistance against rice leaf folder. J Pure Appl Microbiol 8(4):3135–3142
Manikandan R, Naveenkumar A, Stephy RB, Balakrishnan N, Balasubramani V, Sudhakar D, Udayasuriyan V (2014b) Comparative toxicity of chimeric Cry2AX1 Bt protein isolated from recombinant Bt and E. coil hosts against rice leaf folder (Cnaphalocrosis medinalis). Trends Biosci 7(11):1125–1130
Mishra N, Choubey G, Patel DP, Mishra A (2013) Army worm—a serious sporadic insect pest of rice in Rewa region of Madhya Pradesh. Indian J Appl Res 3(9):489–490
Muralidharan K, Pasalu IC (2006) Assessments of crop losses in rice ecosystem due to stem borer damage. Crop Prot 25(5):409–417
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15(3):473–497
Nayak P, Basu D, Das S, Basu A, Ghosh D, Ramakrishnan NA, Ghosh M, Sen SK (1997) Transgenic elite indica rice plants expressing cryIAc delta-endotoxin of Bacillus thuringiensis are resistant against yellow stem borer (Scirpophaga incertulas). Proc Natl Acad Sci USA 94(6):2111–2116
Nester EW, Altosaar I, Stotzty G (2002) 100 years of Bacillus thuringiensis: a critical scientific assessment. American academy of microbiology colloquium report. Based on colloquium, Ithaca
Outchkourov NS, Peters J, de Jong J, Rademakers W, Jongsma MA (2003) The promoter-terminator of Chrysanthemum rbcS1 directs very high expression levels in plants. Planta 216:1003–1012
Pathak MD, Khan ZR (1994) Insect pests of rice. International Rice Research Institute, Manila, p 35
Pillay P, Schlüter U, van Wyk S, Kunert KJ, Vorster BJ (2014) Proteolysis of recombinant proteins in bioengineered plant cells. Bioengineered 5(1):15–20
Qi Y, Chen L, He X, Jin Q, Zhang X, He Z (2013) Marker-free, tissue-specific expression of Cry1Ab as a safe transgenic strategy for insect resistance in rice plants. Pest Manag Sci 69(1):135–141
Qiu C, Sangha JS, Song F, Zhou Z, Yin A, Gu K, Tian D, Yang J, Yin Z (2010) Production of marker-free transgenic rice expressing tissue-specific Bt gene. Plant Cell Rep 29(10):1097–1107
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, vol 3, 3rd edn. Cold Spring Harbor Press, New York, pp A1.1–A1.30 (Appendix 1: Preparation of buffers and stock solutions)
Selvaraj K, Chander Subhash, Sujithra M (2012) Determination of multiple-species economic injury levels for rice insect pests. Crop Prot 32:150–160
Song FS, Ni DH, Li H, Duan YB, Yang YC, Ni JL, Lu XZ, Wei PC, Li L, Yang JB (2014) A novel synthetic Cry1Ab gene resists rice insect pests. Genet Mol Res 13(2):2394–2408
Takeuchi Y, Akagi H, Kamasawa N, Osumi M, Honda H (2000) Aberrant chloroplasts in transgenic rice plants expressing a high level of maize NADP dependent malic enzyme. Planta 211(2):265–274
Tang W, Chen H, Xu C, Li X, Lin Y, Zhang Q (2006) Development of insect-resistant transgenic indica rice with a synthetic cry1C* gene. Mol Breed 18(1):1–10
Taniguchi Y, Ohkawa H, Masumoto C, Fukuda T, Tamai T, Lee K, Sudoh S, Tsuchida H, Sasaki H, Fukayama H, Miyao M (2008) Overproduction of C4 photosynthetic enzymes in transgenic rice plants: an approach to introduce the C4-like photosynthetic pathway into rice. J Exp Bot 59(7):1799–1809
Tsuchida H, Tamai T, Fukayama H, Agarie S, Nomura M, Onodera H, Ono K, Nishizawa Y, Lee BH, Hirose S, Toki S, Ku MS, Matsuoka M, Miyao M (2001) High level expression of C4-specific NADP-malic enzyme in leaves and impairment of photoautotrophic growth of a C3 plant, rice. Plant Cell Physiol 42(2):138–145
Tu JM, Zhang GA, Datta K, Xu CG, He YQ, Zhang QF, Khush GS, Datta SK (2000) Field performance of transgenic elite commercial hybrid rice expressing Bacillus thuringiensis δ-endotoxin. Nat Biotechnol 18(10):1101–1104
Udayasuriyan V (2012) Efficacy of cry2AX1 protein expressed in tobacco plants against Helicoverpa armigera and Spodoptera litura. In: International conference on plant biotechnology for food security (ICPBFS): frontiers, Volume: ST 28—poster presentation, 22 Feb 2012
Udayasuriyan V, Indra Arulselvi P, Balasubramani V, Sudha DR, Balasubramanian P, Sangeetha P (2010) Construction of new chimeric cry2AX1 gene of Bacillus thuringiensis encoding protein with enhanced insecticidal activity. Indian patent no 244427
Visalakshmi V, HariSatyanarayna N, Jyothula DPB, Raju MRB, Ramana Murthy KV (2014) Screening of rice germplasm for resistance to yellow stem borer Scirpophaga incertulas walker. Int J Plant Anim Environ Sci 4(1):130–133
Waddington SR, Li X, Dixon J, Hyman G, de Vicente MC (2010) Getting the focus right: production constraints for six major food crops in Asian and African farming systems. Food Secur 2(1):27–48
Waldbauer GP, Marciano AP (1997) Rice leaf folder: mass rearing and a proposal for screening for varietal resistance in the greenhouse. IRRI research paper series no 27
Wang ZX, Yi ZL, Wang ZC, Jiang JX, Qin JP, Zhang H, Tan YN, Yi Chuan (2007) Quick testing-technology of transgenic rice with npt-screen marker gene. Hereditas 29(4):499–507 (in Chinese)
Wang Y, Zhang G, Du J, Liu B, Wang M (2010) Influence of transgenic hybrid rice expressing a fused gene derived from cry1Ab and cry1Ac on primary insect pests and rice yield. Crop Prot 29(2):128–133
Wang L, Jiang Xingfu, Luo Lizhi, Stanley David, Sappington Thomas W, Zhang L (2013) A cadherin-like protein influences Bacillus thuringiensis Cry1Ab toxicity in the oriental armyworm, Mythimna separate. Environ Microbiol Rep 5(3):438–443
Widner WR, Whiteley HR (1989) Two highly related insecticidal crystal proteins of Bacillus thuringiensis subsp. kurstaki possess different host range specificities. J Bacteriol 171(2):965–974
Wong EY, Hironaka CM, Fischhoff DA (1992) Arabidopsis thaliana small subunit leader and transit peptide enhance the expression of Bacillus thuringiensis proteins in transgenic plants. Plant Mol Biol 20(1):81–93
Wu C, Fan Y, Zhang C, Oliva N, Datta SK (1997) Transgenic fertile japonica rice plants expressing a modified cry1A(b) gene resistant to yellow stem borer. Plant Cell Rep 17(2):129–132
Yang YY, Mei F, Zhang W, Shen Z, Fang J (2014) Creation of Bt rice expressing a fusion protein of Cry1Ac and Cry1I-like using a green tissue-specific promoter. J Econ Entomol 107(4):1674–1679
Ye R, Huang H, Yang Z, Chen T, Liu L, Li X, Chen H, Lin Y (2009) Development of insect-resistant transgenic rice with Cry1C*-free endosperm. Pest Manag Sci 65(9):1015–1020
Yoshida S, Forno DA, Cock JH, Gomez KA (1976) Routine procedures for growing rice plants in culture solution. In: Cock JH, Gomez KA (eds) Laboratory manual for physiological studies of rice. IRRI, Los Banos, pp 61–66
Yun GL, Deng SD, Zang QW, Xu HL, Cai QN (2004) The resistance of Bt corn (MG95) to Pseudaletia separata. Entomol Knowl 41(5):422–426
Zhao HY, Zhang YJ, Wu KM, Zhao KJ, Peng YF, Guo YY (2004) Expression of Cry1Ac/CpTI transgenic rice and its resistance in different stages to Chilo suppressalis. J Agric Biotechnol 12(1):76–79
Zheng SJ, Henken B, Maagd RA, Purwito A, Krens FA, Kik C (2005) Two different Bacillus thuringiensis toxin genes confer resistance to beet armyworm (Spodoptera exigua Hübner) in transgenic Bt-shallots (Allium cepaL.). Transgenic Res 14(3):261–272
Acknowledgments
We are thankful to Mr. Sanjay Gupta, Director J.K Agri. Genetics Ltd. and we acknowledge all type of inputs towards this research from all co-workers in J. K Biotech Lab. This work is supported and Funded by the Department of Biotechnology, Government of India (BIRAC Grant No. BT/BIPP/0320/07/10). The authors wish to thank the anonymous referees for their constructive suggestion.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chakraborty, M., Reddy, P.S., Mustafa, G. et al. Transgenic rice expressing the cry2AX1 gene confers resistance to multiple lepidopteran pests. Transgenic Res 25, 665–678 (2016). https://doi.org/10.1007/s11248-016-9954-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-016-9954-4