Abstract
We describe the use of plant-made β-defensins as effective antimicrobial substances for controlling salmonellosis, a deadly infection caused by Salmonella typhimurium (referred to further as S. typhi). Human β-defensin-1 (hBD-1) and -2 (hBD-2) were expressed under the control of strong constitutive promoters in tobacco plants, and bio-active β-defensins were successfully extracted. In the in vitro studies, enriched recombinant plant-derived human β-defensin-1 (phBD-1) and -2 (phBD-2) obtained from both T1 and T2 transgenic plants showed significant antimicrobial activity against Escherichia coli and S. typhi when used individually and in various combinations. The 2:1 peptide combination of phBD-1:phBD-2 with peptides isolated from T1-and T2-generation plants reduced the growth of S. typhi by 96 and 85 %, respectively. In vivo studies employing the mouse model (Balb/c) of Salmonella infection clearly demonstrated that the administration of plant-derived defensins individually and in different combinations enhanced the mean survival time of Salmonella-infected animals. When treatment consisted of the 2:1 phBD-1:phBD-2 combination, approximately 50 % of the infected mice were still alive at 206 h post-inoculation; the lowest number of viable S. typhi was observed in the liver and spleen of infected animals. We conclude that plant-made recombinant β-defensins (phBD-1 and phBD-2) are promising antimicrobial substances and have the potential to become additional tools against salmonellosis, particularly when used in combination.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Salmonella typhimurium (referred to as S. typhi in text), a Gram-negative bacterium, is capable of causing salmonellosis both in human and animals, resulting significant morbidity and mortality worldwide (Fink and Cookson 2007). S. typhi is a clinical bacterial pathogen that causes food poisoning (Grassl et al. 2008), severe liver infection (Coburn et al. 2007) and acute infections in wounds/burns (Carsiotis et al. 1989). Fluoroquinolones and tetracyclines are the most commonly used antibiotics for treating Salmonella infection. However, a few S. typhi strains are showing resistance to common antibiotics (Stevenson et al. 2007), and the emergence of a multidrug-resistant strain of S. typhi (definitive type 104) is becoming a serious threat to human health worldwide (Threlfall et al. 2000; Poppe et al. 2002; Gebreyes et al. 2004; Perron et al. 2008). Consequently, the formulation of novel therapeutic(s) for controlling drug-resistant bacteria/microbes is currently an urgent research and clinical target. More recently, the focus has been directed towards antimicrobial peptides (AMPs) or molecules resembling AMPs owing to their unique membrane-specific mode of action as source materials for developing unique biologics for controlling drug-resistant bacteria (Hancock and Sahl 2006; Ajesh and Sreejith 2009).
Defensins are antimicrobial peptides that are currently recognized as ‘peptide-based antibiotics’ against a diverse range of microorganisms, including bacteria, fungi and viruses (Schneider et al. 2005). These antimicrobial peptides function as the first-line defense in mucosal surfaces of the eye, gastrointestinal tracts, tracheobronchial tree, lungs and skin (Ganz et al. 1985; Lehrer et al. 1993). Human defensins are small cationic molecules of 3–5 kDa (Zasloff 2002; Ganz 2003) which are classified into two subfamilies, namely, α- and β-defensins, depending on their distribution, sequence homology and the connectivity of the six conserved cysteine residues present in a defensin molecule. The disulphide linkages of cysteine residues in α-defensins are Cys1–Cys6, Cys2–Cys4 and Cys3–Cys5, whereas in β-defensins, the linkages are Cys1–Cys5, Cys2–Cys4 and Cys3–Cys6 (Ganz 2003). The cationic characteristic and amphipathic nature of defensin molecules allow them to interact electrostatically with the negatively charged phospholipid bilayer of the microbial cell membrane, resulting in the formation of pores and subsequent disruptions of the bacterial membrane that ultimately lead to cytoplasmic leakage and cell death (Shimoda et al. 1995; Harder et al. 2001; Shai 2002). In addition to such antimicrobial activities, defensins play a positive role in wound healing (Li et al. 2006) and in controlling immuno-modulatory function (Soruri et al. 2007; Presicce et al. 2009). Furthermore, the overexpression of human defensins in transgenic mice has been shown to provide protection against enteric salmonellosis (Salzman et al. 2003).
The observations of Salzman et al. (2003) led us to explore the role of recombinant plant-derived human defensins in the control of S. typhi growth using both in vitro and in vivo approaches. Since ancient times, plants have been used as sources of food and herbal medicines for human beings (Tiwari 2008; Meena et al. 2009). In more recent times, plants have emerged as a promising alternative to conventional fermentation/cell culture-based systems for the commercial production of life-saving biologics and as a better platform for translational research. The advantages of such plant-based systems are: (1) low costs for the production of recombinant proteins compared to microbial and mammalian cell culture-based systems (Twyman et al. 2003); (2) minimum risks of product contamination(s) with endotoxins or human pathogens; (3) the possibility to trigger the production of recombinant protein in the edible organ of plants, such as the seed and fruit, for easy storage, and allowing them to be administered as edible-vaccines (Mason et al. 2002; Twyman et al. 2003); (4) mass-scale production of recombinant proteins through the cultivation of more plants; (5) downstream processing of plant-made pharmaceuticals (PMPs) is easy and inexpensive particularly, when they are produced exclusively in a particular plant organ (Seon et al. 2002); (6) the plant can impart specific post-translational modifications essential for the stability, bio-activity, immunogenicity and pharmacokinetics of the PMPs (Gomord and Faye 2004).
In the study described here, we developed transgenic tobacco plants expressing human β-defensins hBD-1 (Valore et al. 1998) under control of the MUAS35SCP promoter (Patro et al. 2012) and hBD-2 (Liu et al. 1998; Aerts et al. 2007) under control of the FUASFSCP promoter (Ranjan et al. 2012). We have successfully enriched bio-active plant-derived β-defensin-1 (phBD-1) and β-defensin-2 (phBD-2) from plants. The bactericidal properties of recombinant phBD-1 and phBD-2 (both individually and in combination) were tested against Escherichia coli and S. typhi in vitro and in vivo using the ‘typhoid fever model’ of Balb/c mice.
We report here the production of bio-active beta-defensins in plants and their antimicrobial efficacies individually and in different stoichiometric combinations against E. coli and S. typhi. We presume that such production will be cost effective and easy to scale up. The development of an economically feasible production system for the production of ‘high valued’ defensins in plants may promote antimicrobial peptide-based therapeutics’ for controlling drug-resistant bacteria and facilitate future investigations on the mode of action of these peptides.
Materials and methods
Materials and animals
Restriction enzymes and Pfu DNA Polymerase were purchased from Thermo Scientific/Promega (Madison, WI). The RNA isolation kit, DNaseI digestion kit and other fine chemicals were obtained from Sigma (St. Louis, MO). Baculovirus expression vector-based cDNA clones of human beta defensin-1 (hBD-1) and human beta defensin-2 (hBD-2) were kindly provided by Prof. T. Ganz, University of California. The antibodies against hBD-1 and hBD-2 were procured from Santa Cruz Biotechnology Inc. (Dallas, TX). Commercially available hBD-1 (CYT-564) and hBD-2 (D9690) were obtained from ProSpec (Ness-Ziona, Israel) and Sigma, respectively. S. typhimurium (MTCC No. SL1344) was provided by Dr. D.V. Singh, Institute of Life Sciences (ILS), Bhubaneswar, India.
Balb/c mice (originally imported from the Jackson laboratories, Germany) were obtained from the National Institute of Immunology, New Delhi. Routine breeding and maintenance of mice were carried out in the animal facility at ILS. Male mice aged approximately 8 weeks were used in the study. All experiments with mice were in accordance with the protocols of the animal ethics committee of ILS.
Cloning of hBD-1 and hBD-2 into plant expression vector
The coding sequences of the full-length cDNAs of hBD-1 (207 bp) and hBD-2 (195 bp) were amplified by the PCR from the respective cDNA clones as template using gene-specific primers (hBD-1Fp/hBD-1Rp for hBD-1; hBD-2Fp/hBD-2Rp for hBD-2; Table 1). For cloning, restriction sites for XhoI and SacI were generated at the 5′ and 3′ ends, respectively. The PCR products 5′-XhoI-hBD-1-SacI-3′ and 5′-XhoI-hBD-2-SacI-3′ were purified by gel electrophoresis. The 5′-XhoI-hBD-1-SacI-3′ fragment was cloned into corresponding sites of the pKMUAS35SCPGUS vector (containing the MUAS35SCP promoter; Patro et al. 2012) to generate the pKMUAS35SCP-hBD-1 construct [replacing the β-glucuronidase (GUS) gene]. Likewise, the 5′-XhoI-hBD-2-SacI-3′ fragment was cloned into the pKFUASFSCPGUS vector (containing the FUASFSCP promoter; Ranjan et al. 2012) to generate the pKFUASFSCP-hBD-2 construct (replacing the GUS gene). The sequence integrity of these constructs was verified prior to further use.
Developing transgenic plants expressing hBD-1 and hBD-2
Ten transgenic tobacco plants (D1-1 to D1-10) harboring pKMUAS35SCP-hBD-1 and nine independent transgenic plants (D2-1 to D2-9) corresponding to the pKFUASFSCP-hBD-2 were developed following the standard Agrobacterium-mediated plant transformation protocol (Dey and Maiti 1999). Transgenic plants were grown in the greenhouse until seed setting under the following conditions: a photoperiod of 16/8 h (light/dark), light intensity of 220 µmol/m2/s, temperature of 28 ± 3 °C and humidity of 70–75 %. Segregation analysis was performed using seeds from individual putative transgenic plant lines corresponding to hBD-1 and hBD-2 on solid Murashige and Skoog (1962) (MS) medium containing kanamycin (Kan; 300 mg/l). Approximately 50 seeds (in triplicate) from independent putative transgenic plant lines (T0) corresponding to hBD-1 and -2, respectively, were germinated on the plates. The number of Kan-sensitive (turning yellow) and Kan-resistant (green and healthy) seedlings from each line were counted on each plate, and the data obtained were subjected to chi-square analysis for the expected 3:1 segregation ratio. Independent transgenic lines (T1 generation) with a chi-square value of <1.5 (p ≤ 0.05) were considered to be true transgenic lines showing proper segregation ratio (KanR:KanS = 3:1). Seeds from plants (T1) showing the appropriate segregation ratios were germinated on MS plates for raising T2 plants. Transgenic tobacco plants raised containing the pKYLX vector (Schardl et al. 1987) were used as the transformed control (TC).
Molecular characterization of transgenic plants expressing hBD-1 and hBD-2
Genomic DNA PCR for gene integration analysis
Genomic DNAs were extracted from leaves of both the TC and transgenic plants harboring the hBD-1 and hBD-2 transgene, respectively, following the protocol of Allen et al. (2006). PCR amplifications of hBD-1 and hBD-2 were performed using gene-specific forward (Fp) and vector-specific reverse (Rp) primers (hBD-1Fp and hBD-2Fp as forward primers for hBD-1 and hBD-2, respectively; rbcSE9Rp as the reverse primer; Table 1). The PCR products were analyzed on a 1 % agarose gel.
Reverse transcription-PCR for transcript expression analysis
Total RNA from T1 seedlings corresponding to the TC and transgenic lines (harboring hBD-1 and hBD-2) were extracted separately using the Spectrum™ Plant Total RNA kit (Sigma) and subsequently treated by DNaseI (Sigma). First-strand cDNA synthesis with 1 µg of DNaseI-treated RNA was performed using the First Strand cDNA Synthesis kit of Thermo Scientific/Fermantas (Vilnius, Lithuania). PCR assays were carried out to amplify hBD-1 using first-strand cDNAs as the template in the presence of specific primers (hBD-1Fp, hBD-1Rp; Table 1). Likewise, hBD-2 was amplified by PCR using the corresponding cDNA as the template in the presence of specific primers (hBD-2Fp and hBD-2Rp; Table 1). The integrity of the PCR products was checked on a 2 % agarose gel.
Enrichment of plant-derived hBD-1 and hBD-2 peptides
Enrichment of phBD-1 and phBD-2 from the respective crude protein extracts was performed following the protocol of Kovaleva et al. (2009) with slight modifications. In brief, leaves (20 g) collected from transgenic plants were first cleaned, then ground to a fine powder with liquid nitrogen, homogenized with 60 ml of 50 mM sulfuric acid and finally centrifuged at 14,000 g for 20 min, at 4 °C. The supernatant was collected and the pH adjusted to pH 7.8, following which it was incubated at 4 °C for 30 min and then centrifuged again at 14,000 g for 20 min at 4 °C. Solid ammonium sulfate was added to the supernatant to obtain 35 % relative saturation. The precipitate, formed after stirring for 1 h at 4 °C, was removed by centrifugation at 14,000 g for 30 min, at 4 °C and the supernatant was incubated overnight at 4 °C with constant stirring as an 80 % saturated ammonium sulfate solution. The precipitate was collected as an enriched protein fraction by centrifugation, dissolved in a buffer containing 20 mM Tris–HCl buffer (pH 7.4) and 10 mM NaCl and then dialysed against a buffer containing 20 mM Tris–HCl (pH 7.4), 100 mM NaCl using 2-KDa cutoff dialysis tubing (Sigma). The concentration of the enriched peptides was determined according to Bradford (1976).
Enzyme-linked immunosorbent assay
An enzyme-linked immunosorbent assay (ELISA) was performed using the enriched peptides isolated from transgenic plant lines expressing hBD-1 (D1-3, D1-6, D1-7 and D1-10) and hBD-2 (D2-6 and D2-9) according to Vaazquez et al. (1996) in order to determine the relative accumulation level of hBD-1 and hBD-2 in different plant lines and to estimate the percentage of recombinant defensin present in the enriched product. Briefly, the ELISA plates were coated separately with aliquots containing 5 µg of enriched peptides of phBD-1, phBD-2 and TC plant extract, respectively. Primary antibodies (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) specific to hBD-1 and hBD-2, respectively, were then applied for performing each ELISA separately. Color development reactions were carried out using horse radish peroxidase-conjugated secondary antibody and ortho-phenylenediamine as the substrate. After color development, the optical density (OD) of the reaction products was measured at 492 nm. The standard curves (OD492 vs. concentrations) for β-defensins were prepared by plotting the OD492 of different known concentrations of commercially available pure β-defensin-1 and 2. The purity of phBD-1 and phBD-2 (µg/gm of plant tissue) in corresponding plant-derived enriched fractions were determined using the respective standard curve.
In vitro antimicrobial assays of phBD-1 and phBD-2 against E. coli and S. typhi
The antibacterial activity assays of enriched phBD-1 and phBD-2 (prepared from T1 and T2 plants) against E. coli and S. typhi were performed separately following the protocols described by Krishnakumari et al. (2006). Briefly, freshly grown bacterial cultures of E. coli and S. typhi in phosphate buffered saline (PBS) (50 µl; OD600 = 0.5—containing approximately 105 CFU) were incubated separately with 60 µg of enriched phBD-1 and phBD-2, respectively, in a 100-µl reaction volume at 37 °C with constant shaking (200 rpm). After 1 h of incubation, the treated bacterial mixtures for both E. coli and S. typhi were further diluted by 5,000-fold and a 100-µl sample of each dilution was spread on LB agar plates (in triplicate). After overnight incubation at 37 °C, colony-forming units were counted for E. coli and S. typhi individually.
The antimicrobial assays using 2:1 and 1:1 combinations of phBD-1 and phBD-2 were carried out as described above against E. coli and S. typhi separately. The 2:1 combination contained 40 µg of phBD-1 and 20 µg of phBD-2. Likewise, the 1:1 combination contained 30 µg each of phBD-1 and phBD-2 peptides. Each assay was performed in triplicate.
Mice mortality assay
In vivo efficacy of transgenic plant-derived human β-defensins (phBD-1 and phBD-2) for protecting mice from the challenge of S. typhi was tested in a mouse model of Salmonella-infection following the protocol of Benincasa et al. (2010). Briefly, the mice divided into six treatment groups (4 mice per group): bacterial control (BC), TC, phBD-1, phBD-2, phBD-1:phBD-2 (2:1) and phBD-1:phBD-2 (1:1). First, an inoculum of S. typhi containing 103 CFU (expected to cause 90–100 % mortality in Balb/c mice within 4–6 days) was injected intraperitoneally into each mouse. Subsequently, each infected mouse was injected intraperitoneally with one of the test peptides (100 μg in a 100-μl reaction volume) or one of the combinations [2:1: 66 µg phBD-1 + 33 µg phBD-2; 1:1: 50 µg each of phBD-1 and phBD-2; in a 100-μl reaction volume]. Mice in the TC group were injected with 100 µg of extract obtained from the TC plant, and BC mice were injected with inoculum (103 CFU of S. typhi) only, without any additional peptides. Animal behavior and symptoms of salmonellosis were monitored at regular intervals of 6–8 h until the humane endpoint had been reached, based on sick parameters described by Wu et al. (2010). The time period between the administration of bacterial inoculum and the application of humane endpoint was considered as the survival period of experimental mice. The mean survival periods of experimental mice in each treatment group were recorded. The experiments were repeated three times.
CFU count of viable S. typhi in the liver and spleen of infected mice treated with recombinant peptides and combinations thereof
In a separate experiment, the CFU counts of viable S. typhi in the liver and spleen of infected mice treated with plant-derived defensins either individually and in combination were recorded in each treatment group of the mice (3 mice per group), as described in the previous section. Initially, each mouse in each group was injected with S. typhi (103 CFU) followed by administration of specific peptides, as designated for each treatment group (see preceding section). Mice in the TC group were injected with 100 µg of extract obtained from TC plant, and mice in the BC group were injected with inoculum only, without any peptides.
After 36-h post infection (hpi), the liver and spleen of each experimental mouse were removed; washed three times with PBS, homogenized in double volume (w/v) of PBS separately and centrifuged. The supernatant was diluted 100-fold in PBS and 100-µl samples of diluted culture were plated in duplicate on Salmonella–Shigella agar media (Hi Media Laboratories Pvt. Ltd, Mumbai, India), incubated at 37 °C overnight and CFUs were counted. The results were expressed as the percentage of inhibition obtained from plant-derived defensins with respect to that obtained from the TC plant. The assay was repeated three times.
Statistical analysis
All of the antimicrobial assay data presented in this article are mean values of three independent experiments and given with the respective standard deviation (SD). All the data were also subjected to analysis of variance analysis using GraphPad Prism version 4.0 (GraphPad Software, Inc., La Jolla, CA). The p value of ≤0.05 was considered to indicate significance.
Results
Integration analysis of hBD-1 and hBD-2 in transgenic tobacco plants
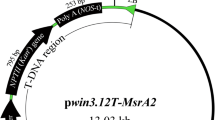
Schematic maps for plant expression vectors pKMUAS35SCP-hBD-1 (expressing hBD-1 under control of MUAS35SCP promoter) and pKFUASFSCP-hBD-2 (expressing hBD-2 under control of FUASFSCP promoter) were presented in Fig. 1.
Schematic presentation of plant expression vectors generated for expressing human defensin genes hBD-1 and hBD-2 in plants. a Expression cassette of the vector pKYLX, b pKMUAS35SCP-hBD-1 carrying hBD-1 directed by the MUAS35SCP promoter, c pKFUASFSCP-hBD-2 carrying hBD-2 under the control of the FUASFSCP promoter. The relative positions of the 3′ terminator sequence of ribulose bisphosphate carboxylase (rbcSE9), the nopaline synthase (NOS)-poly(A) region (Nos Poly A), Kanamycin resistance gene (Kan R) and promoter of the NOS gene (Nos Promoter) are shown. Arrows Directions of gene transcripts. The position of the unique restriction enzyme sites (EcoRI, HindIII, XhoI, SstI, ClaI) used to assemble these constructs are shown. LB Left border, RB right border
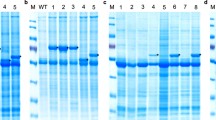
The integration of hBD-1 and hBD-2 into the genome of the transgenic plants was confirmed by PCR amplification using the respective genomic DNA as described in the “Materials and methods” section. We observed discrete PCR-amplified bands of hBD-1 (Fig. 2a) and hBD-2 (Fig. 2b). From among ten T1-transgenic plants of hBD-1 and nine T1-transgenic hBD-2 plants, we identified four PCR-positive hBD-1 transgenic plant lines (D1-3, D1-6, D-7, and D1-10; Fig. 2a) and three-PCR positive hBD-2 transgenic plant lines (D2-2, D2-6 and D2-9; Fig. 2b), respectively.
PCR, reverse transcription (RT)-PCR and enzyme-linked immunosorbent assay of transgenic plants expressing hBD-1 and hBD-2. a Amplification of the expected DNA fragments (847 bp, arrow) containing hBD-1 in combination with the rbcSE9 terminator from different transgenic plant lines (D1-3, D1-6, D1-7, D1-10). b Amplification of hBD-2 in combination with the rbcSE9 terminator were detected (arrow) from independent transgenic plant lines (D2-2, D2-6, D2-9). c RT-PCR analysis showing amplification (arrow) of the hBD-1 transcript from different independent transgenic plant lines (D1-3, D1-6, D1-7, D1-10). d Positive RT-PCR amplification (arrow) of hBD-2 (transcript) from different independent transgenic plant lines (D2-6, D2-9). e Levels of accumulation of plant-derived human β-defensin-1 (phBD-1) in D1-3, D1-6, D1-7 and D1-10 transgenic lines [mean of three independent experiments ± standard deviation (SD]. f Accumulation levels of plant-derived human β-defensin-2 (phBD-2) in D2-6 and D2-9 (mean of three independent experiments ± SD. M StepUp™ 100-bp DNA ladder, TC plant transformed with empty pKYLX71 vector, +ve PCR amplification of hBD-1 or hBD-2 using vectors carrying hBD-1 or hBD-2, respectively, as templates, -ve PCR using empty vector as template
For the transcript analysis, we performed reverse transcription-PCR of hBD-1 from the respective independent transgenic lines (D1-3, D1-6, D-7, D1-10) and of hBD-2 from the respective independent transgenic lines (D2-2, D2-6, D2-9). All of the lines with hBD-1 showed amplification of the hBD-1 transcript (Fig. 2c) while only two lines (D2-6, D2-9) with hBD-2 showed amplification of the hBD-2 transcript (Fig. 2d).
The accumulation level of phBD-1 in the four independent transgenic lines D1-3, D1-6, D1-7 and D1-10) expressing hBD-1 and that of phBD-2 in the two independent transgenic lines D2-6 and D2-9 expressing hBD-2 were measured as described in the “Materials and methods” section and are presented in Fig. 2e, f, respectively. More precisely, the relative accumulation levels of phBD-1 and phBD-2 in the respective transgenic lines were evaluated based on their normalized ELISA data (OD492) obtained by subtracting the normalized ELISA data obtained from TC lines as a control. The accumulation level of phBD-1 in the D1-3, D1-6, D1-7 and D1-10 transgenic lines was measured as 0.52, 0.89, 0.65 and 1.27 µg/g of plant tissues, respectively, and that of phBD-2 in the D2-2 and D2-9 transgenic lines was 1.29 and 1.07 µg/g, respectively. Data presented in Fig. 2e, f are the mean of three independent experiments with the respective SD.
In vitro analysis of antibacterial activity of phBD-1 and phBD-2 derived from T1- and T2- generation plants against E. coli and S. typhi
Assays of antimicrobial activity of the plant-derived phBD-1 and phBD-2 from (the T1 and T2 generations) against E. coli and S. typhi were performed as described in the “Materials and methods” section. The data obtained from these assays are presented in Fig. 3.
Antibacterial activity of phBD-1 and phBD-2 individually and in combination against Escherichia coli and Salmonella typhi. a Percentage inhibition of bacterial population using phBD-1 and phBD-2 isolated from T1-generation plants against E. coli (i) and S. typhi (ii). b Percentage inhibition of bacterial population using phBD-1 and phBD-2 isolated from T2-generation plants against E. coli (i) and S. typhi (ii)
We observed a 58.0, 44.6, 87.3 and 73.8 % reduction in E. coli growth in the presence of phBD-1 and phBD-2 obtained from T1 plants and with the phBD-1:phBD-2 (2:1) and phBD-1:phBD-2 (1:1) combinations, respectively, in comparison to E. coli growth in the presence of enriched peptides obtained from the TC plant [T1 generation; Fig. 3a (i)]. Additionally, we observed a 74.2, 80.8, 96.1 and 91.3 % inhibition of S. typhi growth in the presence of phBD-1 and phBD-2 obtained from T1 plants and with the phBD-1:phBD-2 (2:1) and phBD-1:phBD-2 (1:1) combinations, respectively, in comparison to S. typhi growth in the presence of TC plant extracts [T1 generation; Fig. 3a (ii)]. All of the results presented in Fig. 3a (i, ii) are the mean of three independent experiments ± SD.
Likewise, we evaluated the antimicrobial activities of the above-mentioned peptides isolated from the corresponding T2- generation plants and their combinations against E. coli and S. typhi. We observed a 50.1, 47.3, 83.0 and 70.1 % reduction in E. coli growth in the presence of phBD-1 and phBD-2 isolated from T2-generation plants and their combinations phBD-1:phBD-2 (2:1) and phBD-1:phBD-2 (1:1), respectively [Fig. 3b (i)]. Additionally, we observed a 40.9, 48.4, 84.6 and 76.2 % reduction in the growth of S. typhi in the presence of phBD-1 and phBD-2 isolated from T2- generation plants [Fig. 3b (ii)] and their combinations phBD-1:phBD-2 (2:1) and phBD-1:phBD-2 (1:1), respectively. The results are presented in Fig. 3b (i, ii) as the mean of three independent experiments ± SD.
Mortality assay of Salmonella-infected mice treated with phBD-1, phBD-2 and combinations thereof
We performed a mice mortality assay for evaluating the in vivo efficacy of plant-derived defensins against Salmonella infection as described in "Methods". Overall, the administration of phBD-1 and phBD-2, either individually or in combination (2:1 and 1:1), noticeably enhanced the survival time of S. typhi-challenged mice compared to mice in the control groups (BC and TC). A 100 % mortality was noted among the mice in the BC and TC treatment groups in between 24 and 72 hpi, while a single administration of recombinant peptide (either phBD-1 or phBD-2) increased the survival rate of infected mice to 75 % during the first 72 hpi (Fig. 4a). Furthermore, at 8 days post-injection, it was noted that 50 and 25 % of infected mice were still alive when they had been treated with the 2:1 and 1:1 combinations of defensins, respectively (Fig. 4a). We observed 100 % mortality for the mice receiving the phBD-1:phBD-2 (1:1) combination at approximately 206 hpi; in comparison, 50 % of infected mice injected with the phBD-1:phBD-2 (2:1) combination were still alive (Fig. 4a).
In vivo antibacterial activity assays in transformed and control mice using plant-derived defensins individually and in combinations. a Mortality assay curves of Salmonella-infected mice (Balb/c) treated with phBD-1 and phBD-2, individually and in combination. b Percentage reduction of S. typhi loads in liver and spleen of infected mice treated with phBD-1 and phBD-2, individually and in combination.
Analysis of CFU count of live S. typhi in liver and spleen of infected mice treated with phBD-1, phBD-2 and their combinations
We analyzed the residual load of live S. typhi in the liver and spleen of infected mice treated with recombinant phBD-1, phBD-2, phBD-1:phBD-2 (2:1) and phBD-1:phBD-2 (1:1) at 36 hpi, as described in the “Materials and methods” section. We observed approximately 93.2 and 77.5 % reduction of S. typhi load in the liver and spleen of infected mice treated with the 2:1 combination of phBD-1 and phBD-2, respectively, in comparison to that obtained after the administration of enriched proteins isolated from TC plants. Administration of the 1:1 combination of phBD-1 and phBD-2 resulted in approximately 42.2 and 59.5 % reduction of S. typhi load in the liver and spleen of infected mice, corresponding to the reduction obtained using proteins isolated from the TC plant (Fig. 4b). The data in Fig. 4b are presented as the mean of three independent experiments ± SD.
Discussion
The rapid emergence of drug-resistant S. typhi emphasizes an urgent necessity to develop new classes of antibiotics for challenging such resistant bacteria. During last decade, AMPs have become globally recognized as effective substitutes for conventional antibiotics/fungicides. At the present time, more than 1,500 AMPs with diverse sequences have been characterized from different biological species (Antimicrobial Peptide Database; http://aps.unmc.edu/AR/main.php). Mammalian-defensins, a family of cationic antimicrobial peptides, have also recently emerged as a promising ‘peptide-based’ antibacterial compound against a broad spectrum of microbes (Chen et al. 2006). Furthermore, defensins are involved in (1) controlling human immunodeficiency virus-type 1 (Nakashima et al. 1993; Quiñones-Mateu et al. 2003; Sun et al. 2005); (2) controlling tumor development (Shai 2002; Donald et al. 2003; Young et al. 2003; Markeeva et al. 2005); (3) establishing a link between innate and adaptive immune response during microbial infections; (4) differentiating the subtypes of both renal cell carcinoma and prostate cancer (Donald et al. 2003; Young et al. 2003). Unfortunately, defensin-based therapeutics are extremely costly due to high commercial production costs and inadequate commercial production systems.
We report here our successful production of bio-active recombinant human β-defensins (phBD-1 and phBD-2) in plants. Our study was based on our presumption that plants ensure a comparatively high production yield with less toxicity in a cost-effective way. We prefer the transgenic system over the transient system as the former enables stable inheritance of the transgene and the expression of transgenic proteins over generations (Garabagi et al. 2012) and thus may facilitate translational research. To achieve a high-level production of hBD-1 and hBD-2 in plants, we employed two strong constitutive hybrid promoters, namely, MUAS35SCP (Patro et al. 2012) and FUASFSCP (Ranjan et al. 2012), for expressing hBD-1 and hBD-2, respectively. The MUAS35SCP and FUASFSCP promoters showed about 5.6- and 4.4-fold stronger activity than the most widely used promoter, CaMV35S (Patro et al. 2012; Ranjan et al. 2012). Transgenic plant lines with the pKMUAS35SCP-hBD-1 (D1-3, D1-6, D1-7 and D1-10) and pKFUASFSCP-hBD-2 (D2-2, D2-6 and D2-9) constructs showed the proper segregation ratio, and with the exception of line D2-2, all of these lines showed the presence of the corresponding transcript as detected by RT-PCR. Absence of the hBD-2 transcript in D2-2 transgenic plants may have resulted from transcriptional gene silencing of the inserted transgene. Despite the many merits of plants as a production system for important biologics, the major constraint of this approach is purification of the expressed peptide up to homogeneity, particularly for low-molecular-weight peptides like defensins (3–5 kDa). We enriched the plant-made defensins by eliminating other proteins present in plant crude extract using NH4SO4 precipitation followed by several rounds of dialysis prior to evaluating their antimicrobial activities against E. coli and S. typhi, either individually or in combinations. We evaluated the relative accumulation level of hBD-1 and hBD-2 among the corresponding transgenic lines and found the highest accumulation level of hBD-1 and hBD-2 in lines D1-10 (1.27 µg/g fresh tissue) and D2-6 (1.29 µg/g fresh tissue), respectively. We also evaluated the antimicrobial activities of phBD-1 and phBD-2 in different stoichiometric ratios, as the concept of using multidrug in combination has received global recognition as the most effective approach for controlling resistant or pathogenic microbe. Furthermore, to eliminate the effect of peptides/proteins other than β-defensins which may be present in the enriched protein fraction of transgenic plants, we normalized the antimicrobial activity data of enriched hBD-1 and hBD-2 by subtracting the same data obtained using enriched TC plant extracts both in vivo and in vitro. Our study
demonstrated that plant-derived phBD-1 and phBD-2 were functionally active and had potent bactericidal activity, with approximately 40–50 % bactericidal activity when used individually and about 70–80 % activity when used in combinations against E. coli; against S. typhi, individual usage showed 50–60 % bacterial inhibition and combinations of phBD-1 and phBD-2 showed 80–90 % bacterial inhibition. We also observed that peptides obtained from both T1- and T2-generation plants showed similar antimicrobial activities against these two bacterial species. Analysis of the in vitro antimicrobial activity data revealed that the 2:1 combination of phBD-1 and phBD-2 had the maximum bactericidal activity against both test organisms.
The results of the mice mortality assay demonstrated that both phBD-1 and phBD-2 effectively inhibited the growth of S. typhi and significantly extended the survival of Salmonella-infected mice compared to untreated control mice. Infected mice treated with the 2:1 combination showed a 50 % survival rate, even at 230 hpi. Of all treatments, the 2:1 (phBD-1:phBD-2) combination showed the highest efficacy for extending the survival time of S. typhi-infected mice. We also evaluated the load of S. typhi in two susceptible organs, i.e., the liver and spleen, of infected mice after treatment with plant-derived defensins and their combinations. We detected significantly fewer counts of live S. typhi in the spleen and liver of Salmonella-infected mice treated with the 2:1 (phBD-1:phBD-2) combination compared to the other treatments.
In conclusion, we have demonstrated that plants are capable of producing bio-active defensins. Employing both in vitro and in vivo experiments we confirmed that a combination of plant-derived defensins showed a better efficacy for controlling S. typhi than each defensin used individually. In the future, the economic production of valuable defensins in plants may boost AMP-based treatment for bacterial infection.
References
Aerts AM, Thevissen K, Bresseleers SM et al (2007) Arabidopsis thaliana plants expressing human beta-defensin-2 are more resistant to fungal attack: functional homology between plant and human defensins. Plant Cell Rep 26:1391–1398. doi:10.1007/s00299-007-0329-4
Ajesh K, Sreejith K (2009) Peptide antibiotics: an alternative and effective antimicrobial strategy to circumvent fungal infections. Peptides 30:999–1006. doi:10.1016/j.peptides.2009.01.026
Allen GC, Flores-Vergara MA, Krasynanski S et al (2006) A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc 1:2320–2325. doi:10.1038/nprot.2006.384
Benincasa M, Pelillo C, Zorzet S et al (2010) The proline-rich peptide Bac7(1-35) reduces mortality from Salmonella typhimurium in a mouse model of infection. BMC Microbiol 10:178. doi:10.1186/1471-2180-10-178
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem 72:248–254
Carsiotis M, Stocker BA, Holder IA (1989) Salmonella typhimurium virulence in a burned-mouse model. Infect Immun 57:2842–2846
Chen H, Xu Z, Peng L et al (2006) Recent advances in the research and development of human defensins. Peptides 27:931–940. doi:10.1016/j.peptides.2005.08.018
Coburn B, Grassl GA, Finlay BB (2007) Salmonella, the host and disease: a brief review. Immunol Cell Biol 85:112–118. doi:10.1038/sj.icb.7100007
Dey N, Maiti IB (1999) Structure and promoter/leader deletion analysis of mirabilis mosaic virus (MMV) full-length transcript promoter in transgenic plants. Plant Mol Biol 40:771–782
Donald CD, Sun CQ, Lim SD et al (2003) Cancer-specific loss of beta-defensin 1 in renal and prostatic carcinomas. Lab Invest 83:501–505
Fink SL, Cookson BT (2007) Pyroptosis and host cell death responses during Salmonella infection. Cell Microbiol 9:2562–2570. doi:10.1111/j.1462-5822.2007.01036.x
Ganz T (2003) Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol 3:710–720. doi:10.1038/nri1180
Ganz T, Selsted ME, Szklarek D et al (1985) Defensins. Natural peptide antibiotics of human neutrophils. J Clin Invest 76:1427–1435. doi:10.1172/JCI112120
Garabagi F, McLean MD, Hall JC (2012) Transient and stable expression of antibodies in Nicotiana species. Methods Mol Biol 907:389–408. doi:10.1007/978-1-61779-974-7_23
Gebreyes WA, Thakur S, Davies PR et al (2004) Trends in antimicrobial resistance, phage types and integrons among Salmonella serotypes from pigs, 1997–2000. J Antimicrob Chemother 53:997–1003. doi:10.1093/jac/dkh247
Gomord V, Faye L (2004) Posttranslational modification of therapeutic proteins in plants. Curr Opin Plant Biol 7:171–181. doi:10.1016/j.pbi.2004.01.015
Grassl GA, Valdez Y, Bergstrom KSB et al (2008) Chronic enteric salmonella infection in mice leads to severe and persistent intestinal fibrosis. Gastroenterology 134:768–780. doi:10.1053/j.gastro.2007.12.043
Hancock REW, Sahl HG (2006) Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol 24:1551–1557. doi:10.1038/nbt1267
Harder J, Bartels J, Christophers E, Schroder JM (2001) Isolation and characterization of human beta-defensin-3, a novel human inducible peptide antibiotic. J Biol Chem 276:5707–5713. doi:10.1074/jbc.M008557200
Kovaleva V, Kiyamova R, Cramer R et al (2009) Purification and molecular cloning of antimicrobial peptides from Scots pine seedlings. Peptides 30:2136–2143. doi:10.1016/j.peptides.2009.08.007
Krishnakumari V, Singh S, Nagaraj R (2006) Antibacterial activities of synthetic peptides corresponding to the carboxy-terminal region of human beta-defensins 1–3. Peptides 27:2607–2613. doi:10.1016/j.peptides.2006.06.004
Lehrer RI, Lichtenstein AK, Ganz T (1993) Defensins: antimicrobial and cytotoxic peptides of mammalian cells. Annu Rev Immunol 11:105–128. doi:10.1146/annurev.iy.11.040193.000541
Li J, Raghunath M, Tan D et al (2006) Defensins HNP1 and HBD2 stimulation of wound-associated responses in human conjunctival fibroblasts. Invest Ophthalmol Vis Sci 47:3811–3819. doi:10.1167/iovs.05-1360
Liu L, Wang L, Jia HP et al (1998) Structure and mapping of the human beta-defensin HBD-2 gene and its expression at sites of inflammation. Gene 222:237–244
Markeeva N, Lisovskiy I, Lyzogubov V et al (2005) Expression of beta-defensin-2 in human gastric tumors: a pilot study. Exp Oncol 27:130–135
Mason HS, Warzecha H, Mor T, Arntzen CJ (2002) Edible plant vaccines: applications for prophylactic and therapeutic molecular medicine. Trends Mol Med 8:324–329
Meena A, Bansal P, Kumar S (2009) Plants-herbal wealth as a potential source of ayurvedic drugs. Asian J Tradit Med 4:152–170
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco cultures. Physiol Plant 15:473–497
Nakashima H, Yamamoto N, Masuda M, Fujii N (1993) Defensins inhibit HIV replication in vitro. AIDS 7:1129
Patro S, Kumar D, Ranjan R et al (2012) The development of efficient plant promoters for transgene expression employing plant virus promoters. Mol Plant 5:941–944. doi:10.1093/mp/sss028
Perron GG, Bell G, Quessy S (2008) Parallel evolution of multidrug-resistance in Salmonella enterica isolated from swine. FEMS Microbiol Lett 281:17–22. doi:10.1111/j.1574-6968.2007.01045.x
Poppe C, Ziebell K, Martin L, Allen K (2002) Diversity in antimicrobial resistance and other characteristics among Salmonella typhimurium DT104 isolates. Microb Drug Resist 8:107–122. doi:10.1089/107662902760190653
Presicce P, Giannelli S, Taddeo A et al (2009) Human defensins activate monocyte-derived dendritic cells, promote the production of proinflammatory cytokines, and up-regulate the surface expression of CD91. J Leukoc Biol 86:941–948. doi:10.1189/jlb.0708412
Quiñones-Mateu ME, Lederman MM, Feng Z et al (2003) Human epithelial beta-defensins 2 and 3 inhibit HIV-1 replication. AIDS 17:F39–F48. doi:10.1097/01.aids.0000096878.73209.4f
Ranjan R, Patro S, Pradhan B et al (2012) Development and functional analysis of novel genetic promoters using DNA shuffling, hybridization and a combination thereof. PLoS One 7:e31931. doi:10.1371/journal.pone.0031931
Salzman NH, Ghosh D, Huttner KM et al (2003) Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature 422:522–526. doi:10.1038/nature01520
Schardl CL, Byrd AD, Benzion G et al (1987) Design and construction of a versatile system for the expression of foreign genes in plants. Gene 61:1–11
Schneider JJ, Unholzer A, Schaller M et al (2005) Human defensins. J Mol Med (Berl) 83:587–595. doi:10.1007/s00109-005-0657-1
Seon J, Szarka S, Moloney M (2002) A unique strategy for recovering recombinant proteins from molecular farming: affinity capture on engineered oilbodies. Plant Biotechnol J 4:95–101
Shai Y (2002) Mode of action of membrane active antimicrobial peptides. Biopolymers 66:236–248. doi:10.1002/bip.10260
Shimoda M, Ohki K, Shimamoto Y et al (1995) Morphology of defensin-treated Staphylococcus aureus. Infect Immun 63:2886–2891
Soruri A, Grigat J, Forssmann U et al (2007) beta-Defensins chemoattract macrophages and mast cells but not lymphocytes and dendritic cells: CCR6 is not involved. Eur J Immunol 37:2474–2486. doi:10.1002/eji.200737292
Stevenson JE, Gay K, Barrett TJ et al (2007) Increase in nalidixic acid resistance among non-Typhi Salmonella enterica isolates in the United States from 1996 to 2003. Antimicrob Agents Chemother 51:195–197. doi:10.1128/AAC.00222-06
Sun L, Finnegan CM, Kish-Catalone T et al (2005) Human beta-defensins suppress human immunodeficiency virus infection: potential role in mucosal protection. J Virol 79:14318–14329. doi:10.1128/JVI.79.22.14318-14329.2005
Threlfall EJ, Ward LR, Frost JA et al (2000) Spread of resistance from food animals to man—the UK experience. Acta Vet Scand Suppl 93:63–74
Tiwari S (2008) Plants: a rich source of herbal medicine. J Nat Prod 1:27–35
Twyman RM, Stoger E, Schillberg S et al (2003) Molecular farming in plants: host systems and expression technology. Trends Biotechnol 21:570–578. doi:10.1016/j.tibtech.2003.10.002
Vaazquez F, Gonzaalez EA, Garabal JI et al (1996) Development and evaluation of an ELISA to detect Escherichia coli K88 (F4) fimbrial antibody levels. J Med Microbiol 44:453–463. doi:10.1099/00222615-44-6-453
Valore EV, Park CH, Quayle AJ et al (1998) Human beta-defensin-1: an antimicrobial peptide of urogenital tissues. J Clin Invest 101:1633–1642. doi:10.1172/JCI1861
Wu S, Lu R, Zhang Y, Sun J (2010) Chronic salmonella infected mouse model. J Vis Exp doi: 10.3791/1947
Young AN, de Oliveira Salles PG, Lim SD et al (2003) Beta defensin-1, parvalbumin, and vimentin: a panel of diagnostic immunohistochemical markers for renal tumors derived from gene expression profiling studies using cDNA microarrays. Am J Surg Pathol 27:199–205
Zasloff M (2002) Antimicrobial peptides of multicellular organisms. Nature 415:389–395. doi:10.1038/415389a
Acknowledgments
We are grateful to the Council for Scientific and Industrial Research, Govt. of India for financial support [Project No. 38(1268)/10/EMR-II to ND], Fellowship (20-6/2008 (ii) EU-IV) to SP and ILS/Core fund to ND. We sincerely thank the Director, Institute of Life Sciences, Bhubaneswar, for his constant inspiration and suggestion for pursuing this study. We are grateful to Prof. T. Ganz for providing the cDNA clones of hBD-1 and hBD-2 and to Dr. D.V Singh for providing the bacterial strains. We also thank Mr. Sukumar Purohit for his kind help in performing the mice experiments and Ms. Vineeta Rai for providing the transformed control plant used in this study. The technical support provided by Mr. Abhimanyu Das is gratefully acknowledged.
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Additional information
Sunita Patro and Soumitra Maiti have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Patro, S., Maiti, S., Panda, S.K. et al. Utilization of plant-derived recombinant human β-defensins (hBD-1 and hBD-2) for averting salmonellosis. Transgenic Res 24, 353–364 (2015). https://doi.org/10.1007/s11248-014-9847-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-014-9847-3