Abstract
Plant-mediated RNAi has been developed as a powerful weapon in the fight against agricultural insect pests. The gap gene hunchback (hb) is of crucial importance in insect axial patterning and knockdown of hb is deforming and lethal to the next generation. The peach potato aphid, Myzus persicae (Sulzer), has many host plants and can be found throughout the world. To investigate the effect of plant-mediated RNAi on control of this insect, the hb gene in M. persicae was cloned, plant RNAi vector was constructed, and transgenic tobacco expressing Mphb dsRNA was developed. Transgenic tobacco had a different integration pattern of the transgene. Bioassays were performed by applying neonate aphids to homozygous transgenic plants in the T2 generation. Results revealed that continuous feeding of transgenic diet reduced Mphb mRNA level in the fed aphids and inhibited insect reproduction, indicating successful knockdown of the target gene in M. persicae by plant-mediated RNAi.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
RNA interference (RNAi), first discovered in Caenorhabditis elegans, is machinery for sequence-specific post-transcriptional gene silencing in most eukaryotic organisms (Fire et al. 1998). Double-stranded RNA (dsRNA)-mediated RNAi has been developed as one of the most promising tools to study gene function, particularly in organisms for which stable transgenesis is not available (Hannon 2002; Huvenne and Smagghe 2010). Dicer RNaseIII-type enzymes digest cytoplasmic dsRNAs into small interfering RNAs (siRNA) duplexes composed of 21–23 nucleotides (nt). These siRNA duplexes are incorporated into a multiprotein RNA-inducing silencing complex (RISC) where the antisense strand guide RISC to its homologous target mRNA for endonucleolytic cleavage (Dykxhoorn et al. 2003; Meister and Tuschl 2004). RNAi technology has also become a powerful weapon in biological control of agricultural insect pests (Gordon and Waterhouse 2007; Price and Gatehouse 2008). So far, a variety of feasible methods have been established to deliver exogenous dsRNA to organisms, for example microinjection, feeding, and soaking (Baum et al. 2007; Chen et al. 2010; Liu et al. 2010; Rosa et al. 2012; Tian et al. 2009; Zhang et al. 2010; Zhao et al. 2011; Zhu et al. 2011).
In addition, gene knockdown by expressing dsRNA in plants has also been exploited to control insect pests (Pitino et al. 2011). In 2007, prodigious progress was achieved in constructing transgenic plants expressing insect dsRNA for insect pest management. Transgenic corn plants expressing western corn rootworm (WCR) Diabrotica virgifera virgifera vacuolar ATPase (V-ATPase) subunit dsRNAs showed a significant reduction in WCR feeding damage in a growth chamber assay (Baum et al. 2007). When cotton bollworm larvae (Helicoverpa armigera) were fed transgenic plants expressing double-stranded RNA (dsRNA) specific to CYP6AE14, a cytochrome P450 gene (CYP6AE14) from cotton bollworm, levels of its transcript in the midgut decreased and larval growth was retarded (Mao et al. 2007). The hexose transporter gene NlHT1, the carboxypeptidase gene Nlcar, and the trypsin-like serine protease gene Nltry are highly expressed in the Nilaparvata lugens midgut. When N. lugens nymphs were fed on rice plants expressing dsRNAs of the three targeted genes, RNA interference was triggered but lethal phenotypic effects after dsRNA feeding were not observed (Zha et al. 2011).
Myzus persicae (Sulzer), known as the green peach aphid, is an agricultural pest worldwide. Green peach aphid feeds on hundreds of host plants in more than 40 plant families (Annis et al. 1981). This sap-sucking insect can attain very high densities on young plant tissue, causing water stress, wilting, and reduced growth rate of the plant. Prolonged aphid infestation can cause appreciable reduction in yield of root crops and foliage crops. But, the major damage caused by green peach aphid is by transmission of a variety of plant viruses. Indeed, this aphid is believed by many to be the most important vector of plant viruses, for example potato virus Y and potato leaf roll virus, to members of the nightshade/potato family Solanaceae, and various mosaic viruses to many other food crops throughout the world. Nymphs and adults are equally capable of virus transmission (Namba and Sylvester 1981).
The gap gene hb, which codes for a zinc-finger-containing transcription factor, is a key regulatory gene in the anteroposterior patterning in a number of insects (Jürgens et al. 1984; Lehmann and Nüsslein-Volhard 1987; Liu and Kaufman 2004; Patel et al. 2001; Schröder 2003; Tautz et al. 1987). hb mRNA can be synthesized maternally and zygotically. The maternal RNA is distributed homogeneously in the embryo and is under the control of the posterior maternal factor nanos (nos). Zygotic expression of hb is regulated by the anterior maternal gene bicoid (bcd) (Wolff et al. 1995). In Drosophila, loss-of-function alleles for hb lead to defects in the anterior region, for example deletions of gnathal and thoracic segments (Finkelstein and Perrimon 1990; Tautz et al. 1987). The single depletion of maternal and zygotic hb by parental RNAi in both Tribolium and Nasonia leads to serious defects in the head and thorax. Knockdown of both hb and otd, another gap gene, results in failure to form the head, thorax, and anterior abdomen (Lynch et al. 2006; Schröder 2003). In the milkweed bug Oncopeltus, hb (Of’hb) RNAi depletion results in transformations of gnathal and thoracic regions into an abdominal identity, and impaired posterior elongation and segmentation (Liu and Kaufman 2004). All these reports suggest that knockdown of the hb gene in insects is deforming and lethal to the next generation.
The potential of hb as an RNAi target has been reported by Mao and Zeng (2012). In the study, feeding of hb dsRNA was lethal to the pea aphid, Acyrthosiphon pisum. Here, we first report plant-mediated RNAi of the hb gene in M. persicae. A 1,325-bp fragment of Mphb cDNA was cloned for M. persicae. A vector for in-planta expression of Mphb dsRNA was constructed and transformed into tobacco. DNA blot showed different integration of the transgene. When M. persicae nymphs were fed on Mphb dsRNA-expressing tobacco, Mphb mRNA level was reduced and aphid fecundity was impaired. These results suggested that the hb gene can be used as a plant-mediated RNAi target for biocontrol of sap-sucking insects.
Materials and methods
Insect and plant
Tobacco plants (Nicotiana tobacum cv. Samsun NN) used for insect rearing and transgenic transformation were grown at 28 °C under a 14 h light and 10 h dark cycle. M. persicae aphids obtained from peach trees in Beijing were reared on tobacco plants at 24–25 °C under a 14:10 h light:dark regimen in a greenhouse.
Gene cloning and annotation
Total RNA was extracted from pooled adults by use of Tranzol reagents (Transgene, Beijing, China). DNA contamination was removed by digesting RNA solution with DNase (Ambion, Austin, TX, USA). cDNA was synthesized by use of TransScript First-Strand cDNA Synthesis SuperMix (Transgene, Beijing, China) with anchored Oligo(dT)18 primer.
Degenerate primers (Table 1) were designed according to hb sequence of other insect species to obtain a partial fragment (1,325 bp) of the Mphb cDNA. The PCR was performed at 94 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 57.6 °C for 30 s, and 72 °C for 80 s, finishing with an extension step at 72 °C for 10 min. PCR products were purified by use of the TIANgel Midi Purification Kit (Tiangen, Beijing, China) and then sequenced.
The Mphb cDNA sequence obtained was run blast against GenBank. Amino acid encoded by Mphb cDNA was aligned with Hb proteins of other insect species to analyze conserved motifs.
Vector construction and plant transformation
Construction of plant RNAi vectors pCAMBIA2300-Mphb was based on the pCAMBIA2300 plasmid vector (CAMBIA, Canberra, Australia) and pUCCRNAi vector. Specific primers (Table 1) were designed to amplify a 427-bp target sequence of the Mphb. The PCR products were recovered and inserted at inverted repeats into the Sal I/BamH and Xho I/Bgl II sites of the pUCCRNAi vector to obtain a hairpin RNAi construct, which was then inserted into the Pst I site of the binary vector pCAMBIA2300 to form a plant RNAi vector pCAMBIA2300-Mphb. This final vector and an empty pCAMBIA2300 vector were introduced into Agrobacterium tumefaciens strain EHA105. The plant transformation was performed according to the methods described by Horsch et al. (1985). The well-rooted plantlets were transplanted to soil and grown under greenhouse conditions to obtain homozygous transgenic plants in the T2 generation.
DNA detection
Genomic DNA was isolated from tobacco leaves by use of a DNA extraction kit (Tiangen, Beijing, China). Primers were designed to amplify a 440-bp fragment of the CaMV 35S promoter.
For Southern hybridization, DNAs (20 μg) were digested with EcoR I, subjected to electrophoresis on a 1.0 % agarose gel, and transferred to Hybond-N nylon membranes. An Mphb fragment labeled by the Dig-High Prime method was used as probe. Hybridization and washing processes were carried out according to the instruction manual of the Dig High Primer DNA Labeling and Detection Starter Kit II (Roche, Germany).
Aphid bioassays
T2 homozygous plants (lines 13, 25, and 32) with Mphb dsRNA expression were challenged by neonate aphids and tested for effects on aphid survival, biomass, and fecundity. Empty vector-transformed plants were used as controls. For all experiments, plants were grown at 24 ± 1 °C with 70 % relative humidity under a 16-h light and 8-h dark cycle. At 4–5 leaf stage, 10 neonatal nymphs of M. persicae in instar 1 were deposited on a top leaf of each plant. Aphid mortality was recorded daily. At 14 days post inoculation (dpi), all aphids per plant were weighed and counted. In every experiment, eight plants were used per phenotype. All experiments were repeated three times.
Quantitative real-time PCR
Accumulation level of Mphb mRNA in M. persicae reared on transgenic tobacco was investigated by qRT-PCR using an IQ-5 Real-Time System (Bio-Rad, California, USA). Total RNAs were isolated from feeding aphids at 2, 7, and 14 dpi and cDNA was synthesized according to conventional procedures. qRT-PCR was performed in a final volume of 25 μl containing cDNA produced from 2 μg total RNA, 11.25 μl SYBRH Green Real-time PCR Master Mix (TOYOBO, Japan), and 200 nM each of forward and reverse Mphb specific primers (Table 1). Standard curves were obtained by tenfold serial dilution and three technical replicates of each reaction were performed. M. persicae 18S rRNA (GenBank Accession Number: AF487712.1) was used as internal control and specific primers (Table 1) were designed for normalization. Means and standard errors for each time point were obtained from an average of three independent sample sets. Quantification of the relative changes in gene transcript level was performed according to the 2−ΔΔCt method (Livak and Schmittgen 2001).
Results
Cloning and characterization of the M. persicae hunchback
A 1,325 bp fragment of Mphb (GenBank Accession Number: KC481323) was cloned by using degenerate primers designed to match conserved regions known to flank hb homeodomanis. Mphb cDNA had the highest identity of 90 % with Acyrthosiphon pisum hb (Aphb) mRNA sequence (GenBank Accession Number: NM_001162510.1). Similar to the orthologue Aphb, the 441 amino acids encoded by Mphb also contains four zinc fingers (MF 1–4), a C box, and a Basic box (data not shown). All these motifs are conserved among Hb proteins.
Construction and molecular analysis of transgenic RNAi plants
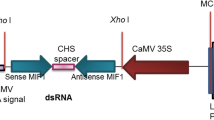
The dsRNA-expressing construct pCAMBIA2300-Mphb was developed for a 427-bp fragment of the Mphb cDNA under the control of CaMV 35S promoter and the nopaline synthase (nos) terminator cassette (Fig. 1). Tobacco leaf discs were infected by A. tumefaciens strain EHA105 harboring pCAMBIA2300-Mphb or empty pCAMBIA2300 vector. A total of 57 independent kanamycin-resistant transformants were regenerated (39 Mphb transformants and 18 empty vector transformants). Most had wild-type morphology and growth.
Construction of the plant RNAi vector. Plant RNAi vector pCAMBIA2300-Mphb was constructed on the basis of the pCAMBIA2300 plasmid vector and pUCCRNAi vector. LB left border; polyA cauliflower mosaic virus terminator; npt II, kanamycin resistance gene; 35S prom, 35S cauliflower mosaic virus promoter; Mphb, M. persicae hunchback gene; OCS, polyA signal from octopine synthase; RB right border
Genomic DNAs were extracted from transgenic plants. Positive transformants were determined by PCR analysis of the 35S promoter. Twenty-eight Mphb transformants and 11 empty vector transformants showed the presence of CaMV 35S promoter sequence, while wild type plants did not show specific amplification (results not shown). These T0 transformants were reproduced to obtain homozygous T2 transgenic lines.
Integration pattern of the Mphb dsRNA-expressing cassette in PCR-positive plants (13, 25, and 32) were investigated by Southern hybridization. The three transgenic transformants showed one integration copy, but no Mphb hybridization band was observed with genomic DNA from empty vector plant (Fig. 2). The unique hybridization patterns indicated that each investigated plant resulted from independent transformation events.
Southern blot analysis of transgenic tobacco. Total DNA (20 μg) from T0 Mphb plants (13, 25, and 32) and empty vector plants (V) was digested with restriction enzyme EcoR I and subjected to 1.0 % agarose gel electrophoresis. DNA samples were transferred to a nylon membrane and hybridized with the Dig-labeled DNA probe. Three transformants showed different integration patterns
Aphid resistance of dsRNA-expressing tobacco
T2 Mphb dsRNA-expressing homozygous plants (lines 13, 25 and 32), together with empty vector plants, were challenged by neonate aphids and tested for effects on aphid survival, growth, and reproduction. Each tobacco plant was inoculated with 10 neonatal nymphs and aphid survival was investigated 7 days post inoculation (dpi). No statistical difference in survival was observed between Mphb dsRNA-expressing lines and control plants (empty vector plants) (results not shown). At 14 dpi, aphids on Mphb dsRNA-expressing plants had statistically smaller populations than those on control plants. The aphid population on each control plant increased from 10 insect per plant to an average of 107 per plant in a 14-day period. But aphid number on the Mphb dsRNA-expressing plant was a maximum of 93 at 14 dpi (Fig. 3a). Aphids on Mphb dsRNA-expressing plants also had lower insect biomass compared with those on empty vector plants (Fig. 3b). On this day, each Mphb dsRNA-expressing plant yielded a biomass of approximately 13 mg. But each empty vector plant produced an average of 15.5 mg.
Inhibition of the transgenic tobacco on population and biomass of M. persicae. a mean population produced on a given plant; b mean insect biomass produced on a given plant. Values are expressed as mean ± SE of eight replicates. Asterisks indicate significant differences between the treatment and the control determined by a t test (*, p < 0.05). Compared with empty vector plants (V), T2 homozygous Mphb plants (lines 13, 25, and 32) showed suppression of fecundity and biomass of the M. persicae aphids
Depletion of Mphb mRNA after ingestion of transgenic tobacco
At 2, 7, and 14 dpi, Mphb mRNA accumulation level after ingestion of transgenic tobacco was analyzed by quantitative real-time PCR. In nymphs fed on Mphb plants, Mphb mRNA level had an obvious decrease and the Mphb transcripts abundance was reduced by approximately 12 % on day 2. The reduction level then increased with elongation of feeding period and reached 31.3 % on the seventh day and 32.4 % on the 14th day (Fig. 4).
Mphb mRNA depletion after feeding on dsRNA-expressing tobacco. Mphb mRNA level was analyzed by qRT-PCR 2, 7, and 14 days after ingestion of transgenic plants. 18S rRNA was used as internal control for normalization. Normalized Mphb expression was expressed as the proportion of that recorded for the vector control plant. Each kinetic point was expressed as mean ± SE of three replicates. Asterisks indicate significant differences in Mphb transcripts levels between the treatment (lines 13, 25, and 32) and the control (V) determined by a t test (*p < 0.05; **p < 0.01)
Discussion
So far, dsRNA-mediated RNAi has been demonstrated in several insect orders, including Coleoptera (Tomoyasu et al. 2008), Diptera (Dzitoyeva et al. 2001), Hymenoptera (Lynch and Desplan 2006), Hemiptera (Chen et al. 2010; Liu et al. 2010; Mao and Zeng 2012; Zhu et al. 2011), Orthoptera (Meyering-Vos and Muller 2007), Lepidoptera (Terenius et al. 2011; Turner et al. 2006) and Isoptera (Zhou et al. 2008). In these reports, dsRNA was usually delivered to insects by artificial feeding or injection. But these two methods are not applicable in farmland. Expressing dsRNA in a plant is another effective approach exploited to control insect pests. Transgenic plants expressing target gene dsRNA had significant resistance against Lepidoptera species, for example western corn rootworm (WCR) D. virgifera virgifera vacuolar (Baum et al. 2007) and cotton bollworm (H. armigera) (Mao et al. 2007). Furthermore, plant-mediated RNAi successfully depleted target gene expression in sap-sucking insect. When N. lugens nymphs were fed on rice plants expressing dsRNAs of the three targeted genes, RNA interference was triggered but lethal effect was not observed (Zha et al. 2011). Aphids (Hemiptera: Aphididae) are exclusive phloem feeders and are among the most economically important pest insects of temperate agriculture. In addition to the effect of feeding, aphids also transmit plant viruses. Now, development of transgenic crops expressing toxins derived from the bacterium Bacillus thuringiensis (Bt) has provided effective plant protection against some insect pests, but Bt toxins have little toxicity against sap-sucking insects (Chougule and Bonning 2012). Here, we report plant-mediated RNAi in M. persicae, a phloem-sucking agricultural pest throughout the world. Transgenic plants expressing of Mphb dsRNA not only suppressed target gene expression, but also impaired insect fecundity. The significance of this study is that it demonstrated the feasibility of plant-mediated RNAi in biocontrol of sap-sucking insect pests.
hunchback, which encodes a zinc-finger-containing transcription factor, is crucial for axial patterning in insects (Lehmann and Nüsslein-Volhard 1987; Liu and Kaufman 2004; Patel et al. 2001; Schröder 2003; Tautz et al. 1987). Acyrthosiphon pisum Hb protein contains six zinc finger domains and two other conserved motifs, C box and Basic box. In this study, C box and Basic box was present in Mphb protein, but we identified only four zinc fingers, not six. It remains possible that Mphb contains two other zinc fingers, because we did not obtain a full length of Mphb cDNA. In addition, sequencing of genomic DNA revealed that the Mphb middle fragment obtained contains no introns (data not shown). This suggested that, similar to its homologs in other insect species, the whole Mphb locus contains no introns. Similarly, M. persicae should have only one copy of hb.
Aside from the crucial functions in the early embryonic anteroposterior patterning, hb is important in sequential cell fate specification within the Drosophila central nervous system (CNS) (Novotny et al. 2002). But up to now, there are no further reports about how the hb functions in CNS development of other insects. Although defective and lethal phenotypes were all absent in our RNAi experiments, we inferred that the reduction of insect population resulted from impairment of the hb functions in the early embryonic anteroposterior patterning, not the role in specifying the CNS development. Further investigation is required to confirm this. In this study, both population and biomass of aphids reared on Mphb RNAi plants were suppressed. We inferred that the reduction of biomass resulted from the inhibited population, because average individual weight of aphids fed on Mphb RNAi plants and control plants was no different (data not shown).
Parental RNAi of hb is deforming and lethal to the next generation (Lehmann and Nüsslein-Volhard 1987; Liu and Kaufman 2004; Patel et al. 2001). In Oncopeltus fasciatus, maternal depletion of hb transcripts resulted in different classes of defect in most progeny (Liu and Kaufman 2004). Continuous feeding of Acyrthosiphon pisum hunchback (Aphb) dsRNA mixed in an artificial diet led to a maximum 50 % reduction of Aphb transcripts and more than doubling of insect lethality, indicating that hb was a good RNAi target for management of insect pests. In this study, ingestion of dsRNA-expressing tobacco only resulted in a 17.6 % reduction of fecundity and 18.4 % suppression of biomass. In addition, defective phenotypes frequently appeared after hb depletion in other insect species but were absent in this feeding bioassay. So, what is the reason? It is less likely that other paralogues in M. persicae compensate for suppression of Mphb, because such insects as A. pisum usually have one hb only (Huang et al. 2010). One explanation is that Mphb dsRNA was not expressed at a high level in transgenic plants and uptake amount of siRNA was not sufficient to trigger defective or lethal effect and to cause high population suppression. Another possible reason is that ingested siRNA is not sufficient to penetrate into target parental and embryonic cells of the M. persicae aphids, because RNAi efficiency may vary for different target genes, different insect species, and different organs. Further work is needed to verify this speculation.
No guarantee of sufficient uptake of dsRNA or siRNA by insects is, maybe, a drawback of plant-mediated RNAi. Neither dsRNA injection nor artificial feeding has this problem, because large amounts of dsRNA synthesized at high concentration can be delivered to insects by these two methods. Another problem we must mention is that it is relatively easier to realize plant-mediated RNAi in chewing mouthparts insects than in stylet mouthparts species. This is because the former insects, for example Lepidoptera, take up all of the plant tissue with chewing mouthparts. Thus, dsRNA expressed in cells can be easily delivered to the insect digestive tract, whereas stylet mouthparts insects, for example aphids, are quite different because they suck sap from plant phloem. Theoretically, plant-mediated RNAi can be transformed into an applicable method for bio-control of stylet mouthparts insects only if three requirements are all met:
-
1.
dsRNA of the target sequence is synthesized at a high level;
-
2.
enough dsRNA is processed into siRNA; and
-
3.
sufficient dsRNA or siRNA is ingested by insects and penetrates the nucleus.
Lack of any of these conditions will lead to failure of RNAi. Transgenic plants expressing dsRNA specific to MjTis11, a transcription factor in a root knot nematode, did not result in a lethal phenotype (Fairbairn et al. 2007). In another study, RNA interference was triggered but lethal phenotypic effects after dsRNA feeding were not observed when N. lugens nymphs were fed on rice plants expressing dsRNAs of three target genes (Zha et al. 2011). These plant-mediated RNAi cases indicated that further work is urgently needed to increase dsRNA level in transgenic plants, to promote dsRNA or siRNA uptake by sap-sucking insects, and to achieve expected phenotypes after target gene depletion.
References
Annis B, Tamaki G, Berry RE (1981) Seasonal occurrence of wild secondary hosts of the green peach aphid, Myzus persicae (Sulzer), in agricultural systems in the Yakima Valley. Environ Entomol 10:307–312
Baum J, Bogaert T, Clinton W, Heck G, Feldmann P (2007) Control of coleopteran insect pests through RNA interference. Nat Biotechnol 25:1322–1326
Chen J, Zhang D, Yao Q, Zhang J, Dong X, Tian H, Chen J, Zhang W (2010) Feeding-based RNA interference of a trehalose phosphate synthase gene in the brown planthopper, Nilaparvata lugens. Insect Mol Biol 19:777–786
Chougule NP, Bonning BC (2012) Toxins for transgenic resistance to Hemipteran pests. Toxins 4:405–429
Dykxhoorn DM, Novina CD, Sharp PA (2003) Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol 4:457–467
Dzitoyeva S, Dimitrijevic N, Manev H (2001) Intra-abdominal injection of double-stranded RNA into anesthetized adult Drosophila triggers RNA interference in the central nervous system. Mol Psychiatry 6:665–670
Fairbairn DJ, Cavallaro AS, Bernard M, Mahalinga-Iyer J, Graham MW, Botella JR (2007) Host-delivered RNAi: an effective strategy to silence genes in plant parasitic nematodes. Planta 226:1525–1533
Finkelstein R, Perrimon N (1990) The orthodenticle gene is regulated by bicoid and torso and specifies Drosophila head development. Nature 346:485–488
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–811
Gordon KH, Waterhouse PM (2007) RNAi for insect-proof plants. Nat Biotech 25:1231–1232
Hannon GJ (2002) RNA interference. Nature 418:244–251
Horsch RB, Fry JE, Fraley RT (1985) A simple and general method for transferring genes into plants. Science 227:1229–1231
Huang T, Cook CE, Davis GK, Shigenobu S, Chen RPY, Chang C (2010) Anterior development in the parthenogenetic and viviparous form of the pea aphid, Acyrthosiphon pisum: hunchback and orthodenticle expression. Insect Mol Biol 19:75–85
Huvenne H, Smagghe G (2010) Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. J Insect Physiol 56:227–235
Jürgens G, Wieschaus E, Nüsslein-Volhard C, Kluding H (1984) Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. Roux’s Arch Dev Biol 193:283–295
Lehmann R, Nüsslein-Volhard C (1987) hunchback, a gene required for segmentation of an anterior and posterior region of the Drosophila embryo. Dev Biol 119:402–417
Liu P, Kaufman TC (2004) hunchback is required for suppression of abdominal identity, and for proper germband growth and segmentation in the intermediate germband insect Oncopeltus fasciatus. Development 131:1515–1527
Liu S, Ding Z, Zhang C, Yang B, Liu Z (2010) Gene knockdown by introthoracic injection of double-stranded RNA in the brown planthopper, Nilaparvata lugens. Insect Biochem Mol Biol 40:666–671
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25:402–408
Lynch JA, Desplan C (2006) A method for parental RNA interference in the wasp Nasonia vitripennis. Nat Protoc 1:486–494
Lynch JA, Brent AE, Leaf DS, Pultz MA, Desplan C (2006) Localized maternal orthodenticle patterns anterior and posterior in the long germ wasp Nasonia. Nature 439:728–732
Mao J, Zeng F (2012) Feeding-based RNA intereference of a gap gene is lethal to the pea aphid, Acyrthosiphon pisum. PLoS ONE 7(11):e48718
Mao Y, Cai W, Wang J, Hong G, Tao X, Wang L, Huang YP, Chen XY (2007) Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotech 25:1307–1313
Meister G, Tuschl T (2004) Mechanisms of gene silencing by double-stranded RNA. Nature 431:343–349
Meyering-Vos M, Muller A (2007) RNA interference suggests sulfakinins as satiety effectors in the cricket Gryllus bimaculatus. J Insect Physiol 53:840–848
Namba R, Sylvester ES (1981) Transmission of cauliflower mosaic virus by the green peach, turnip, cabbage, and pea aphids. J Econ Entomol 74:546–551
Novotny T, Eiselt R, Urban J (2002) Hunchback is required for the specification of the early sublineage of neuroblast 7–3 in the Drosophila central nervous system. Development 129:1027–1036
Patel NH, Hayward DC, Lall S, Pirkl NR, DiPietro D, Ball EE (2001) Grasshopper hunchback expression reveals conserved and novel aspects of axis formation and segmentation. Development 128:3459–3472
Pitino M, Coleman AD, Maffei ME, Ridout CJ, Hogenhout SA (2011) Silencing of aphid genes by dsRNA feeding from plants. PLoS ONE 6:1–8
Price DRG, Gatehouse JA (2008) RNAi-mediated crop protection against insects. Trends Biotechnol 26:393–400
Rosa C, Kamita SG, Falk BW (2012) RNA interference is induced in the glassy winged sharpshooter Homalodisca vitripennis by actin dsRNA. Pest Manag Sci 68(7):995–1002
Schröder R (2003) The genes orthodenticle and hunchback substitute for bicoid in the beetle Tribolium. Nature 422:621–625
Tautz D, Lehmann R, Schürch H, Shuh R, Seifert E, Kienlin A, Jones K, Jäckle H (1987) Finger protein of novel structure encoded by hunchback, a second member of the gap class of Drosophila segmentation genes. Nature 327:383–389
Terenius O, Papanicolaou A, Garbutt JS, Eleftherianos I, Huvenne H, Kanginakudru S, Albrechtsen M, An C, Aymeric JL, Barthel A, Bebas P, Bitra K, Bravo A, Chevalier F, Collinge DP, Crava CM, de Maagd RA, Duvic B, Erlandson M, Faye I, Felföldi G, Fujiwara H, Futahashi R, Gandhe AS, Gatehouse HS, Gatehouse LN, Giebultowicz JM, Gómez I, Grimmelikhuijzen CJ, Groot AT, Hauser F, Heckel DG, Hegedus DD, Hrycaj S, Huang L, Hull JJ, Iatrou K, Iga M, Kanost MR, Kotwica J, Li C, Li J, Liu J, Lundmark M, Matsumoto S, Meyering-Vos M, Millichap PJ, Monteiro A, Mrinal N, Niimi T, Nowara D, Ohnishi A, Oostra V, Ozaki K, Papakonstantinou M, Popadic A, Rajam MV, Saenko S, Simpson RM, Soberón M, Strand MR, Tomita S, Toprak U, Wang P, Wee CW, Whyard S, Zhang W, Nagaraju J, Ffrench-Constant RH, Herrero S, Gordon K, Swevers L, Smagghe G (2011) RNA interference in Lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. J Insect Physiol 57:231–245
Tian H, Peng H, Yao Q, Chen H, Xie Q, Tang B, Zhang W (2009) Developmental control of a lepidopteran pest Spodoptera exigua by ingestion of bacteria expressing dsRNA of a non-midgut gene. PLoS ONE 4:1–13
Tomoyasu Y, Miller SC, Tomita S, Schoppmeier M, Grossmann D, Bucher G (2008) Exploring systemic RNA interference in insects: a genome-wide survey for RNAi genes in Tribolium. Genome Biol 9:R10
Turner CT, Davy MW, MacDiarmid RM, Plummer KM, Birch NP, Newcomb RD (2006) RNA interference in the light brown apple moth, Epiphyas postvittana (Walker) induced by double-stranded RNA feeding. Insect Mol Biol 15:383–391
Wolff C, Sommer R, Schröder R, Glaser G, Tautz D (1995) Conserved and divergent expression aspects of the Drosophila segmentation gene hunchback in the short germband embryo of the flour beetle Tribolium. Development 121:4227–4236
Zha W, Peng X, Chen R, Du B, Zhu L, He G (2011) Knockdown of midgut genes by dsRNA-transgenic plant-mediated RNA interference in the Hemipteran insect Nilaparvata lugens. PLoS ONE 6(5):e20504
Zhang X, Zhang J, Zhu KY (2010) Chitosan/double-stranded RNA nanoparticle-mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles gambiae). Insect Mol Biol 19:683–693
Zhao YY, Liu F, Yang G, You MS (2011) PsOr1, a potential target for RNA interference-based pest management. Insect Mol Biol 20:97–104
Zhou X, Wheeler MM, Oi FM, Scharf ME (2008) RNA interference in the termite Reticulitermes flavipes through ingestion of double-stranded RNA. Insect Biochem Mol Biol 38:805–815
Zhu F, Xu J, Palli R, Ferguson J, Palli SR (2011) Ingested RNA interference for managing the populations of the Colorado potato beetle, Leptinotarsa decemlineata. Pest Manag Sci 7:175–182
Acknowledgments
This work was supported by grants from Project NSFC 31201570 and National Basic Research Program of China (973 Program) (2013CB127602).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mao, J., Zeng, F. Plant-mediated RNAi of a gap gene-enhanced tobacco tolerance against the Myzus persicae . Transgenic Res 23, 145–152 (2014). https://doi.org/10.1007/s11248-013-9739-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-013-9739-y