Abstract
Cotton is an economically important crop worldwide that suffers severe losses due to a wide range of fungal/bacterial pathogens and nematodes. Given its susceptibility to various pathogens, it is important to obtain a broad-spectrum resistance in cotton. Resistance to several fungal and bacterial diseases has been obtained by overexpressing the Non-expressor of Pathogenesis-Related genes-1 (NPR1) in various plant species with apparently minimal or no pleiotropic effects. We examined the efficacy of this approach in cotton by constitutive expression of the Arabidopsis (Arabidopsis thaliana) NPR1 gene. The results show that NPR1-expressing lines exhibited significant resistance to Verticillium dahliae isolate TS2, Fusarium oxysporum f. sp. vasinfectum, Rhizoctonia solani, and Alternaria alternata. Interestingly, the transformants also showed significant resistance to reniform nematodes. Analysis of defense-related, biochemical and molecular responses suggest that when challenged with pathogens or certain systemic acquired resistance-inducing chemicals, the transgenic lines respond to a greater degree compared to the wild-type plants. Importantly, the basal activities of the defense-related genes and enzymes in uninduced transformants were no different than those in their non-transgenic counterparts. The results provide additional evidence supporting the role of NPR1 as an important part of the plant defense system and suggest a means to achieve broad-spectrum resistance to pathogens via genetic engineering.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Systemic acquired resistance (SAR) is a long lasting defense response that is induced in plants by localized infection and provides subsequent protection against a broad spectrum of pathogens (Ryals et al. 1994; Ryals et al. 1996; Grant and Lamb 2006). SAR induction is associated with localized and systemic elevation of salicylic acid (SA). Application of SA or its synthetic analogs 2,6-dichloroisonicotinic acid (INA) and benzo(1,2,3)thiadiazole-7-carbothioic acid-S-methyl ester (BTH) have been shown to induce SAR (Ward et al. 1991; Uknes et al. 1992; Cao et al. 1994; Gorlach et al. 1996).
An Arabidopsis (Arabidopsis thaliana) mutant, npr1 (Non-expressor of Pathogenesis-Related genes-1, also called NIM1 or SAI1), failed to generate SAR in response to SA, INA, BTH or pathogen treatments, indicating that NPR1 plays a critical role in SAR (Cao et al. 1994; Delaney et al. 1995; Shah et al. 1997). However, NPR1-independent induction of disease resistance pathways culminating in the expression of PR genes has also been reported (Kachroo et al. 2000; Shah et al. 2001; Rairdan and Delaney 2002; Shah 2003). In an uninduced state, NPR1 has been shown to be present in the cytosol in its oligomeric form (Mou et al. 2003). SAR-induction triggers changes in the redox status of the cell that is believed to reduce NPR1 to its monomeric form (Mou et al. 2003). The oligomer to monomer reduction of NPR1 can be catalyzed by thioredoxins (Tada et al. 2008). The monomeric form of NPR1 is able to translocate into the nucleus where it activates the expression of PR genes (Kinkema et al. 2000; Mou et al. 2003; Tada et al. 2008) through its interaction with various TGA transcription factors (Fan and Dong 2002; Zhou et al. 2000; Despres et al. 2000; Kesarwani et al. 2007).
Validation for the key role played by NPR1 in various defense networks has been obtained by a number of overexpression studies. Constitutive expression of AtNPR1 in Arabidopsis and other heterologous species conferred a broad-spectrum resistance to various fungal and bacterial pathogens. Transgenic Arabidopsis plants overexpressing NPR1/NIM1 showed resistance to a bacterial pathogen, Pseudomonas syringae and several fungal pathogens, including Hyaloperonospora arabidopsidis, Peranospora parasitica, and Erysiphe cichoracearum (Cao et al. 1998; Friedrich et al. 2001). This gene also conferred resistance to bacterial pathogens, Xanthomonas oryzae and Erwinia chrysathemi and, to two fungal pathogens, Magnaporthe oryzae and Fusarium verticillioides in rice (Chern et al. 2001; Quilis et al. 2008); to F. oxysporum, Stemphylium solani, Ralstonia solanacearum and X. campestris in tomato (Lin et al. 2004); to F. graminearum in wheat (Makandar et al. 2006); and to Botrytis cinerea, Alternaria radicina, Sclerotinia sclerotiorum, Alternaria radicina, Erysiphe heraclei, and Xanthomonas hortorum in carrot (Wally et al. 2009). Transgenic apple, overexpressing a homologous NPR1 gene (MpNPR1), showed increased resistance to the bacterium, E. amylovora and to two fungal pathogens, Venturia inaequalis and Gymnosporangium juniperi-verginianae (Malnoy et al. 2007). Constitutive expression of AtNPR1 or Brassica napus-NPR1 (BnNPR1) in B. napus conferred resistance to the bacterial pathogen, P. syringae (Potlakayala et al. 2007). With the exception of rice, other species in the aforementioned studies did not show any adverse growth or developmental effects as a result of NPR1-overexpression. Results from these studies suggest that NPR1-mediated defense response mechanism is evolutionarily conserved and that overexpression of NPR1 can confer a broad-spectrum resistance to pathogens in other important crop species.
Cotton is a crop of great economic importance worldwide. Cotton lint continues to be the most important source of natural fiber for textile industry, while, the seeds are used to produce edible oil and as a feed for cattle. There is a large array of diseases that inflict the cotton plant and cause significant yield losses. In 2007, various pathogens and nematodes caused a 9% loss in the yield of this crop in the U.S. (Blasingame et al. 2008). While transgenes are widely used in cotton for insect control and herbicide resistance, efforts to confer disease resistance to cotton plant through transgenic means are in their infancy, and there are few published reports showing transgene-mediated disease resistance in cotton. Overexpression of Talaromyces flavus glucose oxidase gene in transgenic cotton showed some protection against Verticillium dahliae, but did not provide protection against F. oxysporum (Murray et al. 1999). Overexpression of an endochitinase gene from Trichoderma virens showed enhanced resistance to Rhizoctonia solani and Alternaria alternata, but failed to provide any protection against F. oxysporum, V. dahliae and Thielaviopsis basicola (Emani et al. 2003). Constitutive expression of an anti-fungal protein from Gastrodia in cotton has been reported to confer resistance to V. dahliae (Wang et al. 2004). In another study, resistance to T. basicola (which causes root rot disease) was observed in transgenic cotton that expressed a gene encoding a synthetic peptide, D4E1 (Rajasekaran et al. 2005). In most of these studies, the protection obtained was a result of the direct effect of the transgene product on the pathogen, thus limiting their efficacy to a narrow spectrum of diseases. In contrast, NPR1/NIM1-mediated protection appears to be due to the activation of plant’s own defense machinery and therefore, the resistance obtained in the transformants was more broad-spectrum (Cao et al. 1994; Cao et al. 1998; Delaney et al. 1995; Lin et al. 2004). The current investigation was undertaken to examine if constitutive expression of AtNPR1 gene can confer a broad-spectrum resistance to diseases in the cotton plant. The results obtained demonstrate that AtNPR1-transformants were resistant to four important fungal diseases of cotton caused by F. oxysporum f. sp. vasinfectum, V. dahliae isolate TS2, R. solani and A. alternata. This study also provides evidence that expression of AtNPR1 in cotton plant confers some protection against the nematode, Rotylenchulus reniformis. In addition to the evidence for disease resistance, we provide results on the transcript levels of some PR genes and the biochemical alterations observed in chemically- or pathogen-induced transgenic plants.
Materials and methods
Vector construction, cotton transformation, and molecular analysis
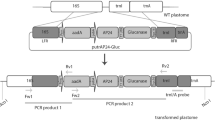
A pCAMBIA2300 based vector containing the GUS expression cassette from pFF19G (Timmermans et al. 1990) constructed earlier in our laboratory was used to assemble the transformation vector for this study. The gusA gene was removed from this vector and replaced with NPR1 cDNA obtained from A. thaliana ecotype Dijon-17 (Zhou et al. 2000), thus placing it under the control of CaMV 35S promoter with the duplicated enhancer element and CaMV 35S polyadenylation signal from plasmid pFF19G (Timmermans et al. 1990).
Gossypium hirsutum cv. Coker 312 was transformed according to the method described by Sunilkumar and Rathore (2001) and Rathore et al. (2006). The primary transformants generated were screened for AtNPR1 transcript accumulation in the leaf tissue by RT-PCR. The PCR primer pair, AtNPR1F: 5′-GAGGACACACTGGTTATACTC-3′ and AtNPR1R: 5′-CCAGATCGAGCAGCGTCATCTT-3′ was used to amplify a 690 bp-size fragment of AtNPR1. Selected transgenic lines identified from this screen were analyzed by Southern hybridization to confirm transgene integration.
Chitinase assay
Endochitinase activity was measured according to the protocol described by Emani et al. (2003) using 4-methylumbelliferyl-β-d-N,N′,N″-triacetylchitotrioside [4-MU-β-(GlucNAc)3] (Sigma, St. Louis, MO, USA) as the substrate. Enzyme activity was examined in total soluble protein (TSP) obtained from the first leaf of a cotton plant with or without F. oxysporum f. sp. vasinfectum infection. The endochitinase specific activity is presented as pmole 4-MU/h/μg TSP.
Glucanase assay
The method published by Abeles and Forrence (1970) was used to estimate glucanase activity in TSP obtained from the first leaf of a cotton plant with or without F. oxysporum f. sp. vasinfectum infection. In this assay, laminarin was used as the substrate and dinitrosalicylic reagent was used to measure the reducing sugars produced in the enzymatic reaction. The enzyme activity is expressed as glucose equivalents, mg/h/mg TSP.
Estimation of total terpenoid aldehydes
The method described by Bell (1967) and Bianchini et al. (1999) was followed to extract and measure terpenoids from the stele tissue at the 5th internode of a cotton plant, and concentrations are presented as ‘gossypol equivalents’. Stele tissue, 2 cm in length, was weighed, chopped into small pieces and then incubated in 3 ml of 95% acetone containing 1% ascorbic acid. Following 10 s of vortex treatment, the samples were kept at 4°C for 2 h. A 500 μl extract was then mixed with 250 μl of 5% phloroglucinol and 500 μl of concentrated HCl and incubated at room temperature for 30 min. The absorbance was measured spectrophotometrically at 550 nm. A standard curve prepared with pure gossypol was used to estimate terpenoids in each sample, which are presented as μg gossypol equivalents/g FW of stele tissue.
Treatment of cotton seedlings with SA
The method of Djonovic et al. (2006) was adapted for the application of SA to cotton seedlings. Cottonseeds, soaked overnight, were germinated vertically within a rolled-up wet filter paper for 2 days at 28°C. The germinated seedling was placed on a filter paper wick support in a test tube (17 × 100 mm, VWR International, LLC, Cat. #60818-667) containing 10 ml of ½ strength MS medium in such a manner that the radicle remained submerged in the medium while the hypocotyl remained above the liquid as the seedling grew. Ten such tube assemblies were kept in a 2-litre glass beaker covered with a loose-fitting plastic Petri dish (150 × 15 mm), sealed with parafilm, and placed under light at 25°C in an incubator. After 24 h, the medium surrounding the root was replaced with 3 mM SA and the seedling was returned to the incubator. After 12, 24 and 48 h of SA treatment, cotyledons were analyzed for chitinase and glucanase activities.
BTH treatment of the cotton plants
Twenty-three-day-old cotton plants were sprayed with aqueous solution of 100 μM BTH (Actiguard, containing 50% active ingredient, Syngenta) in 0.01% silvet till run-off. Cotton plants sprayed with 0.01% silvet (mock-treated) were used as controls. Leaf samples were harvested after 6, 12, 24 h of BTH treatment for the isolation of total RNA.
Reverse transcriptase-polymerase chain reaction (RT-PCR)
The Spectrum Plant Total RNA Kit (Cat # STRN50-1KT, Sigma, St. Louis, MO, USA) was used to extract total RNA from a young healthy leaf of a BTH (100 μM) treated, 3-week-old- or F. oxysporum f. sp. vasinfectum infected, 2-week-old cotton plants. Following DNase treatment (RNase-free DNase Kit, Cat # 79254, Quiagen, USA), Taqman Reverse transcription (RT) reagents kit (Applied Biosystems, Cat # N808-0234) was used to synthesize the complementary-DNA (cDNA) from 400 ng of total RNA. One microliter of the reaction product was used for PCR amplification using a set of gene specific primers (Supplementary Table 1). The PCR conditions were as follows: 94°C for 4 min, initial denaturation, 28 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 60 s for Chitinase, Glucanase, and Osmotin; 94°C for 4 min, initial denaturation, 22 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 60 s for PR1; 94°C for 4 min, initial denaturation, 28 cycles of 94°C for 30 s, 62°C for 30 s, and 72°C for 60 s for AtNPR1 and a final extension at 72°C for 10 min. In the duplex RT-PCR, cotton Histone3 gene was used as an internal control. PCR products were electrophoresed on 1.2% agarose gel.
Rhizoctoniasolani infection assay
These assays with R. solani (strain J1) were conducted following the protocol described in a recent paper from our laboratory (Kumar et al. 2009). Seeds were rolled in a wet filter paper (VWR, Cat. #28298-020) and placed vertically in a beaker, covered with a polythene bag and incubated for 2 days at 28°C. Two-day-old, pre-germinated seedlings were transferred to soil (with or without the fungal inoculum) in a plastic tray. The tray was covered with a clear, plastic Humidome (Acrodome, Agroplastics Ltd, Canada) to maintain humidity and placed in a growth chamber for 6 days at 25°C under a 14:10 h photoperiod to allow disease development.
Fusarium wilt assay
Fusarium oxysporum f. sp. vasinfectum isolate Fov11 (race 1 on cotton; ATCC #46644) was grown on plates containing Difco™ potato dextrose agar (PDA) medium supplemented with chloramphenicol 200 mg/l and tetracycline 20 mg/l for 1 week. A section of agar with sporulating mycelia measuring 1 cm2 was cut from the PDA plate and transferred to 2 ml sterile distilled water and vortexed for 5 s to prepare a conidial suspension. Five drops of this suspension were added to 50 ml Difco™ potato dextrose broth and incubated on a shaker (140 rpm) for 7 days at 23°C. This culture was filtered through paper towel to eliminate mycilial clumps and obtain microconidial suspension. Microconidial count was performed using a hemocytometer and the final count was adjusted to 107 conidia/ml by diluting with sterile distilled water.
Cotton seedlings were grown on washed, sand-loamy clay (75:25) mixture in a greenhouse. The plants were fertilized with 6 g/l of water-soluble, 9-45-15 N-P-K fertilizer (Peters professional® water soluble fertilizer, Scotts-Sierra Horticultural Product Company, Ohio, USA). After 2 weeks, seedlings were removed gently from the soil and roots were washed thoroughly under running tap water. The root portion was immersed in the conidial suspension for 5 min for inoculation and then the seedling was transplanted to a 500 ml plastic cup (with three holes, covered with a plastic screen, at the bottom) containing fresh sand-clay mixture irrigated with 3 g/l of water-soluble 9-45-15 N-P-K fertilizer. Control seedlings were mock-treated with tap water before transfer to the sand-clay mixture. Infected and control seedlings were grown for 3 weeks in a growth chamber at 25/20°C (day/night) temperature, and a 13 h photoperiod. In addition to regular watering, each plant was fertilized once a week with 50 ml of 3 g/l water-soluble 20-20-20 N-P-K fertilizer. Disease severity was scored 3 weeks after inoculation. Leaf disease index (LDI) and total shoot weight were used as the parameters to assess infection severity. LDI was calculated based on the following formula:
Verticillium wilt assay
Verticillium wilt assay and the scoring of results were performed as described by Bolek et al. (2005). V. dahliae Kleb. Isolate TS2 (race 2 on tomato; VCG2A) was grown on Difco™ PDA medium supplemented with chloramphenicol 200 mg/l and tetracycline 20 mg/l and allowed to grow for 1 week at 23°C. An agar block with sporulating mycelia, measuring ~1 cm2, was transferred to a tube containing 3 ml sterile distilled water and vortexed for 5 s to prepare a conidial suspension. Conidial count was performed with a hemocytometer and was adjusted to 107/ml by diluting the suspension with sterile distilled water. Cotton plants were grown for 6 weeks in 500 ml-size plastic cups (with holes in the bottom) containing Metromix® 700 (SUN GRO Horticulture Ltd, Bellevue, WA, USA) plant growth medium supplemented with 6 g/l of water-soluble 9-45-15 N-P-K. A 20 μl drop of conidial suspension was placed on either side of the cotyledonary node of the stem. A small puncture was made into the stem by passing a sterile needle (28.5G Becton-Dickinson & Co., NJ, USA) through the spore suspension droplet. Microwounding in this manner facilitates the movement of spore suspension into the stem that is under negative pressure. Mock-treated plants underwent the same procedure with sterile distilled water. Infected and uninfected plants were grown for 3 weeks in a growth chamber at 27/22°C (day/night) temperature, with a 13 h photoperiod. During this period each plant was fertilized once a week with 50 ml of 3 g/l of water-soluble 20-20-20 N-P-K. Leaf/stem weight ratio, plant height, and total shoot weight at 3 weeks following infection, were used as the parameters to assess disease-severity.
Alternaria alternata infection assay
The method described by Emani et al. (2003) was used to perform A. alternata infection of a detached leaf. First, fully developed leaf from an 18-day-old cotton plant was removed and placed on a wet filter paper soaked with 3 ml of sterile distilled water in a Petri plate. An agar plug containing mycelial mat was placed on each side of the midrib on adaxial surface. The Plates were sealed with parafilm and kept in dark at 28°C. Twelve days following inoculation, the leaves were photographed and infected area from the digital image was measured by using AlphaEase v5.5 software (Alpha Innotech, USA).
Reniform nematode resistance assay
An assay that is based on a count of motile vermiform stages of the nematode in the soil was used to evaluate the resistance of transgenic plants (Robinson et al. 2007). Cotton plants were grown in a medium favorable for growth of cotton and reniform nematodes. The mixture was prepared by mixing 48 l sandi-loam soil, 8 l vermiculite (SUN GRO Horticulture Ltd, Bellevue, WA, USA), 300 g fine dolomite (Carlpool® Products, Gladewater, TX, USA) 100 g Hoe-down™ gypsum soil conditioner (Earl Ricketts Co., FL, USA) and 50 g Esmigran, (micronutrients in sustained released form; Scotts-Sierra Horticultural Product Company, OH, USA) in a cement mixer for 30 min. The soil mixture and washed river sand were pasteurized for 6 h at 70°C. Fifty milliliter of sand was placed in the bottom of a 500 ml plastic cup (with three holes, covered with plastic screen, at the bottom) and the rest filled with the soil mixture described earlier. The soil in each cup was fertilized with 50 ml of 9-45-15 N-P-K (6 g/l) before planting two-day-old pre-germinated cotton seedlings. Nematode inoculum was introduced in the soil around each plant following 2 weeks of growth in a chamber at 28/22°C (day/night) temperature, with a 13 h photoperiod. Baermann funnel extraction method was used to isolate nematodes, consisting of mixed stages of vermiform nematodes, from 100 g of infested soil (Robinson and Heald 1991). A liquid suspension containing 6,000 motile nematodes was injected into the soil around a plant at a distance of two cm from the stem. After inoculation, cotton plants were fertilized once a week with 50 ml of 3 g/l 20-20-20 N-P-K for 5 weeks and then every 2 weeks up to the 10th week. At this time, three soil samples totaling 30 g were removed from each cup using a 12 mm wide cork borer and the nematodes were extracted as described earlier. Nematodes were counted under a microscope (‘Wild Heerbrugg M420 Makroskop’, a non-stereo zoom, 1,25×) and the results are presented as reniform nematodes/g infested soil. In addition to the nematode count, data for yield parameters such as the number of bolls and boll fresh weight (FW) per plant was noted.
Statistical analysis
Treatments were analyzed for significant differences using one-way ANOVA.
Results
Generation of transgenic cotton lines and their molecular analysis
Using the Agrobacterium-mediated transformation method, we generated 144 primary transgenic cotton plants. Ninety-three primary transformants were analyzed by Reverse transcriptase-polymerase chain reaction (RT-PCR) for AtNPR1 transcripts and 68 were found to express the transgene. These AtNPR1-expressing cotton plants appeared normal in their growth and development. Based on consistent transcript levels over several generations, transgenic lines 68L-19 and 68L-20 were selected for detailed analyses (Supplementary Figure 1). Southern blot analysis showed that the transgenic line 68L-19 had a single and line 68L-20 had three transgene copies integrated into their respective genomes (Supplementary Figure 2). T2 or T3 generations obtained from a homozygous parent for both lines were used for disease resistance and other analyses.
Systemic induction of chitinase and glucanase activities in the leaves of transgenic plants infected with F. oxysporum f. sp. vasinfectum
As a measure of the defense response, chitinase and glucanase enzyme activities were examined at various time points in the leaves of plants whose roots were infected with F. oxysporum f. sp. vasinfectum. Without infection, there were no differences in the enzyme activities between transformant, 68L-19 and the wild-type plants confirming that NPR1 expression by itself does not affect the defense response machinery of the plant (Fig. 1a). A small increase in chitinase activity was evident in both sets of plants at 12 h post-inoculation (hpi) with the transgenic plants showing a small but significant induction (P < 0.01) in chitinase activity compared to the mock-treated transformants. Between 12 and 24 hpi, the chitinase activity increased in both sets of infected plants, however, the magnitude of induction was significantly greater in the transformants. At 48 hpi, chitinase activity had decreased in both sets of plants from its peak value, but remained higher in the NPR1-expressing plants compared to the WT. By 72 hpi, the chitinase activity had markedly declined in F. oxysporum f. sp. vasinfectum-infected transgenic plants and was similar to that of infected WT plants (Fig. 1a). Thus, compared to the WT plants, the transgenic line 68L-19 showed a greater increase in chitinase activity in response to F. oxysporum f. sp. vasinfectum infection.
Activities of PR enzymes in the leaves (first true leaf) of the transgenic cotton line, 68L-19 and untransformed WT plant following inoculation of roots with Fusarium oxysporum f. sp. vasinfectum isolate Fov11. a Chitinase activity, and b Glucanase activity. I: Roots were treated with F. oxysporum conidial suspension; M: Roots were mock-treated with tap water. Data represent mean ± SE, n = 10; the induced enzyme activity value for the transgenic line is significantly higher than that of the WT value at * P < 0.05
Glucanase activity remained at the basal level up to 12 h following F. oxysporum f. sp. vasinfectum inoculation. Between 12 and 24 hpi, the glucanase activity increased in both sets of infected plants with the transformants showing a small but significantly higher level at 24 hpi. After this time point, the rate of increase was slightly higher in the WT plants (Fig. 1b). Thus, the transgenic plants do show an early induction of glucanase activity in response to F. oxysporum f. sp. vasinfectum-infection; however, it is overtaken by the WT plants after 48 hpi.
Accumulation of gossypol-related terpenoids in stele tissue of WT and transgenic cotton plants in response to V. dahliae
Gossypol and related terpenoids are known to be induced in cotton plants in response to V. dahliae (Bell 1969; Mace 1983; Mace et al. 1989). Therefore, we were interested in knowing if the terpenoid-based defense mechanism responded differently in the transformants. WT as well as transgenic plants were inoculated with spore suspension of V. dahliae isolate TS2 and the total terpenoids were estimated as gossypol equivalents at various time intervals. The terpenoid levels in the stele tissue of transformants and WT were similar prior to infection. Compared to WT, transgenic tissues accumulated slightly higher levels of terpenoids at 24 hpi. At 48 and 72 hpi, the transgenic plants had 30.1 and 29.8% more terpenoids, respectively, than the WT plants (Fig. 2). The differences were statistically significant (P < 0.05). The terpenoid levels in the mock-treated transgenic as well as WT plants remained at low levels at all time points. The more rapid induction of terpenoids in response to V. dahliae isolate TS2 could be one of the factors that provide added protection to the transgenic plants against this disease.
Induction of terpenoids (gossypol equivalents) at the 5th internode in cotton plants infected with Verticillium dahliae isolate TS2. I: inoculated with V. dahliae conidial suspension; M: treated with sterile distilled water as a mock treatment. The stele tissue obtained from the internode was used to extract terpenoids. Data represent mean ± SE, * P < 0.05; n = 5
SA-induced systemic induction of chitinase and glucanase activities in the cotyledons of transgenic plants
SA has been shown to be an inducer of SAR and its associated PR genes in various plant species, including cotton (Ward et al. 1991; Uknes et al. 1992; Hudspeth et al. 1996; Djonovic et al. 2006). Therefore, we were keen to examine the influence of SA on AtNPR1-expressing cotton lines. After 12, 24, 48 h of treatment of the seedling root with SA, the activities of chitinase and glucanase were determined in the cotyledons of line 68L-19. SA-treatment of the roots induced chitinase and glucanase activities in the cotyledons of both WT and the transgenic plants. However, the induction in the AtNPR1-transformants was faster and significantly higher compared to the untransformed plants (Supplementary Figure 3a, b). Similar inductions were observed in transgenic line 68L-20 at 24 h time point (Supplementary Figure 4a, b). Again, the induced enzyme activities in this line were significantly higher than those observed in WT seedlings in response to SA treatment. Thus, AtNPR1-expressing cotton plants exhibit higher levels of defense-related enzyme activities in response to SA.
BTH- and F. oxysporum-induced expression of PR genes in transgenic lines
We were interested to know whether BTH, a biologically active analog of salicylic acid (Friedrich et al. 1996; Kohler et al. 2002; Makandar et al. 2006), is capable of inducing PR genes in cotton plants, and if so, do the transgenic plants respond differently. Sequences of PR1 (Accession no. CK988133.1), Osmotin (Accession no. CF932065), Glucanase (Accession no. CD486342) and Chitinase (Accession no. CD485880) genes, isolated from cotton, were available. Their expression was monitored at 6, 12 and 24 h post-treatment (hpt) of the leaves with BTH, in WT and the transgenic lines, 68L-19 and 68L-20. At 6 hpt, an induction in the transcripts for PR1, Osmotin and Glucanase was evident in the transgenic lines, 68L-19 and 68L-20 (Fig. 3a). The higher level of transcripts is maintained in line 68L-19 even at 24 hpt. Either a small or no induction in the activities of these genes was observed in the WT plants. The activity of the chitinase gene was induced in WT as well as in the transgenic lines. Overall, the results suggest that AtNPR1-expressing cotton transformants are primed to respond more rapidly and strongly to factors that induce SAR.
a PR gene (PR1, Osmotin, Glucanase, and Chitinase)-expression in WT and AtNPR1-transgenic cotton lines, 68L-19 and 68L-20, after 6, 12 and 24 h of treatment with 100 μM BTH. A young leaf from five plants each were pooled, frozen in liquid N2 and used to extract total RNA. b PR gene (Chitinase, Glucanase, Osmotin and PR1)-expression in the leaves of WT and transgenic line, 68L-19, at 12, 24, 48 hpi with Fusarium oxysporum f. sp. vasinfectum isolate Fov11. A young leaf from ten plants each were pooled together for each time point, frozen in liquid N2 and were used to extract total RNA
Fusarium oxysporum f. sp. vasinfectum can induce SAR related genes in Arabidopsis (Mauch-Mani and Slusarenko 1994). We therefore studied expression of PR genes (Chitinase, Glucanase, Osmotin and PR1) in the leaves of transgenic and WT plants following infection of the roots with F. oxysporum f. sp. vasinfectum. Induction of Chitinase gene activity was observed in both the transgenic line and WT at 12 hpi, but the level appears to be slightly higher in the former than the latter (Fig. 3b). By 24 hpi, the induction had completely subsided in the WT plants while the transgenic line showed transcripts, but at a lower level than at 12 hpi. Transcripts diminished to a very low level at 48 h (Fig. 3b). The results showed that Chitinase gene induction in transgenic plants is greater and the heightened activity lasts a little longer in comparison to the WT plants. F. oxysporum-induced expression of Glucanase gene was observed only at 12 hpi in both sets of plants but the induction was stronger in the transgenic plants. Although induction of Osmotin gene was observed in both transgenic plants and WT plants, the level of transcripts appears to be higher in the transformants. In response to F. oxysporum f. sp. vasinfectum exposure, a progressive increase in transcript levels for this gene was observed in the case of transgenic line (Fig. 3b). No apparent induction of the tested PR1 gene was observed in WT or the transgenic line 68L-19 (Fig. 3b). Plants usually have several members in the PR1 gene family, and once the sequences of the remaining cotton PR1 genes and other PR genes become available, it will be interesting to study their expression in AtNPR1-cotton plants during their interaction with F. oxysporum f. sp. vasinfectum. Taken together, the results suggest that the AtNPR1 transformants express certain PR genes at higher levels compared to their WT counterparts during their interaction with F. oxysporum f. sp. vasinfectum.
AtNPR1-mediated resistance to fungal pathogens in transgenic cotton
Rhizoctonia solani is the most common causative agent of post-emergence damping off and is also responsible for pre-emergence death under cool conditions (Hillocks 1992b). Because of its importance as a cotton pathogen, we were interested in examining the resistance of AtNPR1-expressing cotton lines to R. solani. Disease index, a measure of infection severity, was 79.7% lower (P < 0.001) in transgenic line 68L-20 compared to that of WT. Line 68L-19 also showed enhanced resistance to R. solani (P < 0.05), but at a lower level (Fig. 4a). The collar region in R. solani-infected, WT seedlings was necrotized, while the transgenic seedlings were mostly healthy with very few, if any necrotic spots (Fig. 5a). The results show that overexpression of AtNPR1 gene in cotton confers resistance to R. solani.
a Resistance to Rhizoctonia solani in transgenic cotton lines expressing AtNPR1. Disease indices for WT and transgenic cotton lines infected with R. solani scored after 6 days in a soil-based assay (data represent mean ± SE; mean obtained by averaging results from three sets of seedlings tested with each set consisting of 12 seedlings). The value for the transgenic line is significantly different from the control value at * P < 0.05; ** P < 0.001. b Resistance to Alternaria alternata in transgenic cotton lines expressing AtNPR1. Disease severity was scored on the 12th day following inoculation of detached leaves with the pathogen. Data represent mean ± SE, * P < 0.05; ** P < 0.001, n = 10
Broad-spectrum resistance to fungal diseases in transgenic cotton lines expressing AtNPR1. Images of transgenic seedlings/plants/leaves on the day disease severity was scored: a Rhizoctonia solani; b Alternaria alternata; c Fusarium oxysporum f. sp. vasinfectum isolate Fov11 and d Verticillium dahliae isolate TS2
Under certain conditions, such as injury created by pest attack, adverse weather conditions, moisture, or nutrient stress, A. alternata can infect cotton and cause leaf spots resulting in significant yield losses in cotton (Hillocks 1992a). Transgenic lines were tested for resistance against this foliar pathogen in a detached leaf assay and the results are shown in Fig. 4b. Twelve days following the placement of fungal inoculum, the leaves from transgenic plants were largely unaffected while those of WT showed infection lesions and chlorosis (Fig. 5b). The infected leaf area in lines 68L-19 and 68L-20 were 18 and 37% lower compared to that in the WT plants (Fig. 4b). The transgenic lines were clearly more resistant than the WT against A. alternata.
Wilt diseases are also a serious problem in the production of cotton. Fusarium wilt caused by F. oxysporum f. sp. vasinfectum can affect cotton at any stage of development resulting in dark brown discoloration of the vascular system, wilting, chlorosis and necrosis of leaves, and plant death (Hillocks 1992b; Davis et al. 2006). AtNPR1-expressing lines were examined for their resistance to F. oxysporum f. sp. vasinfectum isolate Fov11. Two parameters were used to measure the degree of disease severity, leaf disease index (LDI) and shoot fresh weight. Both the transgenic lines showed lower disease severity in terms of LDI compared to the WT. The LDI for transgenic lines 68L-19 and 68L-20 were 62% (P < 0.001) and 26.6% lower, respectively, than the WT values (Fig. 6a). The mean shoot fresh weight for transgenic lines 68L-19 and 68L-20 were 2.73 and 1.76 g, respectively, compared to 0.82 g for WT (Fig. 6b). Three weeks following the inoculation of the roots, most of the leaves of WT plants were either senesced/wilted or had dropped, while the transgenic lines showed significantly higher retention of leaves which appeared healthy (Fig. 5c). In fact, line 68L-19 showed a higher degree of resistance to F. oxysporum f. sp. vasinfectum than a commercial cotton variety, DP 444 (Supplementary Figure 5a–c). This variety had shown the highest level of resistance to Fusarium wilt in a comparative study involving 50 different genotypes (Bell, unpublished results). The results from current investigation provide strong evidence that AtNPR1 expression in cotton confers resistance to F. oxysporum f. sp. vasinfectum.
Verticillium wilt caused by V. dahliae and V. albo-atrum is reported in more than two hundred dicotyledonous plant species and affects the production of various economically important crops, including cotton (Fradin and Thomma 2006). Decreased photosynthetic capacity, brown vascular discoloration, wilting and stunting are some of the symptoms of this disease in cotton (Hampton et al. 1990; Fradin and Thomma 2006). We examined the resistance of the AtNPR1-expressing transformants against V. dahliae isolate TS2. Three weeks following the infection, plants were examined for disease severity by measuring three parameters: shoot weight, leaf/stem weight ratio, and plant height. Fresh shoot weights of V. dahliae-infected transgenic lines 68L-19 and 68L-20 were significantly higher at 48.7 g (P < 0.001) and 40.6 g (P < 0.05), respectively, than that of WT at 16.2 g (Fig. 7a). Infected WT plants showed extensive defoliation and the leaves that did remain on the plant were wilted (Fig. 5d). In comparison to the leaf/stem weight ratio of 0.58 for the WT, these values for transgenic lines 68L-19 and 68L-20 were 1.4 (P < 0.001) and 1.11, respectively (Fig. 7b). WT plants were severely stunted in their growth showing an average reduction of 40% in their height as a result of Verticillium infection. The average heights of 68L-19 and 68L-20 plants were 73.8 and 66.3 cm, respectively, significantly higher than the 53.1 cm for the WT plants (Figs. 5d, 7c). Progression of the disease in plants was also monitored by cutting longitudinal sections of the internodes. Severe vascular browning was observed in the stem of Verticillium-infected WT plants at the 5th internode, while this section from the transgenic plants did not show vascular browning indicating suppression of infection (Supplementary Figure 6). In summary, the transgenic lines exhibited strong resistance to V. dahliae isolate TS2.
Resistance to Verticillium dahliae isolate TS2 in transgenic cotton lines expressing AtNPR1. a Shoot weight, b leaf/stem weight ratio, and c plant height were used as parameters to score disease-severity, 3 weeks following stem inoculation with the pathogen. Data represent mean ± SE, * P < 0.05; ** P < 0.001, n = 10
AtNPR1-mediated resistance to reniform nematodes in transgenic cotton
A recent report indicated a possible role for NPR1 in SA-mediated inhibition of parasitism by cyst nematodes (Heterodera schachtii) in Arabidopsis (Wubben et al. 2008). On the basis of this report and encouraged by the broad-spectrum resistance of the AtNPR1-cotton transformants to various fungal pathogens, we examined these plants for their resistance to reniform nematode, an economically important pest of cotton. A preliminary experiment with these nematodes had provided some encouraging results showing better performance of the transformants and significantly lower nematode count in the soil supporting these plants (Supplementary Figures 7a–d, 8). A second experiment was conducted to confirm these results with the same transgenic lines, 68L-19 and 68L-20. Ten weeks after inoculation, the vermiform nematode count for the untransformed WT was 77.9 per g of infested soil. This number is seven- and two-fold higher compared to the counts obtained for the transgenic lines 68L-19 and 68L-20, respectively (Fig. 8a). Cotton plants growing in nematode infested soil under field conditions, suffer from nutritional deficiencies, fruit abortion, and abnormal maturation of the crop (Koenning et al. 2004). Even under controlled environmental conditions in a growth chamber, nutritional deficiencies and the loss of bolls was evident (Fig. 8b–d, Supplementary Figure 8). In addition to the number of bolls per plant, relative position of the bolls on the plant is a good indicator of the health of the plant and provides an indirect measure of the ability of the plant to retain bolls when facing nematode pressure. The data on boll-retention and -position shown in Fig. 8b–d indicates relatively better health of the transgenic plants growing in nematode-infested soil. Interestingly, line 68L-19 demonstrated a stronger resistance to reniform nematodes compared to the line 68L-20 that showed moderate level resistance. Although the degree of resistance was not as high, we observed a similar trend with a different line (68L-16) with respect to vermiform nematode count and boll number/weight (data not shown). Taken together, the results suggest that the AtNPR1-expressing cotton plants are more resistant than WT to the nematode, R. reniformis.
Resistance to reniform nematode Rotylenchulus reniformis in transgenic cotton lines expressing AtNPR1. a Nematode count in soil after 10 weeks of inoculation. b Number of bolls per plant. c Boll weight per plant. d Boll position (number of bolls at given nodes in 10 plants). Data represent mean ± SE, * P < 0.05; ** P < 0.01, n = 10
Discussion
Our results show that the expression of AtNPR1 in cotton can confer increased resistance to two necrotrophic fungal pathogens of cotton, R. solani, (a cause of seedling damping-off) and A. alternata (a cause of foliar disease). Two wilt diseases in cotton, Fusarium wilt caused by F. oxysporum f. sp. vasinfectum and Verticillium wilt caused by V. dahliae, together account for significant losses in yield ranging from 0.5 to 5% during 1952–2008 (National Cotton Council of America—Disease Data Base). In the current study, we show increased resistance to F. oxysporum f. sp. vasinfectum in AtNPR1-expressing transgenic lines compared to WT. Transgene-mediated conferral of resistance to V. dahliae in cotton has been reported. However, only a small reduction in disease severity due to V. dahliae was observed in transgenic cotton plants expressing T. flavus glucose oxidase (Murray et al. 1999) and Gastrodia anti-fungal protein (Wang et al. 2004). In this report, we provide detailed evidence for resistance obtained against a non-defoliating pathovar of V. dahliae isolate TS2 in cotton plants overexpressing the AtNPR1 gene. Thus, results showing protection against four different fungal diseases demonstrate the broad-spectrum nature of the resistance in transgenic cotton plants overexpressing the AtNPR1 gene.
Nematodes are a growing threat to cotton farming in the US and elsewhere. In the U.S., the estimated yield loss due to nematodes was 1–2% in 1950, which increased to 4.78% in 2007. The reniform nematode alone contributed to 2.04% of the loss in total yield in 2007. The lack of resistant cultivars is one of the reasons responsible for the increasing yield losses due to the nematodes (Robinson et al. 1999; Koenning et al. 2004; Blasingame et al. 2008). Thus, there is an urgent need for measures to control losses caused by these parasites. Some studies have shown a reduction in the reproduction of root-knot and reniform nematodes in crop plants such as cowpea, soybean and pineapple by the application of SAR inducer, BTH (Chinnasri et al. 2003, 2006). Nandi et al. (2003) showed that foliar application of SA to okra and cowpea 24 h before the inoculation of roots with M. incognita, induced PR1 and PAL in roots and reduced infestation. Recently, Wubben et al. (2008) have demonstrated inhibition of cyst nematode (H. schachtii) parasitism in Arabidopsis by SA that was shown to be mediated by NPR1. In the current investigation, we examined the role of NPR1 directly, and provide some evidence that constitutive expression of AtNPR1 can enhance resistance to nematode, R. reniformis in the transgenic cotton plants. The protection provided to the transformants by NPR1 overexpression is not only reflected in the lower number of nematodes but is also supported by higher boll retention, boll weight and overall health of the plants.
Earlier studies on overexpression of NPR1 in various plant species including Arabidopsis (Cao et al. 1998), tomato (Lin et al. 2004), wheat (Makandar et al. 2006), B. napus (Potlakayala et al. 2007), apple (Malnoy et al. 2007) did not report any abnormalities in the growth and development of transgenic plants under normal growth conditions. In agreement with above-mentioned studies, we did not observe any obvious abnormal growth behavior in cotton transformants expressing AtNPR1. Importantly, our visual observations on the phenotypic normalcy of the AtNPR1-transformants are also supported by some of the biochemical and molecular data presented in this report showing no significant differences between transgenic and WT control plants in the absence of a challenge from a pathogen or chemical inducer. Besides providing resistance to Xanthomonas oryzae pv. oryzae, NPR1-overexpression in rice resulted in the development of lesion-mimic spots on the leaves under low-light condition of a growth chamber (Fitzgerald et al. 2004; Chern et al. 2005). We did not observe this type of sensitivity in AtNPR1 cotton transformants that were grown under various light levels ranging from 12 to 950 μmol m−2 s−1. In a recent study, AtNPR1 expression in rice, while conferring resistance to fungal and bacterial diseases, had a negative effect on virus infections and the ability to tolerate salt- and drought stress (Quilis et al. 2008). It is important to note that until now, any negative effects of NPR1-overexpression have been observed only in rice, which has very high basal SA level compared to tobacco, Arabidopsis, and cotton (Yang et al. 2004; Raskin et al. 1990; Nawrath and Metraux 1999; Martinez et al. 2000; Silverman et al. 1995). Thus, endogenous, basal SA levels may be an important consideration in selecting a crop species for the development of NPR1-mediated disease resistance.
SA and its synthetic analog, BTH are known to induce SAR and SAR associated genes in various plant species (Ward et al. 1991; Uknes et al. 1992; Hudspeth et al. 1996; Djonovic et al. 2006; Gorlach et al. 1996; Friedrich et al. 1996; Nawrath and Metraux 1999; Colson-Hanks and Deverall 2000; Kohler et al. 2002; Zhu et al. 2003). In some of these studies, SAR was induced by direct bathing of the root or soil-drenching with the chemical. In our investigation, 6 h after treatment with BTH, a clear and strong induction of PR1, glucanase and osmotin genes was observed only in the transgenic cotton lines, 68L-19 and 68L-20. The activities of chitinase and glucanase enzymes were determined in the cotyledons of cotton seedlings following treatment of their roots with SA. Compared to WT, significant elevation in the activities of both the enzymes were observed in the transgenic lines, 68L-19 and 68L-20, following SA treatment. Djonovic et al. (2006) have also reported systemic induction of various defense-related genes in cotton seedlings (cv. Deltapine 50) in response to SA treatment of roots. Taken together, the results suggest that the AtNPR1-expressing cotton plants are primed to respond to SAR-inducers and the levels of gene/enzyme induction is higher in these plants. These factors may be the basis for observed resistance to various fungal pathogens.
In a number of plant species overexpressing NPR1 gene, a greater and faster activation of various PR genes in response to pathogen-challenge has been reported (Cao et al. 1998; Makandar et al. 2006; Malnoy et al. 2007; Quilis et al. 2008). In the current investigation, we studied the expression of various PR genes in transgenic cotton line, 68L-19, following a challenge with F. oxysporum f. sp. vasinfectum. This line had shown a strong resistance to this pathogen. A stronger and longer-lasting induction of Chitinase expression was observed in F. oxysporum-challenged transgenic line, 68L-19 compared to the WT plants (Fig. 3b). A clear and progressive induction of Osmotin was observed only in F. oxysporum-inoculated transgenic line. Our investigation into PR gene activities is limited by of the availability of cotton gene sequences. As additional information on cotton genes that are involved in defense function becomes available, expression of these genes will be examined in the NPR1 transformants in the future.
As a follow up to the PR gene expression study, we determined the activities of chitinase and glucanase in F. oxysporum f. sp. vasinfectum challenged transgenic plants. A sharp induction in the activity of both glucanase and chitinase was observed at 24 hpi in F. oxysporum-inoculated transgenic line 68L-19, which is significantly higher than the values in similarly treated WT. The stronger and early induction of PR genes and enzyme activities in pathogen-challenged transgenic line, 68L-19 suggests an active role of SAR associated PR genes in AtNPR1-mediated resistance to F. oxysporum f. sp. vasinfectum in transgenic cotton.
Induction of gossypol and related terpenoids is believed to be a part of the defense mechanism in cotton plants in response to pathogen infection (Stipanovic et al. 1999; Eldon and Hillocks 1996). In particular, there are several reports showing the involvement of terpenoids in the defense mechanism against Verticillium spp. in cotton. For example, cotton cultivar, Seabrook Sea Island 12B2 (SBSI), which shows resistance to V. dahliae and V. albo-atrum, produces terpenoid aldehydes faster and at higher levels upon infection (Mace 1983; Bianchini et al. 1999; Bell 1969). The higher induction of phytoalexins in stem tissue of cotton during the early phase of infection with V. albo-atrum is positively correlated with resistance in SBSI. In contrast, the susceptible cotton cultivars show a slower induction of terpenoids when challenged with Verticillium (Bell 1969). The NPR1-expressing transgenic lines had shown high degree of resistance to V. dahliae isolate TS2, therefore, we were interested in examining the levels of these compounds in the transformants following a challenge with the pathogen. At 48 and 72 hpi, transgenic line 68L-19 (which shows high level of resistance to V. dahliae) had significantly higher levels of terpenoids compared to the untransformed WT plants. It is possible that the higher terpenoid levels form part of the overall defense response that protects the NPR1-transformants from the disease. Although there are no published reports showing the involvement of NPR1 in the induction of gossypol and related terpenoids in cotton, it stands to reason that activation of the plant’s basal defenses will also involve this important resistance mechanism found in the tribe Gossypeae (Stipanovic et al. 1999).
The data presented in this report show clearly that expression of AtNPR1 in cotton confers resistance to four different fungal pathogens and the reniform nematode. The transformants appeared normal in their growth and development. These results suggest that NPR1 may provide an effective tool to obtain resistance to diverse pathogens by transgenic means with minimal metabolic cost to the plant. Thus, although the current study and several others reported herein show promise in the use of NPR1 gene as a transgenic tool to obtain broad-spectrum disease resistance in important crops, additional research is needed to ensure the overall efficacy of such an approach under field conditions where the plants are exposed to a variety of biotic and abiotic stresses. This is because in addition to being an important mediator of SAR and ISR, cytosolic NPR1 plays some role in modulating the antagonistic cross-talk between SA and jasmonic acid signaling (Dong 2004; Spoel et al. 2003, 2007; Pieterse and Van Loon 2004). The complexity of the role of NPR1 in various signaling networks has been further heightened by a recent study on Arabidopsis showing that ethylene modulates the NPR1 dependency of SA-jasmonic acid antagonism (Leon-Reyes et al. 2009). Regardless, the results presented in this report provide strong evidence supporting the role of NPR1 as a critical component in the defense mechanism that protects plants from certain biotic challenges.
References
Abeles FB, Forrence LE (1970) Temporal and hormonal control of beta-1,3-glucanase in Phaseolus vulgaris L. Plant Physiol 45:395–400
Bell AA (1967) Formation of gossypol in infected or chemically irritated tissues of gossypium species. Phytopathology 57:759–764
Bell AA (1969) Phytoalexin production and Verticillium wilt resistance in cotton. Phytopathology 59:1119–1127
Bianchini GM, Stipanovic RD, Bell AA (1999) Induction of delta-cadinene synthase and sesquiterpenoid phytoalexins in cotton by Verticillium dahliae. J Agr Food Chem 47:4403–4406
Blasingame D, Bank JC, Colyer PD, Davis M, Gazaway WS, Kemerait RC, Kirkpatrick TL, Koenning SR, John Muller, Newman M, Mary Olsen, Phipps PM, Sciumbato GL, Richard Sprenke, Woodward JE, Wrather A, Patel MV (2008) Cotton disease loss estimatecommittee report. In: Procceedings of Beltwide cotton conferences. Nashville, Tennessee. Jan 2008, pp 294–297
Bolek Y, Bell AA, El-Zik KM, Thaxton PM, Magill CW (2005) Reaction of cotton cultivars and an F-2 population to stem inoculation with isolates Verticillium dahliae. J Phytopathol 153:269–273
Cao H, Bowling SA, Gordon AS, Dong X (1994) Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6:1583–1592
Cao H, Li X, Dong X (1998) Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc Natl Acad Sci USA 95:6531–6536
Chern MS, Fitzgerald HA, Yadav RC, Canlas PE, Dong X, Ronald PC (2001) Evidence for a disease-resistance pathway in rice similar to the NPR1-mediated signaling pathway in Arabidopsis. Plant J 27:101–113
Chern M, Fitzgerald HA, Canlas PE, Navarre DA, Ronald PC (2005) Overexpression of a rice NPR1 homolog leads to constitutive activation of defense response and hypersensitivity to light. Mol Plant-Microbe Interact 18:511–520
Chinnasri B, Sipes BS, Schmitt DP (2003) Effects of Acibenzolar-S-methyl application to Rotylenchulus reniformis and Meloidogyne javanica. J Nematol 35:110–114
Chinnasri B, Sipes BS, Schmitt DP (2006) Effects of inducers of systemic acquired resistance on reproduction of Meloidogyne javanica and Rotylenchulus reniformis in pineapple. J Nematol 38:319–325
Colson-Hanks ES, Deverall BJ (2000) Effect of 2,6-dichloroisonicotinic acid, its formulation materials and benzothiadiazole on systemic resistance to alternaria leaf spot in cotton. Plant Pathol 49:171–178
Davis RM, Colyer PD, Rothrock CS, Kochman JK (2006) Fusarium wilt of cotton: population diversity and implication for management. Plant Dis 90:692–703
Delaney TP, Friedrich L, Ryals JA (1995) Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc Natl Acad Sci USA 92:6602–6606
Despres C, DeLong C, Glaze S, Liu E, Fobert PR (2000) The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12:279–290
Djonovic S, Pozo MJ, Dangott LJ, Howell CR, Kenerley CM (2006) Sm1, a proteinaceous elicitor secreted by the biocontrol fungus Trichoderma virens induces plant defense responses and systemic resistance. Mol Plant-Microbe Interact 19:838–853
Dong XN (2004) NPR1, all things considered. Current Opin Plant Biol 7:547–552
Eldon S, Hillocks RJ (1996) The effect of reduced phytoalexin production on the resistance of upland cotton (Gossypium hirsutum) to Verticillium and Fusarium wilts. Ann Appl Biol 129:217–225
Emani C, Garcia JM, Lopata-Finch E, Pozo MJ, Uribe P, Kim DJ, Sunilkumar G, Cook DR, Kenerley CM, Rathore KS (2003) Enhanced fungal resistance in transgenic cotton expressing an endochitinase gene from Trichoderma virens. Plant Biotechnol J 1:321–336
Fan W, Dong X (2002) In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. Plant Cell 14:1377–1389
Fitzgerald HA, Chern MS, Navarre R, Ronald PC (2004) Overexpression of (At)NPR1 in rice leads to a BTH- and environment-induced lesion-mimic/cell death phenotype. Mol Plant-Microbe Interact 17:140–151
Fradin EF, Thomma BPHJ (2006) Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol. Plant Pathol 7:71–86
Friedrich L, Lawton K, Ruess W, Masner P, Specker N, Rella MG, Meier B, Dincher S, Staub T, Uknes S, Metraux JP, Kessmann H, Ryals J (1996) A benzothiadiazole derivative induces systemic acquired resistance in tobacco. Plant J 10:61–70
Friedrich L, Lawton K, Dietrich R, Willits M, Cade R, Ryals J (2001) NIM1 overexpression in Arabidopsis potentiates plant disease resistance and results in enhanced effectiveness of fungicides. Mol Plant Microbe Interact 14:1114–1124
Gorlach J, Volrath S, Knauf-Beiter G, Hengy G, Beckhove U, Kogel KH, Oostendorp M, Staub T, Ward E, Kessmann H, Ryals J (1996) Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell 8:629–643
Grant M, Lamb C (2006) Systemic immunity. Curr Opin Plant Biol 9:414–420
Hampton RE, Wullschleger SD, Ossterhuis DM (1990) Impact of Verticillium wilt on net photosynthesis, respiration and photorespiration in field-grown cotton (Gossypium hirsutum L.). Physiol Mol Plant Pathol 37:271–280
Hillocks RJ (1992a) Fungal diseases of the leaf. In: Hillocks RJ (ed) Cotton diseases. CAB International, Oxon, UK, pp 191–238
Hillocks RJ (1992b) Seedling diseases. In: Hillocks RJ (ed) Cotton diseases. Oxon, CAB International, pp 1–38
Hudspeth RL, Hobbs SL, Anderson DM, Grula JW (1996) Characterization and expression of chitinase and 1,3-beta-glucanase genes in cotton. Plant Mol Biol 31:911–916
Kachroo P, Yoshioka K, Shah J, Dooner HK, Klessig DF (2000) Resistance to turnip crinkle virus in Arabidopsis is regulated by two host genes and is salicylic acid dependent but NPR1, ethylene, and jasmonate independent. Plant Cell 12:677–690
Kesarwani M, Yoo J, Dong X (2007) Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol 144:336–346
Kinkema M, Fan W, Dong X (2000) Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12:2339–2350
Koenning SR, Kirkpatrick TL, Starr JL, Wrather JA, Walker NR, Mueller JD (2004) Plant-parasitic nematodes attacking cotton in the United States—old and emerging production challenges. Plant Dis 88:100–113
Kohler A, Schwindling S, Conrath U (2002) Benzothiadiazole-induced priming for potentiated responses to pathogen infection, wounding, and infiltration of water into leaves requires the NPR1/NIM1 gene in Arabidopsis. Plant Physiol 128:1046–1056
Kumar V, Parkhi V, Kenerley CM, Rathore KS (2009) Defense-related gene expression and enzyme activities in transgenic cotton plants expressing an endochitinase gene from Trichoderma virens in response to interaction with Rhizoctonia solani. Planta 230:277–291
Leon-Reyes A, Spoel SH, De Lange ES, Abe H, Kobayashi M, Tsuda S, Millenaar FF, Welschen RA, Ritsema T, Pieterse CM (2009) Ethylene modulates the role of NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 in cross talk between salicylate and Jasmonate signaling. Plant Physiol 149:1797–1809
Lin WC, Lu CF, Wu JW, Cheng ML, Lin YM, Yang NS, Black L, Green SK, Wang JF, Cheng CP (2004) Transgenic tomato plants expressing the Arabidopsis NPR1 gene display enhanced resistance to a spectrum of fungal and bacterial diseases. Transgenic Res 13:567–581
Mace ME (1983) Elicitation of accumulation of terpenoid aldehyde phytoalexins in Verticillium dahliae infected cotton. New Phytol 95:115–119
Mace ME, Stipanovic RD, Bell AA (1989) Histochemical localization of desoxyhemigossypol, a phytoalexin in Verticillium dahliae-infected cotton stems. New Phytol 111:229–232
Makandar R, Essig JS, Schapaugh MA, Trick HN, Shah J (2006) Genetically engineered resistance to Fusarium head blight in wheat by expression of Arabidopsis NPR1. Mol Plant Microbe Interact 19:123–129
Malnoy M, Jin Q, Borejsza-Wysocka EE, He SY, Aldwinckle HS (2007) Overexpression of the apple MpNPR1 gene confers increased disease resistance in malus x domestica. Mol Plant-Microbe Interact 20:1568–1580
Martinez C, Baccou JC, Bresson E, Baissac Y, Daniel JF, Jalloul A (2000) Salicylic acid mediated by the oxidative burst is a key molecule in local and systemic responses of cotton challenged by an avirulent race of Xanthomonas campestris pv malvacearum. Plant Physiol 122:757–766
Mauch-Mani B, Slusarenko AJ (1994) Systemic acquired resistance in Arabidopsis thaliana induced by a predisposing infection with a pathogenic isolate of Fusarium oxysporum. Mol Plant-Microbe Interact 7:378–383
Mou Z, Fan W, Dong X (2003) Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113:935–944
Murray F, Llewellyn D, McFadden H, Last D, Dennis ES, Peacock WJ (1999) Expression of the Talaromyces flavus glucose oxidase gene in cotton and tobacco reduces fungal infection, but is also phytotoxic. Mol Breed 5:219–232
Nandi B, Kundu K, Banerjee N, Babu SPS (2003) Salicylic acid-inducedsuppression of Meloidogyne incognita infestation of okra and cowpea. Nematology 5:747–752
Nawrath C, Metraux JP (1999) Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11:1393–1404
Pieterse CM, Van Loon L (2004) NPR1: the spider in the web of induced resistance signaling pathways. Current Opin Plant Biol 7:456–464
Potlakayala SD, DeLong C, Sharpe A, Fobert PR (2007) Conservation of non-expressor of pathogenesis-related genes 1 function between Arabidopsis thaliana and Brassica napus. Physiol Mol Plant Pathol 71:174–183
Quilis J, Penas G, Messeguer J, Brugidou C, San Segundo B (2008) The Arabidopsis AtNPR1 inversely modulates defense responses against fungal, bacterial, or viral pathogens while conferring hypersensitivity to abiotic stresses in transgenic rice. Mol Plant-Microbe Interact 21:1215–1231
Rairdan GJ, Delaney TP (2002) Role of salicylic acid and NIM1/NPR1 in race-specific resistance in Arabidopsis. Genetics 161:803–811
Rajasekaran K, Cary JW, Jaynes JM, Cleveland TE (2005) Disease resistance conferred by the expression of a gene encoding a synthetic peptide in transgenic cotton (Gossypium hirsutum L.) plants. Plant Biotechnol J 3:545–554
Raskin I, Skubatz H, Tang W, Meeuse BJD (1990) Salicylic-acid levels in thermogenic and nonthermogenic plants. Ann Bot 66:369–373
Rathore KS, Sunilkumar G, Campbell LM (2006) Cotton (Gossypium hirsutum L.). Methods Mol Biol 343:267–279
Robinson AF, Heald CM (1991) Carbon-dioxide and temperature-gradients in baermann funnel extraction of Rotylenchulus-reniformis. J Nematology 23:28–38
Robinson AF, Cook CG, Percival AE (1999) Resistance to Rotylenchulus reniformis and Meloidogyne incognita race 3 in the major cotton cultivars planted since 1950. Crop Sci 39:850–858
Robinson AF, Bell AA, Dighe ND, Menz MA, Nichols PL, Stelly DM (2007) Introgression of resistance to nematode Rotylenchulus reniformis into upland cotton (Gossypium hirsutum) from Gossypium longicalyx. Crop Sci 47:1865–1877
Ryals J, Uknes S, Ward E (1994) Systemic acquired-resistance. Plant Physiol 104:1109–1112
Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner HY, Hunt MD (1996) Systemic acquired resistance. Plant Cell 8:1809–1819
Shah J (2003) The salicylic acid loop in plant defense. Curr Opin Plant Biol 6:365–371
Shah J, Tsui F, Klessig DF (1997) Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol Plant-Microbe Interact 10:69–78
Shah J, Kachroo P, Nandi A, Klessig DF (2001) A recessive mutation in the Arabidopsis SSI2 gene confers SA- and NPR1-independent expression of PR genes and resistance against bacterial and oomycete pathogens. Plant J 25:563–574
Silverman P, Seskar M, Kanter D, Schweizer P, Metraux JP, Raskin I (1995) Salicylic acid in rice: biosynthesis, conjugation, and possible role. Plant Physiol 108:633–639
Spoel SH, Koornneef A, Claessens SMC, Korzelius JP, Van Pelt JA, Mueller MJ, Buchala AJ, Métraux JP, Brown R, Kazan K, Van Loon LC, Dong X, Pieterse CMJ (2003) NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15:760–770
Spoel SH, Johnson JS, Dong X (2007) Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc Natl Acad Sci USA 104:18842–18847
Stipanovic RD, Bell AA, Benedict CR (1999) Cotton pest resistance: the role of pigment gland constituents. In: Cultler HG, Cultler SJ (eds) Biologically active natural products: agrochemicals. CRC Press, Boca Raton, pp 211–220
Sunilkumar G, Rathore KS (2001) Transgenic cotton: factors influencing Agrobacterium-mediated transformation and regeneration. Mol Breed 8:37–52
Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, Zuo J, Dong X (2008) Plant immunity requires conformational charges of NPR1 via S-nitrosylation and thioredoxins. Science 321:952–956
Timmermans MC, Maliga P, Vieira J, Messing J (1990) The pFF plasmids: cassettes utilising CaMV sequences for expression of foreign genes in plants. J Biotechnol 14:333–344
Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J (1992) Acquired resistance in Arabidopsis. Plant Cell 4:645–656
Wally O, Jayaraj J, Punja ZK (2009) Broad-spectrum disease resistance to necrotrophic and biotrophic pathogens in transgenic carrots (Daucus carota L.) expressing an Arabidopsis NPR1 gene. Planta. doi 10.1007/s00425-009-1031-2
Wang YQ, Chen DJ, Wang DM, Huang QS, Yao ZP, Liu FJ, Wei XW, Li RJ, Zhang ZN, Sun YR (2004) Over-expression of Gastrodia anti-fungal protein enhances Verticillium wilt resistance in coloured cotton. Plant Breed 123:454–459
Ward ER, Uknes SJ, Williams SC, Dincher SS, Wiederhold DL, Alexander DC, Ahl-Goy P, Metraux JP, Ryals JA (1991) Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3:1085–1094
Wubben MJE, Jin J, Baum TJ (2008) Cyst nematode parasitism of Arabidopsis thaliana is inhibited by salicylic acid (SA) and elicits uncoupled SA-independent pathogenesis-related gene expression in roots. Mol Plant-Microbe Interact 21:424–432
Yang Y, Qi M, Mei C (2004) Endogenous salicylic acid protects rice plants from oxidative damage caused by aging as well as biotic and abiotic stress. Plant J 40:909–919
Zhou JM, Trifa Y, Silva H, Pontier D, Lam E, Shah J, Klessig DF (2000) NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol Plant-Microbe Interact 13:191–202
Zhu YJ, Qiu XH, Moore PH, Borth W, Hu J, Ferreira S, Albert HH (2003) Systemic acquired resistance induced by BTH in papaya. Physiol Mol Plant Path 63:237–248
Acknowledgments
We thank Dr. Charles Kenerley for providing Rhizoctonia solani, and Alternaria alternata cultures that were used in this study. This research was supported by funds from Cotton Inc., Texas Higher Education Coordinating Board—Advanced Research Program (#000517-0005-2006), and Texas AgriLife Research.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Parkhi, V., Kumar, V., Campbell, L.M. et al. Resistance against various fungal pathogens and reniform nematode in transgenic cotton plants expressing Arabidopsis NPR1 . Transgenic Res 19, 959–975 (2010). https://doi.org/10.1007/s11248-010-9374-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-010-9374-9