Abstract
Three new reduced amino-acid Schiff base complexes, [Co(HL)2(H2O)2] · 4H2O (1), [Cu(HL)2(H2O)2] · 2H2O (2), and [Cd(HL)2(H2O)3] · 2H2O (3), where H2L is the reduced Schiff-base ligand derived from the condensation of N-(4-hydroxybenzaldehyde) with L-glycine, have been synthesized and characterized by physico-chemical and spectroscopic methods. In these complexes, the two bidentate monoanionic Schiff base ligands coordinate the metal center through the secondary amine N atom and the carboxylate O atom. Water ligands complete a distorted octahedral (1, 2) or a pentagonal bipyramidal coordination geometry (3) around each metal center. The binding interactions of the complexes with CT-DNA have been investigated by UV–visible spectrophotometry and fluorescence quenching methods. The results show that these complexes bind to CT-DNA with an intercalative mode. In addition, DNA cleavage experiments have been also investigated by agarose gel electrophoresis. Complexes 1–3 show oxidative DNA cleavage activity in the presence of H2O2/sodium ascorbate and the reactive oxygen species responsible for the DNA cleavage is most likely singlet oxygen.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The development of artificial nucleases is an important aspect of biotechnology and drug design [1, 2] as well as molecular biology [3]. Over the last few decades, transition metal complexes have been well studied for their application as artificial nucleases, because of their diverse structural features and the possibility to tune their redox potential through the choice of ligands [4–9]. Transition metal complexes containing Schiff base ligands or their reduced products are often used as artificial nucleases and some such complexes have proved to be efficient DNA cleavage reagents [10, 11]. For example, Neves et al. [10] have synthesized and structurally characterized mononuclear cis-dichloro copper(II) complexes containing a tridentate imidazole Schiff base or its reduced counterpart ligand. In aqueous solution, these complexes catalyze phosphodiester hydrolysis with about 100-fold enhancement over the rate of the spontaneous hydrolysis. Most importantly, the Schiff base complex is capable of promoting the cleavage of plasmid DNA in a pH dependent reaction, even in the absence of molecular dioxygen, most probably through a hydrolytic mechanism. Recently, Ferreira et al. [11] obtained some oxindole-Schiff base copper(II) complexes, all of which are able to cause double-strand DNA scission in the presence of hydrogen peroxide.
In this article, we describe a biologically relevant reduced amino-acid Schiff base (H2L = N-(4-hydroxybenzyl)-L-glycine) (Fig. 1) and three of its metal complexes. The interaction between calf thymus DNA and these complexes was investigated by UV spectrophotometry and fluorescence spectroscopy. DNA cleavage experiments in the presence of H2O2/sodium ascorbate are also described. L-Histidine inhibits the oxidative cleavage, suggesting that singlet oxygen is involved in the DNA degradation.
Experimental
All reagents and chemicals were purchased from commercial sources and used as received. Elemental analyses for C, H, and N were obtained on a Perkin-Elmer analyzer model 240. Infrared spectra were recorded on KBr pellets using a Perkin-Elmer FT-IR spectrometer in the range 4,000–400 cm−1. Electronic spectra were measured on a JASCO V-570 spectrophotometer. Fluorescence spectra were obtained on a MPF-4 fluorescence spectrophotometer at room temperature. Calf thymus DNA (CT-DNA), pBR322 DNA, agarose (molecular biology grade), and ethidium bromide (EB) were all purchased from the Sino-American Biotechnol Biotechnology Company. The Tris–HCl buffer solution was prepared using deionized, sonicated triply distilled water. The gel electrophoresis experiments were performed by incubation at 37 °C for 3 h as follows: pBR322 DNA, Co(II), Cu(II), and Cd(II) complexes, H2O2/sodium ascorbate in 50 mM Tris–HCl/18 mM NaCl buffer (pH 7.2). The samples were electrophoresed for 3 h at 70 V on 1% agarose gel using tris–boric acid-EDTA buffer, pH 7.2. After electrophoresis, the gel was stained using 1 mg/mL ethidium bromide and analyzed using a UVITEC automatic gel imaging system.
Synthesis of the reduced Schiff base
H2L was synthesized by reducing the corresponding Schiff base derived from the condensation of L-glycine and 4-hydroxybenzaldehyde with NaBH4 according to a literature method [12].
Synthesis of [Co(HL)2(H2O)2] · 4H2O (1)
A solution of H2L (0.2 mmol) and 0.4 mmol LiOH in EtOH:H2O (1:1 v/v, 10 mL) was added dropwise to a solution of 0.2 mmol CoCl2 · 6H2O in EtOH:H2O (1:1 v/v, 10 mL). After 24 h stirring, the solution was filtered and the filtrate was left in air at room temperature. After several weeks, pale-red well-shaped crystals suitable for X-ray diffraction were obtained. The crystals were collected by filtration, washed with Et2O and dried over silica gel (Yield: 43%). Found: (%) C, 41.2; H, 6.4; N, 5.4. Calcd. (%) for C18H32CoN2O12: C, 41.0; H, 6.1; N, 5.3. FT-IR (KBr phase): 1597, ν as(COO); 1384, ν s(COO); 3130, ν s(N–H); 3495 cm−1, ν s(H2O and/or OH).
Synthesis of [Cu(HL)2(H2O)2] · 2H2O (2) and [Cd(HL)2(H2O)3] · 2H2O (3)
Complexes 2 and 3 were prepared by similar procedures as for complex 1, using Cu(OAc)2 · H2O or Cd(NO3)2 · 9H2O instead of CoCl2 · 6H2O. Complex 2; Yield: 54%. Found: (%) C, 43.6; H, 5.8; N, 5.5. Calcd.(%) for C18H28CuN2O10: C, 43.6; H, 5.7; N, 5.7. FT-IR (KBr phase): 1610, ν as(COO); 1405, ν s(COO); 3200, ν s(N–H) and 3495 cm−1, ν s(H2O and/or OH). Complex 3; Yield: 60%. Found:(%) C, 38.4; H, 5.2; N, 4.9. Calcd.(%) for C18H30CdN2O11: C, 38.4; H, 5.4; N, 5.0. FT-IR (KBr phase): 1590, ν as(COO); 1382, ν s(COO); 3120, ν s(N–H) and 3505 cm−1, ν s(H2O and/or OH).
X-ray crystallography
Diffraction measurements for 1, 2, and 3 were made on a Bruker Smart 1000 CCD area detector equipped with graphite-monochromated Mo-Kα radiation (λ = 0.71073 Å) using the ω-scan technique. Lorentz polarization and absorption corrections were applied using the multiscan program [13]. The structures were solved by direct methods and refined with full-matrix least-squares technique using the SHELXS-97 and SHELXL-97 programs [14]. Anisotropic thermal parameters were assigned to all non-hydrogen atoms. The organic hydrogen atoms were generated geometrically. Analytical expressions of neutral atom scattering factors were employed, and anomalous dispersions were incorporated. A summary of the crystal data is given in Table 1 and selected bond angles and distances are listed in Table 2. Crystallographic data for 1 and 3 have been deposited with the Cambridge Crystallographic Data Centre with CCDC numbers 634151 (1), 672075 (3). Copies of this information may be obtained free of charge from the Director, CCDC, 12 Union Road, Cambridge, CB2 1EZ, UK (fax: +44 1223 336 033; e-mail: deposit@ccdc.cam.ac.uk or www:http://www.ccdc.cam.ac.uk).
DNA-binding and cleavage experiments
All studies on the interaction of the complexes with calf thymus DNA (CT-DNA) were carried out at room temperature in triply distilled water buffer containing 5 mM Tris–HCl/50 mM NaCl and adjusted to pH 7.2 with hydrochloric acid. Relative binding of the complexes to CT-DNA was studied by UV–visible absorption and fluorescence spectroscopy. The solutions of CT-DNA gave a ratio of UV absorbance at 260 and 280 nm, A 260/A 280, of 1.8–1.9, indicating that the DNA was sufficiently free of protein [15]. The stock solution of CT-DNA was prepared in Tris–HCl/NaCl buffer, pH 7.2 (stored at 4 °C and used within 4 days). The concentration of CT-DNA was determined by absorption spectroscopy using the known molar extinction coefficient of 6,600 M−1 cm−1 at 260 nm [16]. UV absorption spectroscopy experiments were conducted by adding CT-DNA solution to solutions of the complexes (1.5 × 10−4 M) at different concentrations. The binding constant K b was determined using the following equation [17]:
Here εa, εf, and εb correspond to A obsd/[complex], the extinction coefficient for the free complex, and the extinction coefficient for the complex in the fully bound form, respectively.
The relative binding of the three complexes to CT-DNA was studied with an EB-bound CT-DNA solution in 5 mM Tris–HCl/NaCl buffer (pH 7.2) by fluorescence spectrophotometry. Fluorescence intensities at 610 nm (510 nm excitation) were measured at different complex concetrations. The emission intensity showed a reduction upon addition of the complex. The relative binding propensity of the complexes to CT-DNA was determined from the slopes of straight lines obtained from plots of the fluorescence intensity versus the complex concentration [18]. The apparent binding constant (k app) was calculated from the equation:
where the complex concentration was the value at a 50% reduction of the fluorescence intensity of EB and K EB = 1.0 × 107 M−1 ([EB] = 4.0 μM).
Oxidative cleavage of supercoiled (SC) pBR322 DNA by the complexes was studied by agarose gel electrophoresis. The reaction was carried out by mixing 4 μL SC DNA (0.1 μg μL−1, 16.5 μM), 8 μL of the complex solution (120 μM), 2 μL 50 mM tris–(hydroxymethylmethane-HCl (Tris–HCl) buffer (pH 7.2) containing 18 mM NaCl with 1 μL H2O2/1 μL sodium ascorbate to yield a total volume of 16 μL. The sample was incubated at 37 °C, followed by the adddition of loading buffer containing 0.25% bromphenol blue, 50% glycerol, 0.61% Tris, and the solution was finally loaded on 1% agarose gel containing 1.0 μg mL−1 ethidium bromide. Electrophoresis was carried out for 3 h at 70 V in TBE buffer (45 mM Tris, 45 mM H3BO3, 1 mM EDTA, pH 8.0). Bands were visualized by UV light and photographed. The extent of cleavage of the SC DNA was determined by measuring the intensities of the bands using the Gel Documentation System [19, 20]. The mechanistic investigation of the cleavage of pBR322 DNA was carried out in the presence of standard radical scavengers and reaction inhibitors. These reactions were carried out by adding scavengers of dimethyl sulfoxide (DMSO), SOD, EDTA, or histidine to SC DNA. Cleavage was initiated by the addition of complex and quenched with 2 μL of loading buffer. Further analysis was carried out by the above standard method.
Results and discussion
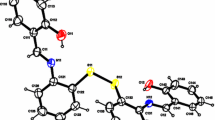
The structures of 1 and 2 are similar and consist of neutral, mononuclear [M(HL)2(H2O)2] (M = Co for 1, Cu for 2) units and four or two lattice water molecules for 1 and 2, respectively. The ORTEP drawings of the mononuclear unit of 1 and 2 are shown in Figs. 2 and 3, respectively. For each mononuclear unit, the central metal atom is located in the symmetric center. HL− has a more flexible backbone than its Schiff base counterpart and serves as a bidentate monoanionic ligand to coordinate the central metal atom. The metal atom is coordinated to two HL− and two water molecules, giving a distorted octahedral geometry with a N2O4 donor set. The cis coordination angles vary from 79.02 to 100.98° for 1 and 81.1 to 98.9° for 2. The M–N and M–O bond distances are within the ranges expected for such species. In complex 2, the relatively short equatorial (Cu1–O2 and Cu1–O2 Å, 1.976 Å) and long axial (Cu1–O4 and Cu1–O4 Å, 2.4007 Å) distances are due to Jahn–Teller distortion of the six-coordinate CuII. From Fig. 4, we see that a hydrogen bonding network exists in the crystals of complex 1, there are complicated intermolecular hydrogen bonds among the water ligands, solvent water molecules, and carboxyl oxygen atoms, giving rise to a 2D hydrogen bonding network in the ab plane. In addition, the structure adopts a 3D hydrogen bonding pattern involving all donors and acceptors.
An obvious difference between 3 and 1 or 2 is the number of coordinated water molecules. In 3, three water molecules (O4, O4A, and O5) coordinate the cadmium(II) center, as shown in Fig. 5, while only two water ligands are present for 1 or 2. Complex 3 crystallizes in the monoclinic system with space group C2/c and has crystallographically imposed C2 symmetry, passing through the center of Cd1 and O5. Each cadmium atom is seven-coordinated with an N2O5 donor set and its coordination polyhedron can be regarded as a highly distorted pentagonal-bipyramid. Cadmium usually has a coordination number between 4 and 7. In 3, the pentagonal plane is composed of atoms of O4, O4A, O3, O3A, and O5 and the axial positions are occupied by atoms N1 and N1A, with the axial bond angle of 165.9º, the degree of distortion from the ideal pentagonal bipyramidal geometry is also reflected in the angles around the Cd atom in the equatorial plane (See Table 2; ideal value 72º).
DNA binding studies
DNA binding is the critical step for DNA cleavage in most cases. Therefore, the binding of complexes 1, 2, and 3 to CT-DNA was studied by measuring their effects on the UV and fluorescence spectra of CT-DNA.
As shown in supplementary Fig. 1, the potential CT-DNA binding ability of the complexes was studied by following the intensity changes of the intraligand π–π* transition bands in the UV spectrum at 218–267 nm. Upon addition of increasing amounts of CT-DNA (0–7.9 × 10−4 M) to the complexes (1.5 × 10−4 M), 9–33% hypochromism and a slight red shift (3–8 nm) were observed, indicating moderate binding of the three complexes to DNA. The extent of hypochromism is consistent with intercalative interaction [21–23]. From the observed spectroscopic changes the values of the intrinsic binding constants K b (3.75 × 103 M−1 for 1, 3.93 × 103 M−1 for 2, and 3.00 × 103 M−1 for 3) were determined by regression analysis using Eq. 1. These K b values are much smaller than those reported for typical classical intercalators (EB-DNA, 3.3 × 105 M−1 in 50 mM Tris–HCl/1.0 M NaCl buffer, pH 7.5) [24].
As a means for further clarifying the binding of these complexes, fluorescence measurements were carried out on CT-DNA at variable complex concentrations. The binding of the compounds to CT-DNA was evaluated by the fluorescence emission intensity of EB bound to DNA as a probe. EB shows reduced emission intensity in buffer because of quenching by solvent molecules and a significant enhancement of the intensity when bound to DNA. Binding of the complexes to DNA decreases the emission intensity and the extent of the reduction gives a measure of the DNA binding propensity of the complexes and intercalation between the adjacent DNA base pairs [25]. The fluorescence quenching of EB bound to DNA by the three complexes is shown in suplementary Fig. 2, in which the fluorescence intensity at 610 nm (λex = 510 nm) of EB in the bound form is plotted against the concentration of complexes. According to the Stern-Volmer equation [26]:
where I 0 and I are the fluorescence intensities in the absence and presence of the quencher, respectively, K is the Stern-Volmer quenching constant, and [Q] is the concentration of the quencher. Plots of I 0/I versus [complex] gave the apparent binding constant (K app) as the ratio of the slope to intercept, which corresponds to the complex concentration [Q] = 1.5 × 10−4 M required for 50% quenching of initial EB fluorescence. The apparent binding constants for complexes 1, 2, and 3 are 3.88 × 104, 4.13 × 104, and 2.81 × 104 M−1, respectively. These results suggest that the interaction of the complexes with DNA is by a moderate intercalative mode. The binding constants of classical interactions are in the order of 107 M−1 [27].
Nuclease activity of the complexes
The ability of the complexes to mediate DNA cleavage was assayed using agarose gel electrophoresis at physiological pH and temperature. When supercoiled circular pBR322 DNA is subjected to electrophoresis, relatively fast migration will be observed for the intact supercoiled form (Form I). If scission occurs on one strand, the supercoiled form will relax to generate a slower-moving open nicked form (Form II), and if both strands are cleaved, a linear form (Form III) that migrates between Form I and Form II will be observed.
Figures 6, 7, and 8 show the gel electrophoretic separations of plasmid pBR322 DNA induced by complexes 1, 2 and 3. All three complexes promote oxidative damage of DNA under physiological conditions (pH 7.2, 37 °C), but with different cleavage activities. The Co(II) and Cd(II) complexes exhibit DNA cleavage activities in the presence of H2O2/sodium ascorbate; however, the Cu(II) complex displays higher DNA damage activity with or without H2O2/sodium ascorbate. As shown in Figs. 6, 7a, and 8a, the DNA cleavage activities of the complexes are obviously concentration-dependent. With the increase of complex concentration, the supercoiled DNA decreases and nicked circular DNA gradually increases. In order to assess whether reactive oxygen species, such as singlet oxygen and hydroxyl radical, were involved in DNA cleavage, several radical scavengers were examined. As shown in the figures, the experimental data indicate that the hydroxyl radical can be ruled out in the DNA cleavage reactions, and singlet oxygen is therefore likely to be the reactive species.
Agarose gel electrophoresis diagram showing the cleavage of pBR322DNA (33 μM) by complex 1 treated with H2O2/sodium ascorbate in Tris–HCl/NaCl buffer (pH 7.2) and incubated for 3 h at 37 °C. Lane 1, DNA control; lane 2, DNA + 1 (33 μM); lane 3, DNA + H2O2/sodium ascorbate (25 μM); lane 4–7, DNA + H2O2/sodium ascorbate + 1 (11, 22, 27.5, 33 μM); lane 8–11: DNA + H2O2/sodium ascorbate + 1 (33 μM) + [DMSO (60 μM); SOD (60 μM); L-Histidine (20 U/mL); EDTA (60 μM), respectively]
a Gel electrophoresis diagrams showing the cleavage of pBR322 DNA (33 μM) by 2 in Tris–HCl/NaCl buffer (pH 7.2) and incubated for 3 h at 37 °C. Lane 1: DNA control; Lane 2–6: DNA + 2 (5; 10; 35; 55; 80 μM). b Lane 1: DNA control; lane 2: DNA + 2 (35 μM); lane 3–6: DNA + 2 (35 μM) + [DMSO (60 μM); SOD (60 μM); L-Histidine (20 U/mL); EDTA (60 μM), respectively]
a Gel electrophoresis diagrams showing the cleavage of pBR322 DNA (33 μM) by 3 in Tris–HCl/NaCl buffer (pH 7.2) and incubated for 3 h at 37 °C. Lane 1: DNA control; Lane 2: DNA + 3 (10 μM); Lane 3–7: DNA + H2O2/sodium ascorbate (25 μM) + 3 (10, 40, 70, 100, 120 μM). b Lane 1, DNA control; lane 2, DNA + 3 (40 μM); lane 3, DNA + H2O2/sodium ascorbate (25 μM); lane 4, DNA + H2O2/sodium ascorbate (25 μM) + 3 (40 μM); lane 5–8, DNA + H2O2/sodium ascorbate (25 μM) + 3 (40 μM) + [DMSO (60 μM); SOD (20 U/mL); L-Histidine (60 μM); EDTA (60 μM), respectively]
Conclusions
Three new complexes of a reduced amino acid Schiff base ligand have been synthesized and structurally characterized. The CT-DNA binding abilities and supercoiled plasmid DNA cleavage activities of the complexes have been studied. The results suggest that all three complexes bind to CT-DNA by an intercalative mode. The agarose gel electrophoresis studies show that all of these complexes can promote the oxidative cleavage of plasmid DNA at physiological pH and temperature in the presence of H2O2/sodium ascorbate, but the Cu(II) complex can also damage plasmid DNA in air (in the absence of H2O2/sodium ascorbate). Our investigation of the DNA cleavage mechanism suggests that singlet oxygen is the reactive oxygen species that leads to DNA cleavage.
References
Fernandez MJ, Wilson B, Palacios M, Rodrigo MM, Grant KB, Lorente A (2007) Bioconjugate Chem 18:121. doi:10.1021/bc0601828
Jin Y, Cowan JA (2005) J Am Chem Soc 127:8408. doi:10.1021/ja0503985
Pitlie M, Croisy A, Carrez D, Boldron C, Meunier B (2005) BioChem 6:686
Maheswari PU, Roy S, den Dulk H, Barends S, van Wezel G, Kozlevcar B, Gamez P, Reedijk J (2006) J Am Chem Soc 128:710. doi:10.1021/ja056970+
Maheswari PU, Lapplainen K, Sfregola M, Barends S, Gamez P, Turpeinen U, Mutikainen I, Weze G, Reedijk J (2007) Dalton Trans 33:3676. doi:10.1039/b704390b
Qian J, Gu W, Liu H, Gao F-X, Feng L, Yan S-P, Liao D-Z, Cheng P (2007) Dalton Trans 11:1060. doi:10.1039/b615148e
Roy M, Pathak B, Patra AK, Jemmis ED, Nethaji M, Chakravarty AR (2007) Inorg Chem 46:11122. doi:10.1021/ic701450a
An Y, Tong M-L, Ji L-N, Mao Z-W (2006) Dalton Trans 16:2006
Tian J-L, Feng L, Gu W, Xu G-J, Yan S-P, Liao D-Z, Jiang Z-H, Cheng P (2007) Inorg Biochem 101:96. doi:10.1016/j.jinorgbio.2006.08.009
Tullius TD, Greenbaum JA (2005) Curr Opin Chem Biol 9:127. doi:10.1016/j.cbpa.2005.02.009
D’Alelio GF, Hofman ET, Zeman JR (1969) J Macromolecular Science Chemistry 3:959. doi:10.1080/10601326908051927
Alam MA, Nethaji M, Ray M (2003) Angew Chem Int Ed Engl 42:1940. doi:10.1002/anie.200250591
Sheldrick GM (2002) SADABS 2.05. University of Gottingen, Germany
SHELXTL 6.10, Bruker analytical Instrumentation. Madison, WI, USA, 2000
Marmur J (1961) J Mol Biol 3:208
Reichmann ME, Rice SA, Thomas CA, Doty P (1954) J Am Chem Soc 76:3047. doi:10.1021/ja01640a067
Wolf A, Shimer GH Jr, Meehan T (1987) Biochem 26:6392. doi:10.1021/bi00394a013
Lee M, Rhods AL, Wyatt MD, Forrow S, Hartley JA (1993) Biochem 28:7268
Bermadou J, Pratviel G, Bennis F, Girardet M, Meunier B (1989) Biochem 28:7268. doi:10.1021/bi00444a019
Pamatong FV, Detmer CA, Bocarsly JR (1996) J Am Chem Soc 118:5339. doi:10.1021/ja953282p
Barton JK, Danishefsky AT, Goldberg JM (1984) J Am Chem Soc 106:2172. doi:10.1021/ja00319a043
Kelly JM, Tossi AB, McConnel DJ, Ohuigin C (1985) Nucleic Acid Res 13:6017. doi:10.1093/nar/13.17.6017
Tysoe SA, Baker AD, Strekees TC (1993) J Phys Chem 97(B):1707
Strothkamp KG, Strothkamp RE (1994) J Chem Educ 71:77
Lepecq JB, Paoletti C (1967) J Mol Biol 27:87. doi:10.1016/0022-2836(67)90353-1
Marsh WE, hatfeild WE, Hodson DJ (1982) Inorg Chem 21:2679. doi:10.1021/ic00137a029
Cory M, Mckee DD, Kagan J, Henry DW, Muller JA (1985) J Am Chem Soc 107:2528. doi:10.1021/ja00294a054
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 20771063).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
These files are unfortunately not in the Publisher's archive anymore:
-
Supplementary material 2 (CIF 14 kb)
-
Supplementary material 3 (CIF 14 kb)
-
Supplementary material 4 (CIF 14 kb)
Rights and permissions
About this article
Cite this article
Ma, XF., Li, DD., Tian, JL. et al. DNA binding and cleavage activity of reduced amino-acid Schiff base complexes of cobalt(II), copper(II), and cadmium(II). Transition Met Chem 34, 475–481 (2009). https://doi.org/10.1007/s11243-009-9219-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11243-009-9219-7