Abstract
In this paper, we derive a pore-scale model for permeable biofilm formation in a two-dimensional pore. The pore is divided into two phases: water and biofilm. The biofilm is assumed to consist of four components: water, extracellular polymeric substance (EPS), active bacteria, and dead bacteria. The flow of water is modeled by the Stokes equation, whereas a diffusion–convection equation is involved for the transport of nutrients. At the biofilm–water interface, nutrient transport and shear forces due to the water flux are considered. In the biofilm, the Brinkman equation for the water flow, transport of nutrients due to diffusion and convection, displacement of the biofilm components due to reproduction/death of bacteria, and production of EPS are considered. A segregated finite element algorithm is used to solve the mathematical equations. Numerical simulations are performed based on experimentally determined parameters. The stress coefficient is fitted to the experimental data. To identify the critical model parameters, a sensitivity analysis is performed. The Sobol sensitivity indices of the input parameters are computed based on uniform perturbation by ± 10% of the nominal parameter values. The sensitivity analysis confirms that the variability or uncertainty in none of the parameters should be neglected.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
A biofilm can be defined as an aggregation of bacteria, algae, fungi, and protozoa enclosed in a matrix consisting of a mixture of polymeric compounds, primarily polysaccharides, generally referred to as extracellular polymeric substance (EPS) (Vu et al. 2009). Biofilms produce biomass, polymers, acids, gases, solvents, and biosurfactants (Sen 2008). Biofilm formation is generally established through three steps (Toyofuko et al. 2015): planktonic cell attachment to the surface, formation of a structured architecture with the assistance of EPS in the maturation stage, and cells leaving the biofilm in the dispersal stage. Different environmental factors affect the biofilm, e.g., temperature and pH (Hoštacká et al. 2010). Biofilms are complex systems, involving different physical, chemical, and biological processes such as bacterial reproduction and decay, attachment and detachment, formation of metabolites, endogenous respiration, erosion, sloughing, and abrasion. Biofilms are present in many systems, with detrimental effect in some cases, but also beneficial applications in some areas, for example in medicine, food industry, and water quality (Parsek and Fuqua 2004; Kokare et al. 2009; Boltz et al. 2017).
In our research, we are interested in studying the biofilm to improve the oil extraction. Microbial enhanced oil recovery (MEOR) is an enhanced oil recovery method relying on microorganisms and their metabolic products to mobilize residual oil in a cost-effective and eco-friendly manner. The particular MEOR mechanism that we are concerned with in this work is called microbial selective plugging. The mechanism consists of growing bacteria in high permeable zones in a reservoir and thereby clogging the preferential water flow paths. Consequently, the water will be forced to flow in new pores and more oil will be recovered. In the current paper, we focus on stimulating endogenous bacterial growth; therefore, only nutrients are injected. MEOR is not yet completely understood, and there is a strong need for mathematical models to be used for improving this technology.
The percentage of water in biofilms constitutes up to 97% (Ahmad and Husain 2017). In the biofilm, cell clusters may be separated by interstitial voids and channels, which create a characteristic porous structure (Wood and Whitaker 1998; Picioreanu et al. 2000). The pore radius in biofilms is of order \(\upmu \mathrm{m}\) (Zhang and Bishop 1994), which is of the same order of pore radius in tight sandstones (Cao et al. 2016). The proportion of EPS in biofilms can comprise approximately 50–90\(\%\) of the total organic matter (Donlan 2002; Vu et al. 2009). Flow velocity near a biofilm changes from a maximum in the bulk solution to zero at the bottom of the biofilm (Lewandowski and Beyenal 2003). In different biofilms, the mechanism of nutrient transport near the biofilm surface and within the biofilm can be dominated by convection or diffusion (Schwarzenbach 1993; Stewart 2003).
Most of the biofilm models are based on simplifying assumptions, e.g., impermeability, a constant biofilm density, and accounting for diffusion but neglecting convection for transport of nutrients (Picioreanu et al. 1998; Duddu et al. 2009; Schulz and Knabner 2016; Tang and Liu 2017). Novel mathematical models must be built to improve accuracy and enhance confidence in numerical results. In Landa-Marbán et al. (2017), a mathematical model for MEOR including the oil–water interfacial area was built. Pore-scale models are used to derive parameters and functional relationships for the core-scale models (van Noorden et al. 2010; Ray et al. 2013; Bringedal et al. 2016; Bringedal and Kumar 2017). In this work, we propose a pore-scale model for a permeable multi-component biofilm including a variable biofilm density, detachment, and transport of nutrients due to convection and diffusion.
The resulting mathematical model involves coupled partial differential equations. Further, the biofilm–water interface location changes over time and therefore is a free boundary problem. Numerical methods for solving free boundary problems are an active research field (Esmaili and Eslahchi 2017; Gallinato and Poignard 2017). The arbitrary Lagrangian–Eulerian (ALE) method is used to track the position of the biofilm–water interface (Donea et al. 2004). In biofilm and reactive flow modeling involving free boundary, it is common to use decoupling techniques to find a numerical solution (Alpkvist and Klapper 2007; Peszynska et al. 2016; Kumar et al. 2013). In our case, a segregated finite element algorithm is used to solve the mathematical equations.
Pore-scale models are important because they aim to describe some of the physical phenomena in detail and one can derive core-scale models through upscaling. Two of the motivations to derive upscaled models are to determine constitutive relationships and to describe the average behavior of the system in an accurate manner with relatively low computational effort compared to fully detailed calculations starting at the microscale (van Noorden et al. 2010).
Due to the cost of performing laboratory experiments to accurately estimate material parameter values, it is of great interest to perform a sensitivity study with respect to the impact of a set of input parameters on certain model output quantities of interest. This ensures that critical parameters are identified. Moreover, for parameters scoring low in sensitivity estimates, less accurate parameter estimates can be justified. Global sensitivity analysis using Sobol indices is a means of quantifying the relative impact of a function of interest in terms of a set of varying input parameters (Sobol 2001). This is computationally prohibitive for problems with a large number of input parameters, but the computational cost can be significantly reduced by computing the Sobol indices using the generalized polynomial chaos framework (Xiu and Karniadakis 2002; Sudret 2008).
In this general context, the objective of the present article is to develop and implement an accurate numerical simulator for biofilm formation.
To summarize, the new contributions of this work are:
-
the development of a multidimensional, comprehensive pore-scale mathematical model for biofilm formation,
-
the inclusion of a biofilm porosity,
-
the inclusion of nutrient transport inside the biofilm due to convection and diffusion, and
-
the calibration of the mathematical model with the laboratory experiments.
We emphasize that the model development here is performed in close relationship with the physical experimental observations. It is through the experiments that we identify the key processes and variables that need to be considered. Accordingly, we compute some of the parameters (but not all due to the limited experimental observations) of the mathematical model through calibration. Finally, we study the sensitivity of the parameters in our model.
The paper is structured as follows. The pore-scale model is defined in Sect. 2, where we introduce the basic concepts, ideas, and equations for modeling biofilms in the pore-scale. In Sect. 3, we describe the computational algorithm to solve numerically the model. In Sect. 4, we present numerical results for some of the unknown model variables using the best available estimates for the input parameters. We perform a sensitivity analysis in Sect. 5 in order to detect the critical model parameters. Finally, in Sect. 6 we present the conclusions.
2 Pore-Scale Model
The modeling of biofilms has been done on different scales and for different metabolic processes. In van Noorden et al. (2010), a pore-scale model for biofilm formation considering the biofilm as impermeable and formed by a single species is built. In Alpkvist and Klapper (2007), a model for heterogeneous biofilm development considering the biofilm formed by different components is built. In this work, we extend these ideas to build a model for biofilm formation which includes the notions of porosity and permeability.
We assume the following:
-
(A1)
The biofilm is a separate phase (as being a porous medium itself), which is modeled by mass conservation and a growth potential.
-
(A2)
The biofilm is modeled as a continuous medium consisting of four components: water, EPS, active bacteria, and dead bacteria.
-
(A3)
The fluid flow and nutrients are in a steady state when we compute the biofilm growth potential and volume fractions at each time step.
-
(A4)
The biofilm growth occurs in the lower substratum.
-
(A5)
There is only one nutrient, which is mobile in both the water and biofilm.
-
(A6)
Temperature is constant (room temperature).
-
(A7)
The gravity effects are neglected.
-
(A8)
The bacterial growth rate is of Monod type (Monod 1949), and the bacterial decay is linear (Bitton 2005).
We comment on these assumptions. Following Alpkvist and Klapper (2007), we need (A1) in order to properly model the dynamics of the biofilm components. Other approaches to model biofilms include cellular automata and individual-based biofilm models (Horn and Lackner 2014). The motivation for (A3) is that the fluid mean flow and biofilm growth velocities are of orders mm/s and \(10^{-5}\) mm/s, respectively (Duddu et al. 2009). Then, water and nutrient displacements due to biofilm growth are neglected (van Noorden et al. 2010). We consider (A4) because a T-microchannel is used to grow the biofilm, where bacteria and nutrients are first injected in the vertical channel and, afterward, only nutrients are injected through the horizontal channel, leading to a greater growth of bacteria on the lower substrate (where the horizontal and vertical channels connect). (A5) is made for simplicity, but the extension to other nutrients can be achieved straightforwardly from the model equations. The experiments were performed at room temperature, hence (A6). We consider (A7) because in the experimental setting, the gravity direction is perpendicular to the plane where the biofilm grows. Due to the lack of data on the studied system, we only consider the two processes in (A8). We refer to Murphy and Ginn (2000) for a quantitative representation of additional biological processes, such as endogenous respiration, metabolic lag, and chemotaxis. We remark that unlike in simple biofilm models, we do not assume a constant density of the biofilm.
2.1 Geometrical Settings
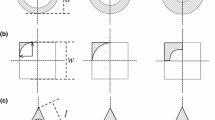
We consider a two-dimensional pore of length L and width W:
We have simplified the geometry in order to focus on the complex processes and upscale the model equations. Furthermore, microbial activity often results in preferential flow paths (Seki et al. 2006; Rubol et al. 2014); therefore, we can approximate a porous medium in the macroscale as a bundle of tubes (van Noorden et al. 2010). Figure 1 shows the water and biofilm domains and boundaries in the pore.
The boundary of the pore consists of the substrate, the inflow, and the outflow:
The domain of the pore consists of the biofilm and the water phase
where d(x, t) is the biofilm thicknesses.
The interface between the water and biofilm phases is denoted by \(\varGamma _\mathrm{wb}(t)\), which mathematically is given by
The inflow and outflow boundaries for the water domain \(\varOmega _\mathrm{w}(t)\) are given by
while the inflow and outflow boundaries for the biofilm domain \(\varOmega _\mathrm{b}(t)\) are given by
The unit normal pointing into the biofilm and the tangential vector are given by
2.2 Equations in the Water Phase
In the water phase \(\varOmega _\mathrm{w}(t)\), equations to describe the water flux and transport of nutrients are needed. The water is assumed to be incompressible. The water flow is described by the Stokes system
where \(\mu \) is the viscosity, \(p_\mathrm{w}\) is the water pressure, and \(\mathbf{q }_\mathrm{w}=(q_\mathrm{w}^{(1)},q_\mathrm{w}^{(2)})\) is the water velocity.
In the water phase, the nutrient concentration (\(c_\mathrm{w}\)) satisfies the convection–diffusion equation
where D and \(\pmb {J}_\mathrm{w}\) are the nutrient diffusion coefficient and nutrient flux in water, respectively.
2.3 Equations in the Biofilm Phase
As mentioned before, the biofilm components are water, EPS, active bacteria, and dead bacteria (\(j=\lbrace w,e,a,d\rbrace \)). Let \(\theta _j(t,\mathbf{x })\) and \(\rho _j(t,\mathbf{x })\) denote the volume fraction and the density (relative to volume fraction) of species j at time t and position \(\mathbf{x }\), respectively. The biomass and water are assumed incompressible (\(\rho _j(t,\mathbf{x })=\rho _j\)). Therefore, the biofilm density in a given position and time is
The volume fractions are constrained to
In the biofilm, the biomass can increase or decrease due to EPS production, bacterial reproduction, and bacterial decay. Let \(\mathbf{u }\) be the velocity of the biomass. Assuming that the biofilm growth is irrotational (Duddu et al. 2009), we can derive the velocity field from a function potential \(\varPhi \),
In Hornung (1997) Chapter 3, the Brinkman model is derived as the Darcy-scale counterpart of the Stokes model at the scale of pores, assuming that the volume of the porous media skeleton is much smaller than the volume of the reference cell. Therefore, recalling that biofilms are mostly water, we assume that the water content is constant (\(\partial _t\theta _\mathrm{w}=0\)) and we describe the water flux in the biofilm by the mass conservation and the Brinkman equation
where \(\mathbf{q }_\mathrm{b}\) and \(p_\mathrm{b}\) are the velocity and pressure of the water in the biofilm, respectively, and k is the permeability.
The conservation of mass for the biomass components (\(l=\lbrace e,a,d\rbrace \)) is given by
where \(R_l\) are the rates on the volume fractions; these rates are discussed in more detail below.
Inside the biofilm, the nutrients are dissolved in the water. The nutrient concentrations satisfy the following convection–diffusion–reaction equations:
where \(c_\mathrm{b}\), \(R_\mathrm{b}\), and \(\pmb {J}_\mathrm{b}\) are the nutrient concentration, reaction term, and flux in the biofilm.
Following Alpkvist and Klapper (2007), summing Eq. 2 over l and using Eq. 1, since \(\theta _\mathrm{w}\) and \(\rho _l\) (for all l) are constants, the growth velocity potential satisfies
2.4 Equations at the Biofilm–Water Interface
Coupling conditions for free flow and flow in porous media is an active research topic, and there are several works that study this problem. One of the first works on this topic was done by Beavers and Joseph (1967), where they proposed an empirical slip-flow condition based on experimental results. Saffman (1971) proposed a modification of the Beavers–Joseph condition based on the finding that the tangential velocity at the interface is proportional to the shear stress. Such results are made mathematically rigorous by Mikelić and Jäger (2000). We refer to Urquiza et al. (2008), Dumitrache and Petrache (2012), and Yang et al. (2017) for an extended description of coupling conditions for free flow and flow in porous media.
We assume that the normal velocity of the interface between the biofilm and fluid is negligible with respect to the velocity of the fluid phase (van Noorden et al. 2010). Then, we choose conditions of continuous velocity and continuity of the normal component of the stress tensor (Dumitrache and Petrache 2012)
Conservation of nutrients is ensured by the Rankine–Hugoniot condition,
The nutrient concentration is assumed continuous across the interface:
We set the growth velocity potential at the interface to zero:
The location of the interface \(\varGamma _\mathrm{wb}(t)\) changes in time due to the production of EPS, active bacteria, bacterial decay, and shear stress produced by the water flux. The review of modeling of biofilm systems in Horn and Lackner (2014) includes a summary of different detachment models. We consider only erosion as a detachment mechanism, i.e., the detachment of biomass due to shear forces at the interface produced by water flow. To incorporate this, we follow Taylor and Jaffe (1990) and van Noorden et al. (2010) and use the following definition for the tangential shear stress:
where \(\left\| \cdot \right\| \) denotes the maximum norm. Then, the normal velocity of the interface is given by
where \(k_\mathrm{srt}\) is a constant for the shear stress. In the above, we ensure that the interface does not cross the strip by taking the positive cut on the right-hand side when \(d=W\), which means that only bacterial decay would lead to the biofilm thickness to decrease. Following van Noorden et al. (2010), the evolution equation for the biofilm thickness reads as
Finally, homogeneous Neumann condition is considered for the biomass components:
2.5 Boundary and Initial Conditions
Boundary and initial conditions are needed to close the system of equations and to have a unique solution. As water is injected, we need to specify either the flux or the pressure at the inflow and outflow. The injected nutrient concentration is kept constant during the experiments; therefore, the nutrient concentration at the inflow is given. The upper and lower boundaries are closed; therefore, no-flux boundary conditions for the water and nutrients are considered.
At the inflow, we specify the pressure and nutrient concentration and we consider homogeneous Neumann condition for the growth velocity potential and volume fractions:
At the outflow, we specify the pressure and we consider Neumann conditions for the concentrations, growth velocity potential, and volume fractions:
At the lower substrate, we consider a no-flux boundary condition for the water, nutrients and volume fractions, and homogeneous Neumann condition for the growth potential:
At the upper substrate, we consider a no-slip boundary condition for the free flow and no-flux for the nutrient concentration:
The initial pressure, nutrient concentrations, growth potential, biofilm height, and volume fractions are given.
2.6 Reaction Terms
The bacteria need to consume nutrients in order to produce EPS and for reproduction. We model the bacterial reproduction using a Monod-type function (Monod 1949). Also, we consider a linear bacterial decay (Bitton 2005). Then, we have the following reaction terms:
where \(Y_\mathrm{e}\) and \(Y_\mathrm{a}\) are yield coefficients, \(\mu _n\) is the maximum rate of nutrient utilization, \(k_n\) is the Monod half-velocity coefficient, and \(k_{\text {res}}\) is the bacterial decay rate. For simplicity, these coefficients are considered as constants. However, processes such as dynamics of energy allotment lead to non-constant yield coefficients and metabolic lag.
2.7 Pore-Scale Model for Permeable Biofilm
For increasing the readability of the paper, we summarize here the mathematical model for permeable biofilm:
Water flow | ||
Stokes equations | \(\nabla \cdot \mathbf{q }_\mathrm{w}=0\quad \mu \varDelta \mathbf{q }_\mathrm{w}=\nabla p_\mathrm{w}\) | \(\varOmega _\mathrm{w}(t)\). |
Continuity velocities | \(\mathbf{q }_\mathrm{w}=\mathbf{q }_\mathrm{b}\) | \(\varGamma _\mathrm{wb}(t)\). |
Continuity stress tensor | \(\pmb {\nu }\cdot (\mu \nabla \mathbf {q}_\mathrm{w}-\mathbb {1}p_\mathrm{w})=\pmb {\nu }\cdot ((\mu /\theta _\mathrm{w})\nabla \mathbf {q}_\mathrm{b}-\mathbb {1}p_\mathrm{b})\) | \(\varGamma _\mathrm{wb}(t)\). |
Brinkman equations | \(\nabla \cdot \mathbf{q }_\mathrm{b}=0\quad (\mu /\theta _\mathrm{w})\nabla \mathbf {q}_\mathrm{b}-(\mu /k)\mathbf {q}_\mathrm{b}=\nabla p_\mathrm{b}\) | \(\varOmega _\mathrm{b}(t)\). |
Nutrient transport | ||
Conservation of mass | \(\partial _t c_\mathrm{w}+\nabla \cdot \pmb {J}_w=0\) | \(\varOmega _\mathrm{w}(t)\). |
Rankine–Hugoniot | \((\pmb {J}_\mathrm{b}-\pmb {J}_\mathrm{w})\cdot \pmb {\nu }=\nu _n(\theta _\mathrm{w}c_\mathrm{b}-c_\mathrm{w})\) | \(\varGamma _{\mathrm{wb}}(t)\). |
Continuity of nutrients | \(\theta _\mathrm{w}c_\mathrm{b}=c_\mathrm{w}\) | \(\varGamma _\mathrm{wb}(t)\). |
Conservation of mass | \(\partial _t(\theta _\mathrm{w} c_\mathrm{b})+\nabla \cdot \pmb {J}_\mathrm{b}=R_\mathrm{b}\) | \(\varOmega _\mathrm{b}(t)\). |
Growth velocity potential | ||
Reference potential | \(\varPhi =0\) | \(\varGamma _\mathrm{wb}(t)\). |
Potential equation | \(-\nabla ^2\varPhi =\varSigma _l(R_l/\rho _l)/(1-\theta _\mathrm{w})\quad \pmb {u}=-\nabla \varPhi \) | \(\varOmega _\mathrm{b}(t)\). |
Volume fractions | ||
Detached component | \(\pmb {\nu }\cdot \nabla \theta _l=0\) | \(\varGamma _\mathrm{wb}(t)\). |
Conservation of mass | \(\partial _t\theta _l+\nabla \cdot (\pmb {u}\theta _l)=R_l/\rho _l\) | \(\varOmega _\mathrm{b}(t)\). |
Biofilm–water interface | ||
Biofilm thickness | \(\partial _t d={\left\{ \begin{array}{ll} -\sqrt{1+(\partial _x d)^2}[\pmb {\nu }\cdot \mathbf {u}]_{+},&{} d=W,\\ -\sqrt{1+(\partial _x d)^2}(\pmb {\nu }\cdot \pmb {u}+k_\mathrm{str}S),&{}0<d<W,\\ 0,&{}d=0,\end{array}\right. }\) | \(\varGamma _\mathrm{wb}(t)\). |
Reaction terms | ||
Nutrient consumption | \(R_\mathrm{b}=-\mu _n\theta _\mathrm{a}\rho _\mathrm{a}c_\mathrm{b}/(k_n+c_\mathrm{b})\) | \(\varOmega _\mathrm{b}(t)\). |
Bacterial decay | \(R_\mathrm{d}=k_{\text {res}}\theta _\mathrm{a}\rho _\mathrm{a}\) | \(\varOmega _\mathrm{b}(t)\). |
Bacterial reproduction | \(R_\mathrm{a}=-Y_\mathrm{a}R_\mathrm{b}-R_\mathrm{d}\) | \(\varOmega _\mathrm{b}(t)\). |
EPS production | \(R_\mathrm{e}=-Y_\mathrm{e}R_\mathrm{b}\) | \(\varOmega _\mathrm{b}(t)\). |
The aforementioned equations define the pore-scale model for permeable biofilm. This is a coupled system of nonlinear partial differential equations with a moving interface.
We use an ALE method for tracking the biofilm–water interface (Donea et al. 2004). We use backward Euler for the time discretization and linear Garlekin finite elements for the spatial discretization. We split the solution process into three sub-steps. A damped version of Newton’s method is used in each of the steps. First, we solve for the pressures and water fluxes. Secondly, we solve for the nutrient concentration. Thereafter, we solve for the volume fractions, growth potential, and biofilm thickness. We iterate between the previous steps until the error (the difference between successive values of the solution) drops below a given tolerance. Then, we move to the next time step and solve again until a given final time. We implement the model equations in the commercial software COMSOL Multiphysics (COMSOL 5.2a, Comsol Inc, Burlington, MA, www.comsol.com).
3 Model Test
Micromodel experiments under controlled conditions have been designed for determination of critical input parameters for the biofilm formation. A glass micromodel (Micronit, Netherland), a camera (VisiCam 5.0), and two syringe pumps (NE-1000 Series, Syringe Pumps) were used to perform the experiments. The bacterium Thalassospira strain A213101 was used for the laboratory studies. This bacterium is facultative anaerobic and uses nitrate as alternative finale electron acceptor. Pyruvate was added as a carbon source to a marine mineral medium to produce a nutrient concentration of \(c_i=0.88 \hbox { kg}/\hbox {m}^3\). Biofilm growth was established in the micromodel by first adding bacteria and then flooding with nutrient medium (Liu et al. 2018). Biofilm was established and monitored by microscope with digital camera (Liu et al. 2018). Differences in biofilm growth profiles were assessed as factor of nutrient concentration and flow velocity (rate). The micromodel used in the laboratory has a width of 100 \(\upmu \)m and thickness/depth of 20 \(\upmu \)m. Figure 2 shows the biofilm formation over time for a flow rate of \(0.2\;\upmu \hbox {l}/\text {min}\), which corresponds to a water velocity injection of \(q_i=1.66 \;\text {mm/s}:=U\) and an entry pressure of \(p_{i}=0.128\;\mathrm{Pa}\). First, microbes and nutrients were injected in the vertical channel for 24 h at a rate of 1 \(\upmu \hbox {l}/\text {min}\). Afterward, the whole system was closed for one day to allow the suspended bacteria to attach to the surface. Then, we started to inject nutrients from the left channel at a rate of 0.2 \(\upmu \hbox {l}/\text {min}\). Bacterial sloughing was not observed during the time the experiments were performed (6–7 days). A detailed description of the performed experiments can be found in Liu et al. (2018).
In order to compare the mathematical model with the laboratory experiments, we perform numerical simulations considering the same experimental input values for flux and nutrient concentration. We study the increase in percentage of biofilm coverage area over time. We consider a space domain of the same width of the microchannel \(W=0.1\) mm and length \(L=0.2\) mm. Recalling that biofilms are mostly composed by water, we set the water volume fraction in the biofilm equal to 90% (\(\theta _\mathrm{w}=0.9\)). Then, the organic matter in the biofilm is equal to 10%. Due to the lack of data, we assume that the biofilm is formed only by water and active bacteria, neglecting the EPS in the early stage (\(\theta _\mathrm{a}(0,x,y)=0.1,\;\theta _\mathrm{e}(0,x,y)=0,\;\text {and}\;\theta _\mathrm{d}(0,x,y)=0\)). We set the initial biofilm thickness to \(d(0,x)=2.5\; \upmu \hbox {m}\). A combination of experimentally determined parameters and values from the literature has been used for the numerical simulations (see Table 1 for details).
For calibration of the stress coefficient, we consider the experimental percentage of biofilm area over time for four different water velocities. Figure 3 shows the experimental and simulated percentage of biofilm area over time. After numerical simulations, the order of stress coefficient that best fits the data is \(k_\mathrm{str}=10^{-10}\;\text {m}/(\text {s Pa})\). Then, we perform a parametric sweep of the stress coefficient in the interval \([10^{-9},10^{-11}]\) with a step of \(10^{-11}\), where we use the method of least squares. The value that best fits the experimental data is \(k_\mathrm{str}=2.6\times 10^{-10}\;\text {m}/(\text {s Pa})\).
4 Numerical Results
We perform numerical simulations with \(c_i=10^{-3}\;\hbox {kg}/\hbox {m}^3\), \(p_{i}=0.128\;\hbox {Pa}\), \(\theta _\mathrm{w}=0.9\), and \(d(0,x)=2.5\; \upmu \hbox {m}\). We consider a smaller nutrient concentration in comparison with the one used in the laboratory experiments to study the biofilm dynamics with nutrient limitation. We consider a heterogeneous biofilm, where initially the biomass on the left half side (\(0<x<\hbox {L}/2\)) is formed by 60\(\%\) of active bacteria and \(40\%\) of EPS and the biomass on the right half side (\(\hbox {L}/2<x<\hbox {L}\)) is formed by \(40\%\) of active bacteria and \(60\%\) of EPS. The remaining input parameters are taken from Table 1, and the calibrated stress coefficient is \(k_\mathrm{str}=2.6\times 10^{-10}\;\text {m}/(\text {s Pa})\). In the next figures, different numerical results at different times are shown.
Figure 4 shows the growth velocity potential \(\phi \) after 120 h and the nutrient concentrations \(c_\mathrm{b}\) and \(c_\mathrm{w}\) after 360 h, respectively. The growth velocity potential is larger on the left lower corner, as a result of the nutrient injection on the left side and the condition of zero potential on the interface. Therefore, the biomass will grow toward the right upper corner. After 360 h of injection of nutrients, we observe that the nutrient concentration in the biofilm decreases from left to right, due to the consumption of nutrients by the active bacteria.
Figure 5 shows the magnitude and the flow direction of the water flux velocity after 360 h, respectively. In the water domain, we observe that the water flux is larger between the wall and the interface. In the biofilm, the water flux decreases from the interface until zero on the wall.
Figure 6 shows the averaged volume fraction after 360 h and the biofilm height profile over time, respectively. We observe that more than 65% of the organic matter in the biofilm is formed by EPS and dead bacteria after 360 h. We also observe that the biofilm height on the left side grows faster over time, due to the nutrients being injected on the left side and also due to the larger initial active bacteria on the left half side, leading to a faster EPS and bacterial production.
5 Sensitivity Analysis
Variability in input parameters may have a significant effect on output quantities of interest, for instance the percentage (0–100%) of biofilm area relative to the area of the whole domain. We perform a global sensitivity analysis (Sobol 2001; Sudret 2008) to quantify the effect of variability or uncertainty in ten material parameters that are assumed to be sensitive with respect to variation in the biofilm area after \(T=50\;\)h of nutrient injection. The initial biofilm thickness is \(d(0,x)=10\;\upmu \)m, the injected nutrient concentration is \(c_i=0.88\;\)kg/m\(^3\), and the entry pressure is \(p_i=0.128\;\)Pa. The input parameters, their range of variation, and total Sobol index are listed in Table 2 (see “Appendix A” for details).
Due to interaction between the parameters and the fact that the total contribution from a given parameter also involves all combinations of this parameter together with the other parameters, the sum of the relative total contribution from the parameters exceeds 1. The relative variability contribution from each parameter is significant for the parameter ranges investigated. The maximum growth rate stands out as more important than the others with respect to total variability, but none of them should be discarded based on this numerical sensitivity study alone. The true value of each of the ten parameters should be estimated with sufficient accuracy to lead to a reliable estimate of the biofilm area.
All previous plots are the result of the parameter values, initial conditions, and input values. The concept of growing a biofilm in the laboratory seems uncomplicated. Nevertheless, the biofilm formation takes up to two weeks and it is very sensitive to the surrounding conditions (e.g., the substrate surface and light conditions). As a result of limitations in the laboratory, we could not estimate all model parameters from the experiments. Then, it is necessary to improve the growth techniques and develop new measurement strategies to give better estimates of the parameters and, in turn, validate the model assumptions.
6 Conclusions
In this work, a pore-scale model for biofilm formation is built considering the biofilm as a porous medium. To our knowledge, the present work is the first study of a permeable multi-component biofilm in a strip geometry including growing, detachment, and transport of nutrients due to convection and diffusion. The stress coefficient \(k_\mathrm{str}=2.6\times 10^{-10}\;\text {m}/(\text {s Pa})\) is selected to match the experimental results. A sensitivity analysis is performed. The sensitivity analysis confirms that the variability or uncertainty in none of the ten parameters investigated should be neglected. In the numerical simulations, we observe a reduction in the biofilm height as the water flux velocity increases. For high flow rates, we must consider the effects of the flow inside the biofilm, which affect the transport of nutrients and, therefore, influence the biofilm thickness.
Abbreviations
- c :
-
Nutrient concentration
- D :
-
Nutrient diffusion coefficient
- d :
-
Biofilm thickness
- J :
-
Nutrient flux
- k :
-
Permeability
- \(k_\mathrm{res}\) :
-
Bacterial decay rate coefficient
- \(k_\mathrm{str}\) :
-
Stress coefficient
- \(k_{n}\) :
-
Monod half-velocity coefficient
- L :
-
Pore length
- p :
-
Pressure
- q :
-
Water velocity
- S :
-
Tangential shear stress
- T :
-
Time
- U :
-
Reference water velocity
- u :
-
Velocity of the biomass
- W :
-
Pore width
- Y :
-
Growth yield coefficient
- \(\mu \) :
-
Dynamic viscosity
- \(\mu _\mathrm{n}\) :
-
Maximum rate of nutrient utilization
- \(\nu \) :
-
Unitary normal vector
- \(\nu _n\) :
-
Interface velocity
- \(\varPhi \) :
-
Growth velocity potential
- \(\rho \) :
-
Density
- \(\tau \) :
-
Unitary tangential vector
- \(\theta \) :
-
Volume fraction
- a :
-
Active bacteria
- b :
-
Biofilm
- d :
-
Dead bacteria
- i :
-
Input
- o :
-
Output
- e :
-
EPS
- w :
-
Water
- ALE:
-
Arbitrary Lagrangian–Eulerian
- EPS:
-
Extracellular polymeric substance
- MEOR:
-
Microbial enhanced oil recovery
References
Ahmad, I., Husain, F.M.: Biofilms in Plant and Soil Health. Wiley, Hoboken (2017). https://doi.org/10.1002/9781119246329
Alpkvist, K., Klapper, I.: A multidimensional multispecies continuum model for heterogeneous biofilm development. Bull. Math. Biol. 69, 765–789 (2007). https://doi.org/10.1007/s11538-006-9168-7
Beavers, G.S., Joseph, D.D.: Boundary conditions at a naturally permeable wall. J. Fluid Mech. 30, 197–207 (1967). https://doi.org/10.1017/S0022112067001375
Bitton, G.: Wastewater Microbiology. Wiley Series in Ecological and Applied Microbiology, p. 315. Wiley, Hoboken (2005)
Boltz, J.P., Smets, B.F., Rittmann, B.E., van Loosdrecht, M.C.M., Morgenroth, E., Daigger, G.T.: From biofilm ecology to reactors: a focused review. J. Fluid Mech. 75, 1753–1760 (2017). https://doi.org/10.2166/wst.2017.061
Bringedal, C., Berre, I., Pop, I.S., Radu, F.A.: Upscaling of non-isothermal reactive porous media flow with changing porosity. Transp. Porous Media 114, 371–393 (2016). https://doi.org/10.1007/s11242-015-0530-9
Bringedal, C., Kumar, K.: Effective behavior near clogging in upscaled equations for non-isothermal reactive porous media flow. Transp. Porous Media 120, 553–577 (2017). https://doi.org/10.1007/s11242-017-0940-y
Cao, Z., Liu, G., Zhan, H., Li, C., You, Y., Yang, C., Jiang, H.: Pore structure characterization of Chang-7 tight sandstone using MICP combined with N2GA techniques and its geological control factors. Sci. Rep. 6, 36919 (2016). https://doi.org/10.1038/srep36919
Crittenden, J.C., Trussell, R.R., Hand, D.W., Howe, K.J., Tchobanoglous, G.: MWH’s Water Treatment: Principles and Design. Wiley, Hoboken (2012). https://doi.org/10.1002/9781118131473
Deng, W., Cardenas, M.B., Kirk, M.F., Altman, S.J., Bennett, P.C.: Effect of permeable biofilm on micro-and macro-scale flow and transport in bioclogged pores. Environ. Sci. Technol. 47, 11092–11098 (2013). https://doi.org/10.1021/es402596v
Donea, J. Huerta, A., Ponthot, J.-P, Rodríguez-Ferran, A.: Arbitrary Lagrangian–Eulerian Methods. Enc. Comput. Mech. (2004). https://doi.org/10.1002/0470091355.ecm009
Donlan, R.M.: Biofilms: microbial life on surfaces. Emerg. Infect. Dis. 8, 881 (2002). https://doi.org/10.3201/eid0809.020063
Duddu, R., Chopp, D.L., Moran, B.: A two-dimensional continuum model of biofilm growth incorporating fluid flow and shear stress based detachment. Biotechnol. Bioeng. 103, 92–104 (2009). https://doi.org/10.1002/bit.22233
Dumitrache, C.A., Petrache, A.: Interface condition for the coupling of a fluid and porous media. Acta Technica Napocensis Ser. Appl. Math. Mech. Eng. 55(2), 1221–5872 (2012)
Esmaili, S., Eslahchi, M.R.: Application of collocation method for solving a parabolic–hyperbolic free boundary problem which models the growth of tumor with drug application. Math. Methods Appl. Sci. 40, 1711–1733 (2017). https://doi.org/10.1002/mma.4092
Gallinato, O., Poignard, C.: Superconvergent second order Cartesian method for solving free boundary problem for invadopodia formation. J. Comput. Phys. 339, 412–431 (2017). https://doi.org/10.1016/j.jcp.2017.03.010
Horn, H., Lacknerk, S.: Modeling of biofilm systems: a review. Adv. Biochem. Eng. Biotechnol. (2014). https://doi.org/10.1007/10_2014_275
Hornung, U.: Homogenization and Porous Media. Interdisciplinary Applied Mathematics, p. 63. Springer, New York (1997)
Hoštacká, A., Čižnár, I., Štefkovičová, M.: Temperature and pH affect the production of bacterial biofilm. Folia Microbiologica 55, 75–78 (2010). https://doi.org/10.1007/s12223-010-0012-y
Kumar, K., Wheeler, M.F., Wick, T.: Reactive flow and reaction-induced boundary movement in a thin channel. SIAM J. Sci. Comput. 35(6), B1235–B1266 (2013)
Kokare, C.R., Chakraborty, S., Khopade, A.N., Mahadik, K.R.: Biofilm: importance and applications. Indian J. Biotechnol. 8, 129–168 (2009)
Landa-Marbán, D., Radu, F.A., Nordbotten, J.M.: Modeling and simulation of microbial enhanced oil recovery including interfacial area. Transp. Porous Media 120, 395–413 (2017). https://doi.org/10.1007/s11242-017-0929-6
Lewandowski, Z., Beyenal, H.: In: Wuertz, S., Bishop, P.L., Wielderer, P.A. (eds.) Biofilms in Wastewater Treatment—An Interdisciplinary Approach, p. 156. IWA Publishing, London (2003)
Liu, N., Skauge, T., Landa-Marbán, D., Hovland, B., Thorbjørnsen, B., Radu, F.A., Vik, B.F., Baumann, T., Bødtker, G.: Microfluidic study of effects of flowrate and nutrient concentration on biofilm accumulation and adhesive strength in a microchannel. J. Ind. Microbiol. Biotechnol. (2018) (Submitted)
Mikelić, A., Jäger, W.: On the interface boundary condition of Beavers, Joseph, and Saffman. SIAM J. Appl. Math. 60, 1111–1127 (2000). https://doi.org/10.1137/S003613999833678X
Monod, J.: The growth of bacterial cultures. Annu. Rev. Microbiol. 3, 371–394 (1949). https://doi.org/10.1146/annurev.mi.03.100149.002103
Murphy, E.M., Ginn, T.R.: Modeling microbial processes in porous media. Hydrogeol. J. 8, 142–158 (2000). https://doi.org/10.1007/s100409900043
Parsek, M.R., Fuqua, C.: Biofilms 2003: emerging themes and challenges in studies of surface-associated microbial life. J. Bacteriol. 186, 4427–4440 (2004). https://doi.org/10.1128/JB.186.14.4427-4440.2004
Peszynska, M., Trykozko, A., Iltis, G., Schlueter, S., Wildenschild, D.: Biofilm growth in porous media: experiments, computational modeling at the porescale, and upscaling. Adv. Water Res. 95, 288–301 (2016). https://doi.org/10.1016/j.advwatres.2015.07.008
Piciorenau, C., van Loosdrecht, M.C.M., Heijnen, J.J.: Mathematical modeling of biofilm structure with a hybrid differential-discrete cellular automaton approach. Biotechnol. Bioeng. (1998). https://doi.org/10.1002/(SICI)1097-0290(19980405)58:1<101::AID-BIT11>3.0.CO;2-M
Piciorenau, C., van Loosdrecht, M.C.M., Heijnen, J.J.: Modelling and predicting biofilm structure. In: Allison, D., Gilbert, P., Lappin-Scott, H., Wilson, M. (eds.) Community Structure and Co-operation in Biofilms (Society for General Microbiology Symposia), pp. 129–166. Cambridge University Press, Cambridge (2000)
Ray, N., van Noorden, T., Radu, F.A., Friess, W., Knabner, P.: Drug release from collagen matrices including an evolving microstructure. ZAMM 93, 811–822 (2013). https://doi.org/10.1002/zamm.201200196
Rubol, S., Freixa, A., Carles-Brangarí, A., Fernàndez-Garcia, D., Romaní, A.M., Sanchez-Vila, X.: Drug release from collagen matrices including an evolving microstructure. J. Hydrol. 517, 317–327 (2014). https://doi.org/10.1016/j.jhydrol.2014.05.041
Saffman, P.G.: On the boundary condition at the surface of a porous medium. Stud. Appl. Math. 50, 93–101 (1971). https://doi.org/10.1002/sapm197150293
Schulz, R., Knabner, P.: Derivation and analysis of an effective model for biofilm growth in evolving porous media. Math. Method Appl. Sci. 40, 2930–2948 (2016). https://doi.org/10.1002/mma.4211
Schwarzenbach, R.P., Gschwend, P.M., Imboden, D.M.: Environmental Organic Chemistry. Wiley, New York (1993)
Seki, K., Thullner, M., Hanada, J., Miyazaki, T.: Moderate bioclogging leading to preferential flow paths in biobarriers. Groundw. Monit. Remediat. 26, 68–76 (2006). https://doi.org/10.1111/j.1745-6592.2006.00086.x
Sen, R.: Biotechnology in petroleum recovery: the microbial EOR. Prog. Energy Combust. Sci. 34, 714–724 (2008). https://doi.org/10.1016/j.pecs.2008.05.001
Smolyak, S.: Quadrature and interpolation formulas for tensor products of certain classes of functions. Sov. Math. Dokl. 4, 240–243 (1963)
Sobol, I.M.: Global sensitivity indices for nonlinear mathematical models and their Monte Carlo estimates. Math. Comput. Simul. 55, 271–280 (2001). https://doi.org/10.1016/S0378-4754(00)00270-6
Stewart, P.S.: Diffusion in Biofilms. J. Bacteriol. 185, 1485–1491 (2003). https://doi.org/10.1128/JB.185.5.1485-1491.2003
Sudret, B.: Global sensitivity analysis using polynomial chaos expansions. Reliab. Eng. Syst. Saf. 93, 964–979 (2008). https://doi.org/10.1016/j.ress.2007.04.002
Tang, Y., Liu, H.: Modeling multidimensional and multispecies biofilms in porous media. Biotechnol. Bioeng. 114, 1679–1687 (2017). https://doi.org/10.1002/bit.26292
Taylor, S., Jaffe, P.R.: Substrate and biomass transport in a porous medium. Water Resour. Res. 26, 2181–2194 (1990). https://doi.org/10.1029/WR026i009p02181
Toyofuko, M., Inaba, T., Kiyokawa, T., Obana, N., Yawata, Y., Nomura, N.: Environmental factors that shape biofilm formation. Biosci. Biotechnol. Biochem. 80, 7–12 (2015). https://doi.org/10.1080/09168451.2015.1058701
Urquiza, J.M., N’Dri, D., Garon, A., Delfour, M.C.: Coupling Stokes and Darcy equations. Appl. Numer. Math. 58, 525–538 (2008). https://doi.org/10.1016/j.apnum.2006.12.006
van Noorden, T.L., Pop, I.S., Ebigbo, A., Helmig, R.: An upscaled model for biofilm growth in a thin strip. Water Resour. Res. (2010). https://doi.org/10.1029/2009WR008217
Vu, B., Chen, M., Crawford, R.J., Ivanova, E.P.: Bacterial extracellular polysaccharides involved in biofilm formation. Molecules 14, 2535–2554 (2009). https://doi.org/10.3390/molecules14072535
Wood, B.D., Whitaker, S.: Diffusion and reaction in biofilms. Chem. Eng. Sci. 53, 397–425 (1998). https://doi.org/10.1016/S0009-2509(97)00319-9
Xiu, D., Karniadakis, G.E.: The Wiener–Askey polynomial chaos for stochastic differential equations. SIAM J. Sci. Comput. 24, 619–644 (2002). https://doi.org/10.1137/S1064827501387826
Yang, G., Weigand, B., Terzis, A., Weishaupt, K., Helmig, R.: Numerical simulation of turbulent flow and heat transfer in a three-dimensional channel coupled with flow through porous structures. Transp. Porous Media 122, 145–167 (2017). https://doi.org/10.1007/s11242-017-0995-9
Zhang, T.C., Bishop, P.L.: Density, porosity, and pore structure of biofilms. Water Res. 28, 2267–2277 (1994). https://doi.org/10.1016/0043-1354(94)90042-6
Acknowledgements
The work of DLM, NL, KK, PP, GB, TS, and FAR was partially supported by GOE-IP and the Research Council of Norway through the projects IMMENS No. 255426 and CHI No. 255510. ISP was supported by the Research Foundation-Flanders (FWO), Belgium, through the Odysseus programme (Project G0G1316N) and the Akademia grant of Equinor.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix A: Sensitivity Analysis Method
Appendix A: Sensitivity Analysis Method
In this Appendix, we describe the theory behind the performed sensitivity analysis. The variation is assumed uniform in the sense that each parameter varies within a range where all values are equally likely. The sensitivity analysis relies on the Hoeffding or Sobol decomposition of the quantity of interest, here denoted q, as a series expansion in subsets of all possible combinations of the n input parameters \(\pmb {y} = (y_1,...,y_n)\),
The Sobol decomposition terms are defined recursively as integrals over subsets of the range of \(\pmb {y}\), denoted \(\pmb {Y}\). We introduce a uniform weight function \(w(\pmb {y})=w_1(y_1)...w_{n}(y_n)\) with \(w_i = 1/(\max (y_i)-\min (y_i))\) and the subscript notation \(\sim i\) to denote all parameters except parameter i. The decomposition terms are then determined by
and so on for higher-order terms.
The Sobol index for the s-parameter combination \(\{ y_{i_1},y_{i_2},...,y_{i_s}\}\) is given by
The total variability of variable i is obtained by summing over all subsets of parameters including parameter i, which yields the total Sobol index for parameter i,
In this work, the Sobol decomposition terms are computed from a generalized polynomial chaos expansion in Legendre polynomials (Sudret 2008), where the expansion coefficients are obtained from sparse quadrature rules using the Smolyak algorithm (Smolyak 1963). This quadrature rule is very sparse but assumes high regularity on the quantity of interest as a function of the input parameters.
Rights and permissions
About this article
Cite this article
Landa-Marbán, D., Liu, N., Pop, I.S. et al. A Pore-Scale Model for Permeable Biofilm: Numerical Simulations and Laboratory Experiments. Transp Porous Med 127, 643–660 (2019). https://doi.org/10.1007/s11242-018-1218-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11242-018-1218-8