Abstract
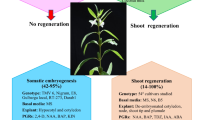
Sesamum indicum is an important oilseed crop with beneficiary nutrients such as essential fatty acids, proteins, and lignan. S. indicum crop improvement using conventional breeding methods was disastrous. Moreover, no reproducible regeneration and transformation protocol for large-scale generation of independent primary transformed plant lines of S. indicum virtually exists. Therefore, in the present study, a reproducible regeneration and transformation method was developed for S. indicum. Direct shoot regeneration (94.44%) was achieved using plumule tips as an explant source and when maintained on regeneration medium (BM11) an amalgamation of half the strength of MS macronutrients and B5 macronutrients together with MS micronutrients and vitamins. Maximum number of shoots (~ 16–19 shoots/explant) was obtained on BM11 medium supplemented with TDZ (1 mg/L) and BAP (0.1 mg/L). High root induction efficiency (~ 97%) was observed on RM4 medium consisted of half MS macronutrients, SH micronutrients and vitamins. The developed protocol was found effective for other commercially valuable genotypes such as Co 1, Phule til and Tilottama as well. Successful regeneration was subsequently extended to transformation of reporter gene (gfp: green fluorescent protein) as revealed by molecular analyses includes Southern hybridization, northern hybridization, real time PCR and histological study of the transformants developed in the present study. Based on the Southern analysis the calculated transformation frequency was found to be 1.33%. The presently developed regeneration and transformation system might facilitate any desired improvement of sesame crop.
Key message
The present study demonstrated an improved regeneration and transformation method for S. indicum, which is genotype independent and can undergo desired modifications.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sesame (Sesamum indicum L.), an oldest oilseed crop, is widely cultivated in tropical and subtropical regions belong to the Sesamum genus of the Pedaliaceae family (Ashri 2010), is rich in oil (48.5%), proteins (20%), carbohydrate (7.78%) and fibre (9.4%) (Nzikou et al. 2009). Sesame is considered as a valuable source of an essential fatty acid including linoleic acid. Recently, it has gained attention as a rich source of dietary lignans includes sesamin, sesamolin and sesaminol, with nutraceutical and antioxidative property (Nakai et al. 2003; Zimik and Arumugam 2017). Studies indicate that sesame consumption helps to lower blood cholesterol and hypertension, prevents atherosclerosis, coronary heart diseases and cancers (Hirose et al. 1991; Miyahara et al. 2001; Sankar et al. 2005; Tsuruoka et al. 2005; Nakano et al. 2008; Siao et al 2015). In addition to food consumption, sesame has many potential applications such as manufacturing paints, soap, perfumes, pharmaceutical products and insecticides (Oplinger et al. 1990). However, despite its nutritional, medicinal and industrial applications the crop productivity worldwide is low due to various biotic and abiotic stresses (Pathak et al. 2014). To surmount these restrictions, developments of a new sesame stress resistant variety are of urgent need. Furthermore, there remains certain possibility for nutritional improvement in its oil quality that could contribute to human health.
Due to the sexual incompatibility, the sesame crop improvement via conventional breeding method between the cultivated and wild species was of limited success (Tiwari et al. 2011; Kulkarni et al. 2017). Genetic modification of crops by introducing desired genes has evolved as an alternative biotechnology approach and has been practiced for decades. Reproducible regeneration and transformation systems form the foundation for genetic engineering strategy. However, genetic engineering of sesame is mostly limited due to unavailability of suitable and reproducible regeneration and transformation system (Zimik and Arumugam 2017). In recent times, many researchers attempted to establish in vitro regeneration protocol for sesame without much success. Indirect regeneration via intervening callus phase was tried using various explants such as cotyledon and/or hypocotyls (Younghee 2001; Baskaran and Jayabalan 2006; Were et al. 2006; Chakraborti and Ghosh 2009; Rao and Honnale 2011; Shashidhara et al. 2011a; Pusadkar et al. 2015), embryo (Saravanan and Nadarajan 2005), shoot tip (Baskaran and Jayabalan 2006). All these studies claimed induction of good callus growth; however, in most cases the induced calluses were turned out to be non-regenerative. Shoot regeneration from callus remained a barrier in indirect regeneration approaches in sesame (Pusadkar et al. 2015). Direct shoot regeneration from de-embryonated cotyledon isolated from mature seeds (Seo et al. 2007; Lokesha et al. 2012; Malaghan et al. 2013; Chowdhury et al. 2014; Pusadkar et al. 2016), cotyledon (Yadav et al. 2010; Al-Shafeay et al. 2011; Honnale and Rao 2013) and shoot tip taken from the germinated seedlings (Baskaran and Jayabalan 2006; Chakraborti and Ghosh 2009) was achieved with varying degree of success. However, cotyledonary explants were found to undergo necrosis on MS medium only or on cytokinins supplemented medium (Baskaran and Jayabalan 2006; Were et al. 2006). The variation in regeneration response was found to be due to the differences in selected genotypes. Successful regeneration through internodal transverse thin cell layer culture was reported by Chattopadhyaya et al. (2010) but at low frequency (29%). Overall, sesame genotypes, explant type and age of the explants were detected as a crucial factor that influenced in vitro shoot regeneration (Malaghan et al. 2013; Zimik and Arumugam 2017).
Despite of lack of good regeneration system, few researchers reported Agrobacterium-mediated transformation in sesame. For instance, the first Agrobacterium-mediated transformation was reported by Taskin et al. (1999), however, their study failed in generating fertile transgenic lines. Thereafter, few successful attempts were reported but the frequency (< 2%) was less (Yadav et al. 2010; Al-shafeay et al. 2011). Chowdhury et al. (2014) reported transformation efficiency of 42.66% in sesame cv. VRI 1 using de-embryonated cotyledon explants. However, all these studies emphasised the significance of age of explant, bacterial cell density, co-cultivation period and genotypic variations on the transformation efficiency. Hence, development of genotype independent regeneration and transformation system is of foremost necessity. Keeping the above in view, an approach was taken to develop a reproducible regeneration and Agrobacterium-mediated transformation system for S. indicum in the present study. Different explant type, media compositions and growth regulators were assessed to develop a suitable regeneration system that could be applicable for wide range of genotypes. Likewise, various physical parameters such as bacterial culture density and co-cultivation period were studied to achieve a suitable Agrobacterium-mediated transformation system. Combination of MS and B5 media components was established to be effective in increasing regeneration efficiency (~ 94%) in S. indicum. In addition, supplementation of medium with BAP (0.1 mg/L) and TDZ (1 mg/mL) was found to facilitate the shoot multiplication tendency (~ 16–19 shoots/explant). PCR followed by Southern and RT-PCR analyses were performed to confirm the integration and expression of gfp gene. The expression of the gene in T0 transformants was finally validated by green confocal microscopy.
Materials and methods
Plant material
Seven cultivars of sesame viz. Rama, Lambasuti, Co1, Tillottama, DS1, CST 2001 and Phule til were used for the present study. These are widely cultivated varieties in different regions of India. Among them cv. Rama is high yielding variety and its seed contains high oil content, therefore, considered for detailed study (Sagar and Ganesh 2004; Chongdar et al. 2015). Initially, culture conditions were optimized for cv. Rama. Seeds were surface sterilized by washing with 0.2% HgCl2 for 2 min and then rinsed thrice with sterile distilled water. Seed coat was removed with the help of a scalpel and placed on MS medium to assist germination. Mature seed and 2 days older germinated seedlings were considered as a source of explant.
Explant preparation and optimization of media composition for indirect regeneration
Embryos from mature seeds, hypocotyls and cotyledons from 2 days older germinated seedlings were used as explants for callus induction. The explants were incubated on different basal media includes MS (Murashige and Skoog 1962), B5 (Gamborg et al. 1968), N6 (Chu et al. 1975) and SH (Schenk and Hildebrandt 1972). All the media was supplemented with fixed concentration of 2, 4-D (0.25 mg/L) to determine the suitable basal medium and explant for callus induction. Subsequently, 2, 4-D (0.5, 1, 2, 3 and 4 mg/L), NAA (0.5, 1, 2, 3 and 4 mg/L) alone or in combination with BAP/Kinetin (0.25, 0.5, 1, 1.5 mg/L) was tested to assess their effect on selected explants for callus induction. Callus induction frequency was recorded for every explant type in all the treatments. Callus obtained from various explants were tested for shoot regeneration on MS basal medium supplemented with varying concentrations of BAP, zeatin, kinetin and TDZ (0.5, 1, 1.5 and 2 mg/L) alone or in combination with 2, 4-D (0.5 mg/L).
Explant preparation and optimization of media composition for direct regeneration
De-embryonated cotyledon, embryonic axis from mature seed, plumule tip and cotyledons from 2 days older germinated seedlings were used as explants for direct regeneration study. The apical meristematic region of embryonic axis and plumule tip was wounded by making a longitudinal incision. Preliminary studies were conducted to select explant type able to regenerate on MS medium containing BAP at 0.25 mg/L. Consequently, the selected explants were used to examine the effect of basal salt composition on regeneration frequency. Four different basal media components namely MS, B5, N6 and SH media were used further. For shoot multiplication, cytokinins such as BAP (0.5, 1, 2, 3 mg/L), Kinetin (0.5, 1, 2, 3 mg/L), TDZ (0.5, 1, 2, 3 mg/L), Zeatin (0.5, 1, 2, 3 mg/L) and NAA (0.1 and 0.5 mg/L) were tested. All the cultures were incubated in growth chamber at 25 °C with moderate relative humidity (50%) and with a photoperiod of 16:8 (light:dark) cycle. The optimized media were further assessed for regeneration and multiple shooting capacities of other genotypes used in the present study.
Construction of gene cassette
The gfp gene encoding green fluorescent protein was used for transformation method optimization. cDNA sequence of gfp was amplified from pX6 vector by performing PCR using the gfp gene specific primers (forward: 5′-AGTAGGTACCATGGGTAAGGGAGAAGAAC-3′ and reverse: 5′-ACGCTGGAGCTCTTATTTGTATAGTTCATCC-3′). containing restriction sites KpnI and SacI in forward and reverse direction respectively. 35S promoter (HindIII/BamHI), cDNA sequence of gfp (KpnI/SacI) and nos terminator (SacI/EcoRI) were cloned in pCAMBIA1300 vector. The construct developed was designated as pCAMBIA/gfp (Fig. 1). Prior to use in plant transformation, the plasmid pCAMBIA/gfp isolated from transformed E.coli cells was digested with different restriction enzymes available within the expression construct in order to confirm the proper alignment of the genetic elements. Analysis of the bands generated by restriction digestion of the isolated recombinant DNA showed that the genetic elements were arranged as per the designed order (Fig. 2). The cassette (pCAMBIA/gfp) was finally transformed into Agrobacterium EHA105 strain for its utilization in Agrobacterium-mediated plant transformation in S. indicum.
Schematic diagram of plant expression cassette of gfp gene (pCAMBIA/gfp). The relevant restriction sites for construction were shown. hptII indicates hygromycin phosphotransferase resistant gene. nos indicates nopaline synthase transcriptional terminator. LB indicates Left border. RB indicates Right border
Assessment of antibiotic sensitivity of the untransformed plants
The T-DNA region of pCAMBIA/gfp containing hptIIgene (hygromycin phosphotransferase IIgene: confers resistance to hygromycin) was helpful in screening transformed shoots in presence of hygromycin avoiding large population of false positives. Different concentrations of hygromycin (30, 35, 40 mg/L) were applied to untransformed shoots in order to determine the minimal dosage required for complete elimination. Mortality rates were recorded after 10 days of treatment.
Transformation and selection of shootlets
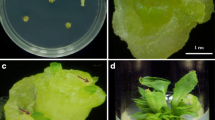
EHA105 cells harboring pCAMBIA/gfp vector were cultured in LB medium in presence of Rifampicin (10 mg/L) and Kanamycin (50 mg/L) for overnight. The next day, Agrobacterium cells were harvested from bacterial culture of different OD600 (0.2–0.8) and resuspended in fresh medium containing acetosyringone (20 µM). Plumule tips were isolated from 2 days old germinated seedlings and wounded at tip by making a longitudinal incision (Fig. 3a). Plumule tip explants were incubated along with Agrobacterium cell suspension and kept idle for 1 h (Fig. 3b). Instantaneously, the explants were picked up, blotted to eliminate excess bacteria and transferred to co-cultivation medium (BM11 containing 20 µM acetosyringone) following incubation in dark for 48 to 72 h (Fig. 3c). Explants regenerated after Agrobacterium treatments (Fig. 3d) were subcultured on multiple shoot induction medium (BM11-TB1) containing cefotaxime at every 10 days of interval for 6–8 weeks to induce multiple shoots (Fig. 3e). Cefotaxim at 250 mg/L concentration was used to avoid bacterial contamination. Shoots formed were separated and placed on selection medium containing optimum dosage of hygromycin for 2 weeks (Fig. 3f). The green healthy shoots recovered on selection medium were incubated on RM4 medium for rooting (Fig. 3g). Plantlets with well-developed roots were acclimatized to sand and soil (1:1) mixture (Fig. 3h).
Representative figures depicting steps involved in Agrobacterium mediated transformation a The apical meristematic region of plumule tip was wounded by making a longitudinal incision. b Plumule tips explants immersed inAgrobacterium suspension culture. c Blot dried infected explants placed on cocultivation medium(BM11 medium containing 20 µM acetosyringone). d Shoots regenerated from infected explants after cocultivation. e Multiple shoots formed after 6–8 weeks of subculture on multiple shoot induction medium (BM11-TB1 containing 250 mg/L of cefotaxim). f Shoots formed were screened on BM11 medium containing hygromycin for 4 weeks: green shoots showing resistance to hygromycin antibiotic. g The green healthy shoots recovered from hygromycin screening formed roots on RM4 medium. h Plantlets with roots were hardened to sand soil (1:1) mixture and established in green house
Molecular analyses of generated putative transformants
Total genomic DNA was extracted from fresh leaves of hygromycin resistant sesame plantlets by following modified CTAB method (Doyle and Doyle 1990).Untransformed line and pCAMBIA/gfp vector were taken as negative and positive control respectively.Transgene integration in the infected plantlets was initially detected by PCR usinghptII gene specific primers (Forward: 5′-ATGAAAAAGCCTGAACTCACCGCGACGTCT-3′ and reverse: 5′-GCATCAGCTCATCGAGAGCCTGCGCGACGG -3′). PCR was carried out under the following conditions: initial denaturation at 94 °C for 4 min; 30 cycles consisted of denaturation at 94 °C for 30 s, annealing at 60 °C for 40 s, extension at 72 °C for 1 min; final extension of 72 °C for 10 min. Further, for detection of presence of gfp gene the plants were subjected to PCR analysis usinggfp gene specific primers (forward: 5′-AGTAGGTACCATGGGTAAGGGAGAAGAAC-3′ and reverse: 5′-ACGCTGGAGCTCTTATTTGTATAGTTCATCC-3′). PCR was carried out under the following conditions: initial denaturation at 94 °C for 4 min; 30 cycles consisted of denaturation at 94 °C for 30 s, annealing at 62 °C for 40 s, extension at 72 °C for 1 min; final extension of 72 °C for 10 min.
To validate the transgene integration, Southern hybridization was performed. Genomic DNA of the S. indicum plant was digested with PstI enzymes andelectrophoresed on 0.8% agarose gel. Genomic DNA (~ 12 μg) isolated from both the untransformed control and transformed lines were digested with PstI enzyme (Since the cassette does not contain any internal restriction site for PstI enzyme). pCAMBIA/gfp vector (~ 10 ng)digested with HindIII was taken as positive control. The DNA thus prepared were electrophoresed on 0.8% agarose gel followed by transfer to nylon membrane. The blot was hybridized with gfp gene specific radiolabelled probes.
Transcript expression of gfp transgene was detected by northern hybridization technique. Total RNA was extracted from seeds of S. indicum using the Trizol reagent (Ambion) following the method described by Chomczynski and Sacchi (1987). The uniformity in amount of RNA loaded in the northern gel was confirmed by rehybridizing the blot with actin specific radiolabelled probe. To quantitate the transcript expression, real time PCR was conducted using gfp and actin gene specific primers: GFP qRTFP: 5′ GGCCAACACTTGTCACTA 3′ and GFP qRTRP: 5′ TTCCTGCACGTATCCCTCAGGCA 3′ for amplification of GFP cDNA and RT ACT FP: 5′ CTGGATTTGCTGGTGATGATGC 3′ and RT ACT RP: 5′ CATGTCATCCCAGTTGCTGACT 3′ for amplification of actin cDNA. The PCR conditions are as follows: initial denaturation at 95 °C for 2 min followed by 40 cycles of denaturation at 95 °C for 15 s, primer annealing at 58 °C for 15 s, extension at 72 °C for 30 s. All reactions were carried out in triplicates.The 2−ΔΔCT method for relative quantification was followed in the present study to estimate the expression levels of gene of interest in all transgenic plants (Livak and Schmittgen 2001). Knowing the fact that sesame genome does not contain endogenous gfp gene, the least expressing transgenic line (analysed by northern blotting technique) was taken as control for relative quantification of rest of the transformants.
Detection of green fluorescence by confocal microscopy
Transverse sections of fresh leaf petiole were treated with 80% ethanol and were kept overnight to remove chlorophyll. Following chlorophyll removal, the tissue sections were placed under confocal microscope. The DIC images of the tissue were captured. Subsequently, the tissue sections were exposed under UV light (488 nm) to detect green fluorescence using 505–530 nm band pass filter. The DIC and confocal images were overlapped to identify the fluorescence in tissue sections.
Statistical analysis
All the media optimization experiments were performed in three replicates with minimum of 30 explants per experiment. Data for callus induction, shoot regeneration, shoot multiplication and root induction were recorded and analyzed using one way ANOVA test. Transformation efficiency was determined for the positive plants detected in Southern hybridization analysis with respect to total number of explants taken for infection.
Results
Effect of growth regulators and media on indirect regeneration
Among the basal media, MS basal medium supplemented with 2, 4-D was found to be more prominent for callus induction (Supplementary Table S1). Callus induction potential varied for different explants. Cotyledons showed higher frequency of callus induction in the MS basal medium compared to hypocotyl and embryo. Hence, cotyledon was considered as an ideal explant for callus induction in the present study. The evaluation of auxins (2, 4-D and NAA) for callus induction showed that MS basal medium fortified with 2, 4-D profoundly influenced callus induction, while NAA was found to have comparatively less significant influence (Supplementary Table S2). A high incidence of callus induction (~ 71%) was observed under 2 mg/L of 2, 4-D supplementation. Apart from auxins, cytokinins also had positive impact on callus induction. Callus induction frequency was effectively enhanced by supplementing cytokinins along with auxins rather than auxin alone (Table 1). Highest callus induction frequency (~ 89%) was exhibited on MS medium supplemented with 2, 4-D (2 mg/L) and BAP (1.5 mg/L). However, the calluses induced on different combinations of growth regulators were found to be of different morphological patterns including green friable callus, yellowish green friable callus, green compact callus and white watery callus (Table 1; Supplementary Fig. S1). Watery calluses (Supplementary Fig. S1b) were observed to turn brown faster with prolonged subculturing, whereas green compact calluses (Supplementary Fig. S1c), green friable calluses (Supplementary Fig. S1d) and yellowish green friable calluses (Supplementary Fig. S1e) were found to be growing normally. The healthy growing calluses were considered for organogenesis and therefore transferred to MS basal medium fortified with different types of cytokinins alone or in combination with 2, 4-D for shoot regeneration. None of the callus type was found to regenerate shoots, rather each type of callus behaved differently on regeneration media. The green compact calluses were found to grow profusely maintaining similar nature up to 3–4 cycles of subculturing. Likewise, green and yellowish green friable calluses were found to undergo necrosis on regeneration media in most of the cases. In contrast, green friable calluses showed occasional shoot bud like structures (indicated with arrows) on medium supplemented with TDZ, which eventually did not develop into shoots (Supplementary Fig. S1f). On the other hand, root induction from yellowish green friable calluses was observed on addition of TDZ and 2, 4-D rather than shoot regeneration (Supplementary Fig. S1g).
Effect of growth regulators and media on direct regeneration
Among the explants tested, embryonic axis and plumule tips were found to produce shoots, whereas cotyledon and de-embryonated cotyledon did not show any positive response. However, plumule tips were observed as competent explant for shoot regeneration (~ 78%) than embryonic axis (Supplementary Table S3). Therefore, plumule tips were considered further for direct regeneration capacity. Based on the basal media composition assessment MS was found to be more effective in inducing shoot generation (~ 78.89%) than the other basal media (Table 2). However, combining media components with B5 macronutrients and MS micronutrients (BM6 medium) resulted in more positive shoot regeneration response (~ 86.67%). Interestingly, the shoot regeneration frequency was further enhanced (~ 94.44%), when MS and B5 macronutrient components were diluted to half of its original strength (BM11). Approximately, ~ 5 shoots per explants were observed under the said condition. Henceforth, BM11 medium was considered suitable for shoot regeneration capability from plumule tips.
Effect of growth regulators and media on shoot multiplication
The BM11 medium supplemented with varying concentrations of cytokinins from different sources were assessed for multiple shoot induction capacity (Table 3; Fig. 4). BM11 medium containing BAP or zeatin was found to induce approximately 4–7 shoots from each explant. On the other hand, BM11 medium containing kinetin induced lesser number (~ 2–6 shoots) of explants. In contrast, BM11 medium containing TDZ alone was observed to produce high number of shoots (~ 7–12 shoots/explant). The shoot induction efficiency was found to increase further to ~ 12–19 shoots/explants on addition of BAP (0.1 mg/L) along with TDZ (BM11-TB1). Furthermore, addition of NAA along with TDZ negatively impacted on multiple shoot induction efficiency of the explants.
Representative pictures showing multiple shoot induction efficiency of plumule tips on multiple shooting medium. a Explants observed to produced only ~ 5 shoots on BM11 medium supplied with 0.25 mg/L BAP. b BM11 medium supplemented with BAP (> 0.25 mg/L) was observed to induced ~ 4–7 shoots from each explants, that were succulent in nature. c BM11 medium containing zeatin induced ~ 4–7 shoots per explants, that were healthy in nature. d Kinetin supplemented BM11 medium showed least multiple shoot induction efficiency (~ 2–6 shoots per explant). e TDZ was found to be efficiently produce ~ 7–12 shoots/explants. f Highest multiple shoot (~ 16–19 shoots) formed on BM11 medium supplemented with TDZ (1 mg/L) and BAP (0.1 mg/L)
Shoots obtained on TDZ, zeatin and kinetin supplemented media were healthy in nature whereas shoots produced on BAP (> 0.25 mg/L) containing media were found to become vitrified. However, low concentration of BAP (0.1 mg/L) in combination with TDZ in medium (BM11-TB1) was found to have no adverse effect on the physiology of regenerated shoots rather enhanced the multiple shoot induction efficiency of the explants (Table 3). The results indicated that TDZ (1 mg/L) along with BAP (0.1 mg/L) was found to be appropriate for growth regulators that actually promoted large number of healthy shoots per explant.
Effect of growth regulators and media on root induction from the shootlets
The effect of SH medium on induction of multiple roots can be observed in Table 2. Full strength of SH medium without any aid of auxin (RM1) was found to promote root growth from shootlets, initiated around 2–3 roots per shoot with ~ 62% frequency (Supplementary Table S4). However, modification of SH medium by supplementing with MS macronutrients (RM2) or B5 macronutrients (RM3) in place of SH macronutrients was noticed to enhance the root formation efficiency by ~ 76% and ~ 65% respectively. Significant increment in root induction efficiency (~ 97%) was observed in RM4 medium prepared on diluting the MS macronutrients salt concentration to half of its original strength.
Different types of auxins at varying concentrations were tested for further enhancement of root induction efficiency (Data not shown). However, auxins were not found to have any significant effect on root induction from shootlets. Instead, addition of NAA and IAA at a concentration of 0.5–1.5 mg/L concentrations resulted in growth of basal callus that interfered with the shoot growth. Furthermore, higher concentration (2–4 mg/L) of all the three auxins affected the shootlets to become succulent, followed by tissue necrosis leading to shoot fatality. Thus, it could be suggested that RM4 medium alone is suitable for root induction. Well-developed multiple roots were observed after 10–14 days of culture on root induction medium (SupplementaryFig. S2a). Shoots with multiple roots were transferred to glass bottles containing same medium and allowed to grow full plantlets until 6—7 cm height was attained (Supplementary Fig. S2b).
Effect of the optimized media on shoot regeneration and multiplication in different S. indicum varieties
The optimized culture conditions were applied to different S. indicum genotypes to test their regeneration and multiple shoot induction efficiency (Supplementary Table S5). It was observed that with the newly developed modified regeneration condition helped in shoot induction in genotypic independent way. However, varied degrees of response were observed. Apart from Rama, CO1, Phule til and Tilottama varieties were found to be more effective in regenerating and shoot multiplication capability. However, least regeneration and multiple shoot formation were observed for CST2001 variety of S. indicum.
Effect of hygromycin on the growth of untransformed shoots
Hygromycin concentration up to 35 mg/L was found insufficient to completely suppress shoot growth. Few of them were able to survive in presence of hygromycin (Supplementary Fig. S3a-c), while medium containing 40 mg/L of hygromycin was observed to drastically affect shootlets health that resulted in complete elimination of shootlets (Supplementary Fig. S3d).
Influence of Agrobacterium cell density and co-cultivation period on transformation
In the present study, bacterial culture density and co-cultivation period were considered as the crucial factors affecting transformation efficiency. However, it was observed that bacterial cell density had remarkable effect on regeneration frequency. Probably, the increasing bacterial culture density decreased the shoot regeneration frequency of treated explants. Least regeneration frequency was observed for explants infected with Agrobacterium culture of 0.8 OD600 (Supplementary Fig. S4a). In contrast, increase in bacterial density upto 0.6 OD600 was found to increase transformation efficiency (number of hygromycin resistance shoots with respect to number of explants regenerated upon cocultivation). Explants infected with bacterial density of 0.8 OD600 could not survive due to bacterial overgrowth, hence no transformants were recovered. Similarly, co-cultivation period was found to play an important role in increasing transformation efficiency. The co-cultivation of infected explants (explants infected with Agrobacterium culture of OD600 0.6) for 72 h was found to increase the transformation efficiency (Supplementary Fig. S4b).
Following the optimized condition for Agrobacterium mediated transformation method, 44, 32 and 38 putative transformed shoots were recovered from three independent transformation attempts respectively (Table 4; Fig. 3). All the shoots produced roots on rooting media (RM4) and successfully transferred to soil. The putative transformant lines were subjected to molecular and histological analysis.
Molecular analyses of putative T 0 transformants
On PCR analysis, amplification of 1026 bp in six (GT01, GT03, GT04, GT05, GT06 and GT08)of the transformants indicated the presence of hptII gene (Fig. 5a; Table 4). Moreover, rest of the putative transformants (GT02 and GT07) did not show any amplification. As expected, amplification of 717 bp fragmentwas observed in six transformants (GT01, GT03, GT04, GT05, GT06 and GT08) in PCR analysis (Fig. 5b). However, identical amplifications were absent in untransformed control in both cases (Fig. 5a, b). Southern hybridization using gfp gene specific probe further proved the presence of gfp transgene in the genome of all PCR positive transgenic lines (Fig. 5c). Among them, 3 plants (GT03, GT04 and GT08) showed single integration, while 2 plants (GT01 and GT05) exhibited double and one plant (GT06) displayed multiple integrations. Furthermore, individuality of transformants was identified by different transgene integration pattern. Based on Southern analysis results, the average transformation frequency of the three independent transformation attempts is (Number of transformants showing transgene integration with respect to total explants taken for transformation experiments) calculated to be 1.33% (Table 4). The transgenic lines with single integration (GT03, GT04 and GT08) were considered for further analysis in order to avoid any possibility of co-suppression (Fig. 6).
Representative figures depicting detection of the transgene in transformants. a Ethidium bromide stained agarose gel showing amplification of hptII gene using gene specific primers. UC denotes untransformed control plant, GT01-GT08 indicates putative transgenic lines, PC denotes positive control and M denotes molecular size marker (pUC18 digested with HinfI). b Ethidium bromide stained agarose gel showing amplification of gfp gene using gene specific primers. UC denotes untransformed control plant, GT01-GT08 indicates putative transgenic lines, PC denotes positive control and M denotes molecular size marker (pUC18 digested with HinfI). c Autoradiogram of Southern blot analysis showing the presence of transgene as well as difference in integration pattern in T0 putative transgenic sesame lines. UC denotes untransformed control, GT01-GT08 indicates putative transformants, PC denotes positive control and M denotes molecular size marker (EcoRI/HindIII digested λ DNA)
Analysis of gfp gene expression at transcript level in sesame
The northern autoradiogram showed the existence of transcripts of gfp in varied proportions in transgenic plants whereas no such expression was detected in untransformed line (Fig. 7a). The variation in expression of gfp transgene among transformants could possibly be due to variation in site of integration of the transgenes which is evident from Southern blot analysis. High level of expression was found in GT03 followed by GT04 and GT08 respectively as revealed by real time PCR (Fig. 7b). There was no such amplification in untransformed control sample; therefore, for relative quantification of transgene in transformants the least expressing line GT08 (evident from northern blot analysis) was taken as control.GT03 transgenic line was found to show 4.9-fold higher expressions with respect to GT08 line. The least expressing transgenic line (GT08) was considered as control to determine relative fold expression in other two lines (GT03 and GT04). The GT03 line was found to show higher expression followed by GT04 and GT08 respectively. Therefore, the expression levels in figure representing real time data (Fig. 7b) are shown in reverse order. The transgenic line with higher transcript expression compared to other lines was considered for histological analyses.
Analyses of transcript expression of gfp in untransformed and T0 transgenic lines. a Northern blot analysis showing variation in transcript expression of gfp among T0 transgenic lines. UC indicates untransformed control plant. GT03, GT04 and GT08 denote transgenic lines. The blots were subsequently stripped and re-hybridized with β-actin gene probe to demonstrate uniform loading of total RNA. b Bar diagram representing relative gfp transcript expression in the transformants. The actin gene was used as the internal control for normalization
Histological analysis of GFP fluorescence by confocal microscopy
The tissues of transformed lines were found to show green fluorescence confirming the expression of GFP protein (Fig. 8). In contrast, the untransformed tissue did not show green fluorescent signals. These results suggest successful expression of gfp transgene in transformed sesame lines.
Expression of GFP in petiole tissue of the transgenic sesame lines. Upper panel a–d represents DIC image, middle panel e–h showing green fluorescence signal and lower panel i–l showing overlay projection of DIC and GFP fluorescence. a, e and i represent tissue of untransformed control (scale bar in red—40 µm). b, f and j represent tissue of GT03 (scale bar in red—30 µm). c, g and k represent tissue of GT04 (scale bar in red—40 µm). d, h and l represent tissue of GT08 (scale bar in red—40 µm)
Discussion
Regeneration and Agrobacterium-mediated transformation has been successfully achieved in several oilseed crop species, viz., sunflower (Sujatha et al. 2012), Safflower (Shilpa et al. 2010; Belide et al. 2011), rapeseed (Boszoradova et al. 2011), groundnut (Tiwari et al. 2015) and sesame (Yadav et al. 2010; Al-Shafeay et al. 2011). However, almost all the studies suggested about genotype dependent regeneration and transformation system. Genotype dependent regeneration and transformation is the major constraint in genetically modified crops including sesame. Although, these studies have discussed the effect of factors such as genotype, explant type, explant age, growth regulator type and concentration, Agrobacterium bacterial cell density, cocultivation period etc., on regeneration and transformation efficiency. Therefore, an attempt was made in the present study to standardize regeneration and transformation system for S. indicum.
Regeneration through indirect organogenesis under the influence of media and variable concentration of growth regulators resulted in better induction of calli formation from cotyledons compared to other explants on MS medium containing 2, 4 D (0.25 mg/L) (Supplementary Table S1). MS medium was observed to have influential effect on callus initiation and growth of tissues in several plant spices including different varieties of sesame (Zouzou et al. 2000; Kouakou, 2003; Baskaran and Jayabalan 2006; Michel et al. 2008; Głowacka et al. 2010; Shashidhara et al. 2011a; Yuan et al. 2013; Pusadkar et al. 2015; Gerszberg et al. 2016; Osman et al. 2016). The induction aggravated further around 71% with 2, 4-D at 2 mg/L concentration (Supplementary Table S2). The finding was in agreement with the earlier observations in callus induction in other sesame varieties (Jeyamary and Jayabalan 1997; Chakraborti and Ghosh 2009). On the contrary, few studies showed NAA to be the suitable auxin for callus induction in S. indicum (Baskaran and Jayabalan 2006; Rao and Honnale 2011). The variation in callus response could possibly be due to difference in endogenous auxin among the genotypes (Khemkladngoen et al. 2011; Zimik and Arumugam 2017). Furthermore, supplementation of the medium with cytokinins was shown to have callus promoting effect in different plant species including Malus domestica (Caboni et al. 2000) and Holostemmaada-kodien (Martin 2002) as well as in S. indicum cv. SVPR—1 (Raja and Jayabalan 2010). In consistent with previous findings, addition of BAP along with 2, 4-D in MS basal medium was found to enhance callus induction frequency (~ 89%) remarkably from cotyledons in the present study (Table 1). However, the combination of cytokinins and 2, 4-D was found to produce calluses of varied texture (Supplementary Fig. S1a–e). In sesame, induction of various kinds of calluses in presence of 2, 4-D and BAP was already established in previous studies by Baskaran and Jayabalan (2006) and Pusadkar et al. (2015). Unfortunately, none of the callus types was found to regenerate shoots in cytokinin supplemented regeneration medium except the green friable calluses, those were found to show shoot bud like structures occasionally. Eventually, the shoot buds failed to develop into shoots (Supplementary Fig. S1f). In addition, yellowish green friable callus regenerated roots rather than shoots were observed (Supplementary Fig. S1g). The present finding was in concurrence with the earlier studies wherein the efficiency of regeneration through indirect organogenesis was limited by genotypic variation (Rao and Honnale 2011; Pusadkar et al. 2015). This unsuccessful event of shoot regeneration from calluses led to adopt direct regeneration strategy. Direct regeneration from various explants of immature inflorescence, cotyledonary node, leaf, apical shoot bud and cotyledon was achieved in other crops includes giant miscanthus, soybean, pistachio and sesame (Ma and Wu 2008; Tilkat et al. 2009; Yadav et al. 2010; Perera et al. 2015). In direct organogenesis, plumule tip was found to result in high regeneration frequency than embryonic axis, while cotyledon explants were failed to produce shoots. In contrast, it underwent necrosis when cultured for long hours on MS supplemented medium with BAP (0.25 mg/L) (Supplementary Table S3). Likewise, necrosis of cotyledon tissue on cytokinins supplemented media was also observed in previous studies (Baskaran and Jayabalan 2006; Were et al. 2006). The plumule tip was observed to be the most potential explant undergoing regeneration experiments in many plant species such as pigeon pea, cowpea, peanut, lentil and chick pea (Surekha et al. 2007; Aasim et al. 2009, 2013; Singh and Hazra 2009; Aasim 2012). By culturing plumule tip explants on different media helped in assessing the effect of basal media composition on regeneration frequency. It was observed that by replacing MS micronutrients with B5 macronutrient in MS basal media had encouraged in shoot regeneration from explant up to ~ 86% as opposed to full MS basal media ~ 78% (Table 2). However, the shoots emerged were turning yellowish and the growth was stunted after 5–6 days of regeneration. This was probably due to the presence of high nitrate ions that made the media less acidic that interfered with nitrate uptake and resulted in stunted plant growth (George et al. 2008).Therefore, a modified basal medium was formulated by combining half the concentration of MS and B5 macronutrients of their original strength (BM11) and was used to check their effect on regeneration frequency. Surprisingly, the media was observed to enhance regeneration frequency up to ~ 94%, while managing good plant growth (Table 2). These results supported the earlier observation that the presence of moderate proportion of NO3−/NH4+ could be an essential factor to retain good plant growth (Ramage and Williams 2002; Were et al. 2006). Furthermore, combination of MS and B5 media components has already been proven to be effective in increasing regeneration efficiency in Psidium guava (Chandra et al. 2004) and in Cichoriumintybus (Nandagopal and Kumari 2006).
Cytokinins are known to have an inducing effect on regeneration of shoots from explants of S. indicum (Shashidhara et al. 2011b; Malaghan et al. 2013). Among the cytokinins tested in the present study, TDZ (1 mg/L) was found to be superior over other cytokinins for inducing maximum number of shoots from excised plumule tip explant (~ 7–12 shoots/explant) (Table 3; Fig. 4). Furthermore, addition of BAP (0.1 mg/L) at low concentration along with TDZ was found to enhance the shoot multiplication tendency of plumule tip (~ 16–19 shoots/explant). A combinatorial effect of TDZ and BAP in promoting shoot multiplication has already been documented in different varieties of S. indicum (Shashidhara et al. 2011b; Malaghan et al. 2013; Chowdhury et al. 2014).
Apart from shoot formation, root regeneration is also an essential part of tissue culture to develop a complete plant. Among the defined basal media, MS and SH basal media were used for root regeneration from various plant species and indicated that the efficiency of root generation in SH was better over MS media (Vooková and Kormuťák 2001; Baque et al. 2010; Cui et al. 2010; Chae 2014). High root induction efficiency of SH medium was suggested to be due to high thiamine concentration compared to other basal media. High thiamine concentration was found to have root promoting effect in other plants (Chee 1995; Sepahvand et al. 2012). However, the root regeneration efficiency in S. indicum cv. Rama was found to be moderate (~ 62%) in SH medium (Supplementary Table S4), while the shoot growth was affected inversely. Furthermore, in the present study the retardation of shoot growth was probably due to the effect of high nitrate concentration. Like shoot regeneration study, to overcome the shoot growth problem, amalgamation of SH media components with other basal media components were tested for root induction in S. indicum cv. Rama (Supplementary Table S4). It was found that the SH media modified by replacing its own macronutrient content with MS macronutrients was found to increase the root induction efficiency up to ~ 76%. Further, dilution of MS macronutrient salt concentration to half of its original strength remarkably enhanced root regeneration efficiency (~ 98%) (Supplementary Fig. S2a). The results indicated that macronutrient composition had greatly influenced the root regeneration. This showed that salt strength of the media could be another factor responsible for root induction. The result was in agreement with the previous findings for Chrysanthemum sp. (Verma 2012). Variation in shoot regeneration efficacy among different genotypes was reported in previous studies on sesame (Were et al. 2006; Al-Shafeay et al. 2011; Malaghan et al. 2013). In the present study, it was observed that direct regeneration was suitable for in vitro tissue culture of sesame than indirect regeneration. The optimized direct regeneration protocol was found to influence shoot regeneration and multiplication in different varieties of S. indicum, but with varying extent depending on the varieties (Supplementary Table S5). Thus, it could be suggested that the developed tissue culture system can be a better method in comparison to previous studies reported for other sesame varieties as well.
Agrobacterium-mediated transformation in plant species is a widely accepted method of choice as it results in integration of lower copy number and least rearrangement of transgene (Karami 2008). Thus, a reliable and reproducible Agrobacterium-mediated transformation system for S. indicum cv. Rama was established using pCAMBIA/gfp cassette (Fig. 1). However, a remarkable effect of density of Agrobacterium culture harboring pCAMBIA/gfp cassette and co-cultivation period, on regeneration as well as transformation frequency was observed.The regeneration frequency was found to decrease with the increasing density of the Agrobacterium culture, while transformation frequency was found to gradually increase with increasing density of Agrobacterium suspension up to 0.6 OD600 and then started to decrease (Supplementary Fig. S4a). The transformation frequency was found to increase further with the increase of co-cultivation period to 72 h (Supplementary Fig. S4b). The results were in agreement with the previous observations that suggested high bacterial inoculum concentrations and longer cocultivation period had resulted into rapid tissue necrosis and cell death due to hypersensitive response around the infection site (Hansen 2000; Richter and Ronald 2000; Chakrabarty et al. 2002; Das et al. 2002; Opabode 2006; Hasnatet al. 2008; Yadav et al. 2010; Al-shafeay et al. 2011; Sun et al. 2011; Ziemienowicz 2014). Finally, Agrobacterium-mediated transformation of pCAMBIA/gfp cassette resulted in generation of only eight prospective hygromycin resistant plants. The transformants were preliminary analysed by PCR (Fig. 5a and 5b) followed by Southern analysis (Fig. 5c). Integration of the gfp transgene into the genome of S. indicum was found to vary in number and in loci as revealed by Southern blot analysis. Single integration was observed in GT03, GT04 and GT08 while GT01 and GT05 displayed double integration and GT06 displayed multiple integrations as evidenced by Southern blot analysis. Moreover, the integration pattern of the transgene was found to be different among all the transformants as well as among the transformants with single integration.
The transgenic lines with single integration (GT03, GT04 and GT08) were considered for further analysis in order to avoid any possibility of co-suppression (Fig. 6). In spite of having single gfp transgene, northern blot analysis of the transformants revealed different level of expression of gfp gene in the independent T0 transformants and 4.9-fold higher expression was observed in GT03 line compared to GT08 line as revealed by real time PCR data analysis (Fig. 7). The occurrence of variation in the expression of gfp transgene, in the present study, could possibly be explained by site of integration of the transgenes as has been suggested previously (Matzke and Matzke 1998; Day et al. 2000). The finding was further validated by histological analysis of the transversely dissected leaf petiole tissue of transgenic lines and wild type plants. Green fluorescence was observed in tissue sections of transgenic lines while such fluorescence was absent in control plant (Fig. 8). This confirmed the presence of GFP protein in the transgenic line only.
Based on the Southern analysis data the calculated transformation frequency in the present study was found to be 1.33% (number of plants showing positive response in Southern analysis with respect to number of explants infected). However, previous studies claimed transformation efficiencies determined for the transformants against the number of explants regenerated after infection (Yadav et al. 2010; Chowdhury et al. 2014). The present result is in agreement with the previous findings observed in S. indicum (Kapoor et al. 2015) but in contrast with the findings of Chowdhury et al. (2014). Chowdhury et al. (2014) suggested high Agrobacterium culture density (OD6001.6) and cocultivation of infected explants for one day resulted in high transformation efficiency, however following these conditions failed to produce any transgenic in the present study. Low Agrobacterium culture density and longer co-cultivation duration was found to be suitable for transformation of the genotype used in the present study and in case of other sesame genotypes as well (Yadav et al. 2010; Al-shafeay et al. 2011).This indicated that the transformation efficiency in S. indicum is highly variety specific (Chowdhury et al. 2014). Factors influencing transformation efficiency varies for different genotypes.
In conclusion, the present study could be considered as a major breakthrough in sesame research with respect to establishment of regeneration and transformation protocol of sesame which is highly recalcitrant in nature. The genotypes chosen in the present study are widely cultivated varieties, among them cv. Rama is high yielding variety and also possess high oil content. This is the first study where regeneration and transformation protocols have been developed for cultivars of commercial importance. Most of the genotypes was found to be responded to the optimized regeneration media, hence could be used for further biotechnological studies. The transformation system developed in the present study is efficient and easy. Transformation efficiency reported here might not be too high but could be of help for transformation of similar genotypes or more recalcitrant species of sesame.
References
Aasim M, Khawar KM, Özcan S (2009) In vitro micropropagation from plumular apices of Turkish cowpea (Vigna unguiculata L.) cultivar Akkiz. Sci Hortic 122:468–471
Aasim M (2012) Micropropagation of lentil (Lens culinarisMedik.) using pulse treatment of immature plumular apices. Pak J Agric Sci 49:149–154
Aasim M, Day S, Rezaei F, Hajyzadeh M (2013) Multiple shoot regeneration of plumular apices of chickpea. Turk J Agric For 37:33–39
Al-Shafeay AF, Ibrahim AS, Nesiem MR, Tawfik MS (2011) Establishment of regeneration and transformation system in Egyptian sesame (Sesamum indicum L.) cv Sohag1. GM Crops 2:182–192
Ashri A (2010) Sesame breeding. Plant breeding reviews. Wiley, Hoboken, pp 179–228
Baskaran P, Jayabalan N (2006) In vitro mass propagation and diverse callus orientation on Sesamum indicum L.-an important oil plant. J Agric Technol 2:259–269
Baque MA, Hahn E, Paek K (2010) Induction mechanism of adventitious root from leaf explants of Morindacitrifolia as affected by auxin and light quality. In Vitro Cell Dev Biol Plant 46:71–80
Belide S, Hac L, Singh SP, Green AG, Wood CC (2011) Agrobacterium-mediated transformation of safflower and the efficient recovery of transgenic plants via grafting. Plant Methods 7:12
Boszoradova E, Libantova J, Matusikova I, Poloniova Z, Jopcik M, Berenyi M, Moravcikova J (2011) Agrobacterium-mediated genetic transformation of economically important oilseed rape cultivars. Plant Cell Tissue Organ Cult 107:317–323
Caboni E, Lauri P, Angell SD (2000) In vitro plant regeneration from callers of shoot apices in apple shoot culture. Plant Cell Rep 19:755–760
Chae SH (2014) Influence of media on in vitro root regeneration and micropropagation of Chrysanthemummorifolium Ramat cv. Hwiparam. Life Sci J 11:797–799
Chakrabarty R, Viswakarma N, Bhat SR, Kirti PB, Singh BD, Chopra VL (2002) Agrobacterium-mediated transformation of cauliflower: optimization of protocol and development of Bt-transgenic cauliflower. J Biosci 27:495–502
Chakraborti P, Ghosh A (2009) Variation in callus induction and root shoot bud formation depend on seed coat of sesame genotypes. Res J Bot 5:14–19
Chandra R, Bajpai A, Gupta S, Tiwari RK (2004) Embryogenesis and plant regeneration from mesocarp of Psidium guava L. Indian J Biotechnol 3:246–248
Chattopadhyaya B, Banerjee J, Basu A, Sen SK, Maiti MK (2010) Shoot induction and regeneration using internodal transverse thin cell layer culture in Sesamum indicum L. Plant Biotechnol Rep 4:173–178
Chee PP (1995) Stimulation of adventitious rooting of Taxus species by thiamine. Plant Cell Rep 14:753–757
Chomczynski P, Sacchi N (1987) Single-step method RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159
Chongdar S, Chhetri B, Mahato SK, Saha A (2015) Production potentials and economic feasibility of improved sesame (Sesamum indicum L.) cultivars under varying dates of sowing in prevailing agro-climatic condition of North Bengal. Int J Agric Sci 7:434–439
Chowdhury S, Basu A, Kundu S (2014) A new high-frequency Agrobacterium-mediated transformation technique for Sesamum indicum L. using de-embryonated cotyledon as explant. Protoplasma 251:1175–1190
Chu CC, Wang CS, Sun CC, Hsu C, Yin KC, Chu CY (1975) Establishment of an efficient medium for another culture of rice through comparative experiments on the nitrogen sources. Sci Sinica 18:659–668
Cui X, Chakrabarty D, Lee E, Paek K (2010) Production of adventitious roots and secondary metabolites by Hypericum perforatum L. in a bioreactor. Bioresour Technol 101:4708–4716
Das DK, Reddy MK, Upadhyaya KC, Sopory SK (2002) An efficient leaf-disk culture method for the regeneration via somatic embryogenesis and transformation of grape (Vitisvinifera L.). Plant cell Rep 20:999–1005
Day DC, Lee E, Kobayashi J, Holappa DL, Albert H, Ow DW (2000) Transgene integration into the same chromosome location can produce alleles that express at a predictable level, or alleles that are differentially silenced. Genes Dev 14:2869–2880
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Gamborg OL, Miller RA, Ojima K (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50:151–158
George EF, Hall MA, Klerk GD (2008) The components of plant tissue culture media I: macro- and micro-nutrients. In: George EF, Hall MA, De Klerk GJ (eds) Plant propagation by tissue culture, 3rd edn. Springer, New York, pp 65–113
Gerszberg A, Hnatuszko-Konka K, Kowalczyk T, Kononowicz AK (2016) Efficient in vitro callus induction and plant regeneration protocol for different polish tomato cultivars. NotulaeBotanicaeHortiAgrobotanici Cluj-Napoca 44:452–458
Głowacka K, Jeżowski S, Kaczmarek Z (2010) The effects of genotype, inflorescence developmental stage and induction medium on callus induction and plant regeneration in two Miscanthus species. Plant Cell Tissue Organ Cult 102:79–86
Hansen G (2000) Evidence for Agrobacterium-induced apoptosis in maize cells. Mol Plant-Microbe Interact 13:649–657
Hasnat R, Abbasi NA, Hafiz IA, Ahmad T, Chudhary Z (2008) Effect of different bacterial dilutions on transformation efficiency of hot chilli (Capsicumfrutescens L.) varieties. Pak J Bot 40:2655–2662
Hirose N, Inoue T, Nishihara K, Sugano M, Akimoto K, Shimizu S, Yamada H (1991) Inhibition of cholesterol absorption and synthesis in rats by sesamin. J Lipid Res 32:629–638
Honnale H, Rao S (2013) Direct somatic embryogenesis in Sesamum indicum (L.) Cv-E8 from cotyledon and hypocotyl explants. Int J Appl Biol Pharm Technol 4:120–127
Jeyamary R, Jayabalan N (1997) Influence of growth regulators on somatic embryogenesis in Sesamum indicum L. Plant Cell Tissue Organ Cult 46:67–70
Kapoor S, Parmer SS, Yadav M, Chaudhary D, Sainger M, Jaiwal R, Jaiwal PK (2015) Sesame (Sesamum indicum L.). In: Wang K (ed) Agrobacterium protocols. Methods in molecular biology. Humana Press, Totowa, pp 37–45
Karami O (2008) Factors affecting Agrobacterium-mediated transformation of plants. Transgenic Plant J 2:127–137
Khemkladngoen N, Cartagena J, Shibagaki N, Fukui K (2011) Adventitious shoot regeneration from juvenile cotyledons of a biodiesel producing Plant, Jatropha curcas L. J Biosci Bioeng 111:67–70
Kouakou TH (2003) Contribution à l’étude de l’embryogenèsesomatique chez le cotonnier: évolution de quelquesparamètresbiochimiques au cours de la callogenèse et de la culture de suspension cellulaires. Thèse de doctorat N° 023/2003, Université de Cocody, Abidjan, Côte d’Ivoire
Kulkarni VV, Ranganatha CN, Shankergoud I (2017) Interspecific crossing barriers in Sesame (Sesamum indicum L.). Int J Curr Microbiol Appl Sci 6:4894–4900
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408
Lokesha R, Rahaminsab J, Ranganatha ARG, Dharmaraj PS (2012) Whole plant regeneration via adventitious shoot formation from de-embryonated cotyledon explants of sesame (Sesamum indicum L.). World J Sci Technol 2:47–51
Ma XH, Wu TL (2008) Rapid and efficient regeneration in soybean (Glycine max L. Merrill) from whole cotyledonary node explants. Acta Physiologiae Plant 30:209–216
Malaghan SV, Lokesha R, Savitha R, Ranganatha ARG (2013) Adventitious shoot regeneration in Sesame (Sesamum indicum L.) (Pedaliaceae) via deembryonatedcotyledonary explants. Res J Biol 1:31–35
Martin KP (2002) Rapid propagation of Holostemmaada-kodienSchult., a rare medicinal plant, through axillary bud multiplication and indirect organogenesis. Plant Cell Rep 21:112–117
Matzke AJ, Matzke MA (1998) Position effects and epigenetic silencing of plant transgenes. Curr Opin Plant Biol 1:142–148
Michel Z, Hilaire KT, Mongomaké K, Georges AN, Justin KY (2008) Effect of genotype, explants, growth regulators and sugars on callus induction in cotton (Gossypium hirsutum L.). Aust J Crop Sci 2:1–9
Miyahara Y, Hibasami H, Katsuzaki H, Imai K, Komiya T (2001) Sesamolin from sesame seed inhibits proliferation by inducing apoptosis in human lymphoid leukemiaMolt 4b cells. Int J Mol Med 7:369–371
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plant 15:473–497
Nakai M, Harada M, Nakahara K, Akimoto K, Shibata H, Miki W, Kiso Y (2003) Novel antioxidative metabolites in rat liver with ingested sesamin. J Agric Food Chem 51:1666–1670
Nakano D, Kurumazuka D, Nagai Y, Nishiyama A, Kiso Y, Matsumura Y (2008) Dietary sesamin suppresses aortic NADPH oxidase in DOCA salt hypertensive rats. Clin Exp Pharmacol Physiol 35:324–326
Nandagopal S, Kumari BDR (2006) Adenine sulphate induced high frequency shoot organogenesis in callus and in vitro flowering of Cichoriumintybus L. cv. Focus - a potent medicinal plant. Acta agriculturaeSlovenica 87:415–425
Nzikou JM, Matos L, Bouanga-Kalou G, Ndangui CB, Pambou-Tobi NPG, Kimbonguila A, Silou Th, Linder M, Desobry S (2009) Chemical composition on the seeds and oil of Sesame (Sesamum indicum L.) grown in Congo-Brazzaville. Adv J Food Sci Technol 1:6–11
Opabode JT (2006) Agrobacterium-mediated transformation of plants: emerging factors that influence efficiency. Biotechnol Mol Biol Rev 1:12–20
Oplinger ES, Putnam DH, Kaminski AR, Hanson CV, Oelke EA, Schult EE, Doll JD (1990) Sesame, Alternative field crops manual. www.hort.purdue.edu/sesame.html
Osman NI, Sidik NJ, Awa A (2016) Effects of variations in culture media and hormonal treatments upon callus induction potential in endosperm explant of Barringtoniaracemosa L. Asian Pac J Trop Biomed 6:143–147
Pathak N, Rai AK, Saha S, Walia S, Sen SK, Bhat KV (2014) Quantitative dissection of antioxidative bioactive components in cultivated and wild sesame germplasm reveals potentially exploitable wide genetic variability. J Crop Sci Biotechnol 17:127–139
Perera D, Barnes DJ, Baldwin B, Reichert N (2015) Direct and indirect in vitro regeneration of Miscanthus × giganteus cultivar Freedom: effects of explant type and medium on regeneration efficiency. In Vitro Cell Dev Biol Plant 51:294–302
Pusadkar PP, Kokiladevi E, Aishwarya V, Gnanam R, Sudhakar D, Balasubramanian P (2015) Efficacy of growth hormone for callus induction in sesame (Sesamum indicum L.). J Cell Tissue Res 15:5067–5071
Pusadkar P, Kokiladevi E, Shilpa B (2016) Efficacy of plant growth hormones for shoot induction and regeneration in Sesame (Sesamum indicumL.). Res J Biotechnol 11:27–30
Raja A, Jayabalan N (2010) Callus induction and plantlet regeneration from leaf explant of sesame (Sesamum indicum L. cv. SVPR-1). J Swamy Bot Club 27:93–98
Ramage CM, Williams RR (2002) Mineral nutrition and plant morphogenesis. In Vitro Cell Dev Biol Plant 38:116–124
Rao S, Honnale HN (2011) Callus induction and organogenesis in Sesamum indicum L. Cv. E 8. Curr Trends Biotechnol Pharm 5:1462–1468
Richter TE, Ronald PC (2000) The evolution of disease resistance genes. Plant Mol Biol 42:195–204
Sagar R, Ganesh C (2004) Frontline demonstration on sesame in West Bengal. Agric Ext Rev 16:7–10
Sankar D, Sambandam G, Ramakrishna RM, Pugalendi KV (2005) Modulation of blood pressure, lipid profiles and redox status in hypertensive patients taking different edible oils. Clin Chemica Acta 355:97–104
Saravanan S, Nadarajan N (2005) Effect of media supplements on in vitro response of sesame (Sesamum indicum L.) genotypes. Res J Agric Biol Sci 1:98–100
Schenk RU, Hildebrandt AC (1972) Medium and techniques for induction and growth of monocotyledonous and dicotyledonous plant cell cultures. Can J Bot 50:199–204
Seo HY, Kim YJ, Park TI, Kim HS, Yun HS (2007) High-frequency plant regeneration via adventitious shoot formation from deembryonated cotyledon explants of Sesamum indicum L. In Vitro Cell Dev Biol Plant 43:209–214
Sepahvand S, Ebadi A, Kamali K, Ghaemmaghami SA (2012) Effect of myo-inositol and thiamine on micropropagation of GF677 (Peach × Almond hybrid). J Agric Sci 4:275–280
Shashidhara N, Ravikumar H, Ashoka N, Santosh DT, Pawar P, Lokesha R, Janagoudar BS (2011a) Callus induction and subculturing in sesame (Sesamum indicum L.): a basic strategy. Int J Agric Environ Biotechnol 4:153–156
Shashidhara N, Ravikumar H, Ashoka N, Santosh DT, Pawar P, Lokesha R, Janagoudar BS (2011b) Thidiazuron (TDZ) induced shoot regeneration in sesame (Sesamum indicum L.). Int J Agric Environ Biotechnol 4:41–44
Shilpa KS, Kumar VD, Sujatha M (2010) Agrobacterium-mediated genetic transformation of safflower (Carthamustinctorius L.). Plant Cell Tissue Organ Cult 103:387–401
Siao AC, Hou CW, Kao YH, Jeng KC (2015) Effect of sesamin on apoptosis and cell cycle arrest in human breast cancer MCF-7 cells. Asian Pac J Cancer Prev 16:3779–3783
Singh S, Hazra S (2009) Somatic embryogenesis from the axillary meristems of peanut (Arachis hypogaea L.). Plant Biotech Rep 3:333–340
Sujatha M, Vijay S, Vasavi S, Reddy PV, Rao SC (2012) Agrobacterium-mediated transformation of cotyledons of mature seeds of multiple genotypes of sunflower (Helianthus annuus L.). Plant Cell Tissue Organ Cult 110:275–287
Sun Q, Zhao Y, Sun H, Hammond RW, Davis RE, Xin L (2011) High efficiency and stable genetic transformation of pear (Pyrus communis L.) leaf segments and regeneration of transgenic plants. Acta Physiologiae Plant 33:383–390
Surekha C, Arundhati A, Seshagiri Rao G (2007) Differential response of Cajanus cajan varieties to transformation with different strains of Agrobacterium. J Biol Sci 7:176–181
Taskin KM, Ercan AG, Turgut K (1999) Agrobacterium tumefaciens-mediated transformation of Sesame (Sesamum indicum L.). Turk J Bot 23:291–295
Tilkat E, Onay A, Yıldırım H, Ayaz E (2009) Direct plant regeneration from mature leaf explants of pistachio, Pistacia vera L. Sci Hortic 121:361–365
Tiwari S, Kumar S, Gontia I (2011) Biotechnological approaches for sesame (Sesamum indicum L.) and niger (GuizotiaabyssinicaLf Cass.). Asia-Pac J Mol Biol Biotechnol 19:2–9
Tiwari V, Chaturvedi AK, Mishra A, Jha B (2015) An efficient method of Agrobacterium-mediated genetic transformation and regeneration in local Indian cultivar of groundnut (Arachis hypogaea) Using Grafting. Appl Biochem Biotechnol 175:436–453
Tsuruoka N, Kidokoro A, Matsumoto I, Abe K, Kiso Y (2005) Modulating effect of sesamin, a functional lignan in sesame seeds, on the transcription levels of lipid and alcohol-metabolizing enzymes in rat liver: a DNA microarray study. Biosci Biotechnol Biochem 69:179–188
Verma OP (2012) Standardization of auxin concentration for root induction in Chrysanthemum morifolium. Adv Appl Sci Res 3:1449–1453
Vooková A, Kormuťák B (2001) Effect of sucrose concentration, charcoal and indole 3-butyric acid on germination of Abiesnumidica somatic embryos. Biologia Plant 44:181–184
Were BA, Gudu S, Onkware AO, Carlsson AS, Welander M (2006) In vitro regeneration of sesame (Sesamum indicum L.) from seedling cotyledon and hypocotyl explants. Plant Cell Tissue Organ Cult 85:235–239
Yadav M, Chaudhary D, Sainger M, Jaiwal PK (2010) Agrobacterium tumefaciens-mediated genetic transformation of sesame (Sesamum indicum L.). Plant Cell Tissue Organ Cult 103:377–386
Younghee K (2001) Effects of BA, NAA, 2, 4-D and AgNO3 treatments on callus induction and shoot regeneration from hypocotyl and cotyledon of sesame (Sesamum indicum L.). J Korean Soc Hortic Sci 42:70–74
Yuan JL, Yue JJ, Wu XL, Gu XP (2013) Protocol for callus induction and somatic embryogenesis in Moso Bamboo. PLoS ONE 8:e81954
Ziemienowicz A (2014) Agrobacterium-mediated plant transformation: factors, applications and recent advances. Biocatal Agric Biotechnol 3:95–102
Zimik M, Arumugam N (2017) Induction of shoot regeneration in cotyledon explants of the oilseed crop Sesamum indicum L. J Genet Eng Biotechnol 15:303–308
Zouzou M, Kouadio YJ, Koné M, Kouakou TH, Dénézon DO (2000) Callogenèse chez Gossypium hirsutum L.: effets cultivar, conditions de culture et type de matériel. Biot Rev Int Sci Vie Terre 1:48–56
Acknowledgements
Financial assistance in the form of the grant support to this laboratory from the Indian Council of Agricultural Research (NAIP/ICAR), Government of India, is acknowledged.
Author information
Authors and Affiliations
Contributions
TG and AB conceived and designed research, conducted experiments, analysed data and wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by K X Tang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gayatri, T., Basu, A. Development of reproducible regeneration and transformation system for Sesamum indicum. Plant Cell Tiss Organ Cult 143, 441–456 (2020). https://doi.org/10.1007/s11240-020-01931-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-020-01931-1