Abstract
WRKY transcription factors have been suggested to play crucial roles in the response to biotic and abiotic stresses. However, previous studies concerning WRKYs have primarily focused on model plants, and fairly limited research has been performed with cotton. In the present study, we functionally characterized a stress-responsive IId WRKY gene (GhWRKY39) from cotton. GhWRKY39 is present as a single copy gene, and subcellular localization analysis indicated that GhWRKY39 localizes to the nucleus. Additionally, some cis-acting elements associated with the environmental stress response were observed in the promoter region of this gene. Consistently, a β-glucuronidase activity assay and quantitative PCR analysis revealed that GhWRKY39 expression could be induced by bacterial and fungal infection or NaCl treatment. Furthermore, the constitutive overexpression of GhWRKY39 in Nicotiana benthamiana conferred greater resistance to bacterial and fungal pathogen infections, and the expression of several pathogenesis-related (PR) genes was significantly increased. The transgenic plants also exhibited less H2O2 accumulation than wild-type plants following pathogen infection. Moreover, GhWRKY39-overexpressing plants displayed enhanced tolerance to salt and oxidative stress and increased transcription of antioxidant enzyme genes, including ascorbate peroxidase (APX), catalase (CAT), glutathione-S-transferase (GST) and superoxide dismutase (SOD). Importantly, overexpression of GhWRKY39 improved the activities of the antioxidant enzymes SOD, POD and CAT after pathogen infection and salt stress treatment. Overall, our data suggest that the overexpression of GhWRKY39 may positively regulate the plant response against pathogen infection and salt stress, likely through the regulation of the reactive oxygen species system via multiple signaling pathway.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants are constantly challenged by a variety of biotic and abiotic stresses, such as pathogen infection and excessive salt stress. These stresses can occur simultaneously and affect plant growth and development or even alter plant-species distribution. To combat these challenges, plants have developed intricate mechanisms to perceive external signals and respond with the proper physiological and morphological changes (Tuteja 2007). Generally, plants regulate the expression of many stress-related genes by activating or repressing their transcription upon signal perception and transduction of the external stimuli (Yang et al. 1997; Wang et al. 2007). During response regulation, WRKY transcription factors play a role in the adaptive plasticity of plants in highly variable environments by forming a network (Eulgem and Somssich 2007). These transcription factors are involved in various plant processes but most notably play important roles in the response to diverse biotic and abiotic stresses (Pandey and Somssich 2009).
The WRKY transcription factors compose a large family of plant-specific regulatory proteins. Since the first member was isolated from sweet potato, increasing numbers of WRKY members have been identified, including 74 members in Arabidopsis thaliana and 109 members in Oryza sativa (Eulgem et al. 2000; Ross et al. 2007). The typical characteristic of WRKY transcription factors is their DNA-binding domain, which is known as the WRKY domain and comprises a highly conserved WRKYGQK sequence at its N-terminus and a zinc finger motif (either CX4–5CX22–23HXH or CX7CX23HXC) at its C-terminus (Eulgem et al. 2000; Rushton et al. 2012). Moreover, the conserved WRKY domain is essential for recognizing and binding the W-box in the promoter regions of target genes (Sun et al. 2003). Based on the number of conserved WRKY domains and the features of the zinc finger motif, the WRKY superfamily can be divided into three distinct groups: I, II and III. Group II can be further subdivided into five distinct subgroups (IIa–IIe) according to the presence of additional short, conserved structural motifs (Eulgem et al. 2000).
Previous loss- and gain-of-function studies have demonstrated that WRKY transcription factors act as either positive or negative regulators in defense responses (Eulgem and Somssich 2007). Constitutive overexpression of CaWRKY40 (Capsicum annuum) resulted in increased resistance to Ralstonia solanacearum. In contrast, the silencing of CaWRKY40 enhanced the plant’s susceptibility to the pathogen (Dang et al. 2013). GhWRKY15-overexpressing tobacco plants exhibited more resistance to viral and fungal infections than wild-type plants and showed increased expression of several pathogen-related (PR) genes, including NONEXPRESSOR OF PR1 (NPR1) and two genes encoding enzymes involved in ethylene biosynthesis (Yu et al. 2012). However, when CaWRKY58 in pepper plants was silenced by virus-induced gene silencing (VIGS), the plants exhibited elevated tolerance and the expression levels of defense-related pepper genes were increased. These results suggest that CaWRKY58 functions as a negative regulator of the primary defense response (Wang et al. 2013b). Similarly, virulent Pseudomonas syringae displayed restrained growth in Atwrky48 mutants, and these mutants showed increased induction of PR1; AtWRKY48 overexpressors exhibited the opposite phenotypes. This observation suggests that AtWRKY48 negatively influences basal resistance to bacterial infection (Xing et al. 2008). Interestingly, a dual function in defense signaling has been observed for AtWRKY53 and AtWRKY41 (Murray et al. 2007; Higashi et al. 2008). Atwrky53 mutants displayed delayed symptom development against R. solanacearum; however, these plants were more susceptible to P. syringae (Murray et al. 2007; Hu et al. 2008). A large body of evidence indicates that WRKY transcription factors increase the expression levels of PR genes and the NPR1 gene in the defense response via specific binding to the W-box element in the promoter regions of these genes (Yang et al. 1999; Yu et al. 2001). PR proteins, such as antimicrobial chitinases and glucanases, can directly destroy the cell wall of the fungal pathogen upon infection (Boland et al. 1990). A previous study indicated that NPR1 was necessary for salicylic acid (SA)-induced expression of the PR genes and systemic acquired resistance; however, loss-of-function mutations in NPR1 did not confer a complete loss of PR gene expression or disease resistance (Shah et al. 2001). These data suggest that the regulation mechanism of the defense response mediated by WRKY transcription factors is fairly complex, and further research is needed.
In addition, accumulating evidence has demonstrated that WRKYs play vital roles in various abiotic stresses, including high salinity. Salinity is a major problem that reduces agricultural crop production and negatively affects irrigated soils. Cellular ion homeostasis and osmotic potential can usually be disrupted under highly saline conditions, and these problems are generally accompanied by secondary effects, which include the increased generation of active oxygen species and oxidative stress (Wang et al. 2003a, b; Tausz et al. 2004). A recent study has demonstrated that transgenic Arabidopsis overexpressing TaWRKY2 or TaWRKY19 (Triticum aestivum L.) exhibited improved salt tolerance compared with controls (Niu et al. 2012). Analogously, the overexpression of DgWRKY3 (Dendranthema grandiflorum) enhances the tolerance to salt stress in transgenic plants, and DgWRKY3-overexpressing plants displayed increased levels of proline, higher activities of antioxidant enzymes and enhanced expression of stress-related genes. All of these data revealed that DgWRKY3 functions as a positive regulator in mediating salt-stress tolerance (Liu et al. 2013). Generally, multiple pathways function when plants respond to high salinity, specifically the ionic and osmotic homeostasis, detoxification and growth regulation pathways (Shinozaki 1999; Hasegawa et al. 2000; Munns et al. 2008). Furthermore, microarray analysis of Arabidopsis roots under NaCl stress indicated that the regulatory networks for salt tolerance were fairly complicated and that numerous genes involved in different signaling pathways may function jointly (Jiang et al. 2006). These studies suggest that the integrated pathway underlying the regulation of salt stress is highly complicated and needs to be explored further.
Cotton (Gossypium hirsutum) is one of the oldest and most important fiber and oil crops, and its growth and yield are severely impaired when it is exposed to various biotic and abiotic stress conditions. To date, few WRKYs have been identified in cotton, and their functions have not been well studied. In our present work, we isolated a putative IId WRKY gene, GhWRKY39, from cotton and showed that it was activated by pathogens and salt stress. The ectopic expression of GhWRKY39 in transgenic plants enhanced the resistance to bacterial and fungal infections. GhWRKY39-overexpressing plants also displayed high salt tolerance and increased ability to scavenge ROS. These results suggest that GhWRKY39 may play a significant role in the regulation of abiotic and biotic stress tolerance.
Materials and methods
Plant materials and treatments
Cotton (Gossypium hirsutum L. cv. lumian 22) seeds were grown in a chamber at 26 ± 1 °C under a 16 h light/8 h dark cycle with a light intensity of ~200 μmol m−2 s−1 and a relative humidity of 60–75 %. Seven-day-old cotton seedlings were collected for use in different treatments. For pathogen infections, the roots of the seedings were dipped into suspensions of the bacterial pathogen R. solanacearum (OD600 = 0.6–0.8) or conidial suspensions of the fungal pathogen Rhizoctonia solani (105 conidia/ml) in 1 % glucose. For hormone treatments, the leaves of uniformly developed seedlings were sprayed with 2 mM salicylic acid (SA), 100 μM methyl jasmonate (MeJA), 100 μM H2O2 and 10 μM methyl viologen (MV). For salt treatment, the roots of the seedlings were cultured in solutions containing 200 mM NaCl.
Isolation of the full-length GhWRKY39 cDNA and its promoter
According to Shi et al. (2011), GhWRKY39 was isolated using specific primers, and the sequences of these primers are provided in Supplementary Table 1. The 5′-flanking region of GhWRKY39 was amplified from the genomic DNA of cotton using a high-efficiency TAIL-PCR method (hiTAIL-PCR) (Liu and Chen 2007). Three TAIL-PCR reactions were performed. First, TAIL-PCR was “pre-amplified” with cotton genomic DNA as the template using primer P1 with four LAD primers: LAD1-1, LAD1-2, LAD1-3 and LAD1-4. Then, primary TAIL-PCRs were performed using the pre-amplification products as templates with the primers P2 and AC1. Secondary TAIL-PCRs were carried out using the products of the primary TAIL-PCRs as templates with the primers P3 and AC1. The thermal conditions for all hiTAIL-PCRs were based on those described by Liu and Chen (2007). The final PCR product was cloned into the pMD18-T vector (TaKaRa, Dalian, China) according to the manufacturer’s instructions and was then sequenced. The PlantCARE database (Lescot et al. 2002) was used for nucleotide sequence analysis.
Vector construction and plant transformation
The GhWRKY39 coding region was subcloned into the binary vector pBI121 under the control of the Cauliflower mosaic virus (CaMV) 35S promoter, and the resulting plasmid was electroporated into Agrobacterium tumefaciens strain LBA4404. N. benthamiana was transformed using the leaf disc method (Horsch et al. 1985), and transgenic plants were selected based on resistance to kanamycin (100 mg/l) resistance and further confirmed by PCR. To test the promoter activity of GhWRKY39, a 1,377-bp fragment of the GhWRKY39 promoter was fused to the GUS reporter gene and was used to replaced the CaMV35S promoter in the pBI121 binary vector. The recombinant plasmid ProGhWRKY39::GUS was induced into the Agrobacterium tumefaciens strain GV3101, which was used to transform Arabidopsis (ecotype Col-0) via the floral dip method (Clough and Bent 1998). Transgenic seedlings were selected on MS agar medium containing 35 μg/ml kanamycin and further confirmed by PCR. The transgenic T3 generation was used in all experiments.
Subcellular localization of GhWRKY39
The open reading frame (ORF) of GhWRKY39 was PCR amplified using the specific primers QC1 and QC2 (Supplementary Table 1). The fragment was fused to the N-terminus of green fluorescent protein (GFP), which was controlled by the cauliflower mosaic virus (CaMV) 35S promoter. For transient expression, the recombinant plasmid pBI121-GhWRKY39-GFP or the pBI121-GFP control plasmid was transformed into onion epidermal cells using the particle bombardment method, as described by Varagona et al. (1992). After bombardment, the tissues were incubated on MS agar medium in the dark at 25 °C for 12 h. DAPI (100 μg/ml, Solarbio, Beijing, China) was used for nuclei staining, and the onion epidermal cells were visualized using a fluorescence microscope (Olympus, Tokyo, Japan).
Disease resistance of the transgenic plants
For bacterial infection, the detached leaves of 8-week-old plants were inoculated with 10 μl R. solanacearum bacterial suspensions (OD600 = 0.6–0.8), which were cultured overnight at 37 °C in Luria–Bertani (LB) broth, harvested by centrifugation and resuspended in sterile tap water. For fungal infection, R. solani was cultured on potato dextrose agar (PDA) medium at 28 °C for 2 weeks, and the spores were then suspended in 1 % glucose. R. solani spore suspensions (105 spores ml−1) were infiltrated into leaves that had been detached from 8-week-old T3 transgenic and WT plants using a needleless syringe. The inoculated plants were maintained in a growth chamber at 25 °C for 24 h in the dark and subsequently incubated at 25 °C under a 16 h light/8 h dark photoperiod. At least three independent experiments were performed for each pathogen.
Analysis of transgenic plants under the salt treatment
For salt treatment, WT and T3 generation transgenic seeds were surface-sterilized and sown on MS medium containing different concentrations of NaCl (0, 50, 100 and 200 mM), and their germination rates were determined. To examine the post-germination response, seeds that were sown on the MS medium an incubated for 3 days that showed radicle emergence were transferred onto MS medium with various concentrations of NaCl (0, 50, 100 and 200 mM), and the growth of the seedlings was observed. In addition, 8-week-old WT and transgenic plants that had been sown in soil were irrigated with a solution of 200 mM NaCl for 2 weeks.
3,3′-Diaminobenzidine (DAB), nitro blue tetrazolium (NBT) and GUS histochemical staining assays
For H2O2 staining, leaves that had been treated with 200 mM NaCl at different time points were incubated in DAB solution (1 mg/ml, pH 3.8) for 24 h at 25 °C in the dark. After staining, the leaves were soaked in 95 % ethanol overnight to remove chlorophyll (Thordal-Christensen et al. 1997). Moreover, the accumulation of H2O2 in 8-week-old plants that had been inoculated with R. solanacearum or R. solani for 7 days was also detected by DAB staining. NBT staining was also performed to detect superoxide anion radicals, as described by Jabs et al. (1996). For this assay, treated leaves were incubated in NBT solution (0.1 mg/ml) at 25 °C for 24 h in the dark, and after staining, 95 % ethanol was used to remove the chlorophyll. A GUS histochemical staining assay was performed to test the activity of the GhWRKY39 promoter. Seven-day-old T3 seedlings were exposed to R. solanacearum bacterial suspensions, R. solani spore suspensions and 200 mM NaCl solution, and GUS histochemical staining assays were performed as described by Baumann et al. (1999).
Oxidative stress tests and enzyme activity assays
For oxidative stress analyses, uniform leaves that had been detached from robust and fully expanded WT and transgenic plants were floated in 10 ml of a solution containing different concentrations of MV (0, 10 and 20 μM) for 72 h. Subsequently, the leaves were immersed in 95 % ethanol for 24 h to extract chlorophyll. Chlorophyll a and b contents were determined by spectrophotometric measurement. The experiment was repeated at least twice. For the enzyme activity assays, the leaves of transgenic and WT plants were inoculated with R. solanacearum or R. solani, and 8-week-old seedlings were treated with 200 mM NaCl for 2 weeks. The leaves (0.5 g) were then collected to test for SOD, POD and CAT activity as previously described (Yang et al. 2008).
RNA extraction and quantitative PCR
Total RNA was extracted from the cotton seedlings using the modified cetyltrimethyl ammonium bromide (CTAB) method according to Lu et al. (2013), while TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was used to isolate the RNA from N. benthamiana. Quantitative PCR was carried out using cDNA that had been synthesized from the above-mentioned RNA as the template. The primers used in the qPCR analyses are shown in Supplementary Tables 1 and 2. Quantitative PCR was performed using the SYBR Prime-Script RT-PCR Kit (TaKaRa, Dalian, China) and a CFX96TM Real-Time System (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s instructions. The PCR amplifications were conducted as follows: a pre-denaturation step at 95 °C for 30 s; 40 cycles of 95 °C for 30 s, 54 °C for 15 s and 72 °C for 15 s; and a melting curve from 65 to 95 °C. The polyubiquitin (UBI) and N. benthamiana β-actin genes were used as internal controls. The Statistical Analysis System (SAS), version 9.1 (SAS Institute, USA), was used to determine significant differences, and the copy number of GhWRKY39 was confirmed using the qPCR protocol of Mason et al. (2002).
Results
Isolation and sequence characterization of GhWRKY39
Due to the important function of WRKYs in plant responses to diverse stresses, the degenerate primers M1 and M2 (Supplementary Table 1) were designed and synthesized to isolate the conserved internal fragment of WRKY. Next, rapid amplification of cDNA ends PCR (RACE-PCR) was performed, and the full-length sequence was retrieved. The full-length cDNA sequence consisted of 1,348 nucleotides containing a 256-bp 5′-untranslated region (UTR), 120-bp 3′ UTR and 972-bp open reading frame, which encoded a 324-amino acid protein with a predicted molecular mass of 36.225 kDa and an isoelectric point of 10.26. The putative WRKY clone from cotton was highly homologous to WRKY39 of A. thaliana and B. napus; thus, it was designated as GhWRKY39 (GenBank: KF220643).
Multiple sequence alignments of WRKYs from various plants were performed using the DNAman software (5.2.2; Lynnon Biosoft, Canada). Consistent with other plant WRKYs, the GhWRKY39 protein exhibits a similar family signature, which includes an approximately 60-amino-acid WRKY domain composed of the conserved amino acid sequence WRKYGQK and zinc-finger motif C–X4–5–C–X22–23–H–X1–H, as well as a conserved primary motif (C-region: VSSFK[K/R]VISLL) that is a distinct characteristic of WRKY Group IId (Park et al. 2005). Overall, these results indicate that GhWRKY39 belongs to Group IId of the WRKY family.
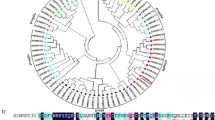
To investigate the evolutionary relationship between GhWRKY39 and WRKYs from different plant species, we constructed a phylogenetic tree using the amino acid sequences derived from the GenBank database using the neighbor-joining method and MEGA version 4.1. Software (Fig. 1b). Generally, three groups of WRKY proteins could be classified, and Group II was further divided into five subgroups (IIa, IIb, IIc, IId and IIe). Phylogenetic analysis indicated that GhWRKY39 was highly similar to Group IId WRKY family members, such as AtWRKY39, BnWRKY39, AtWRKY74 and AtWRKY21. These results strongly imply that GhWRKY39 is a member of WRKY Group IId.
Characterization and sequence analysis of GhWRKY39. a Alignment of the deduced amino acid sequence of GhWRKY39 with AtWRKY39 (NP_566236), AtWRKY74 (AF426255_1), BnWRKY39 (ACN89259) and ZmWRKY21 (NP_001147091). Identical amino acids are shaded black, and the approximately 60-amino acid WRKY domain and C and H residues that are located in the zinc-finger motif (C–X4–5–C–X22–23–H–X1–H) are indicated by a two-headed arrow and inverted triangle, respectively. The conserved primary motif (domain C) and highly conserved amino acid sequence WRKYGQK are boxed. b Phylogenetic relationship of GhWRKY39 with other plant WRKY proteins. Numbers above or below the branches indicate bootstrap values (>50 %) from 500 replicates. GhWRKY39 is highlighted in the box, and the gene name is followed by the protein ID. The species of origin of the WRKYs are indicated by the abbreviations before the gene names: At Arabidopsis thaliana, Bn Brassica napus, Ca Capsicum annuum, Gh Gossypium hirsutum, Pt Populus tomentosa, Vv Vitis vinifera and Zm Zea mays
Additionally, we examined the copy number of the GhWRKY39 locus in cotton by qPCR analysis. RNA-dependent RNA polymerase 6 (GhRDR6), which is present in the cotton genome as a single copy gene, was utilized as an internal standard (Wang et al. 2012). The standard curves of GhRDR6 and GhWRKY39 are shown in Supplementary Fig. 1, and highly significant correlation coefficients between the standard curves of these genes were observed. As shown in Supplementary Table 3, our results indicated that GhWRKY39 is likely a single-copy gene in cotton.
Subcellular localization of GhWRKY39
Bioinformatics analysis using the Plant-mPLoc program predicted that GhWRKY39 localizes to in the nucleus; however, the CELLO program (version 2) predicted the extracellular localization of GhWRKY39. To confirm these predictions, a biolistic transformation system was used to perform a transient assay in onion epidermal cells. The overexpression vectors 35S-GhWRKY39::GFP and 35S-GFP (Fig. 2a) were individually introduced into onion epidermal cells to investigate the localization of GhWRKY39, and the fluorescence signals of 35S-GhWRKY39::GFP and 35S::GFP were observed using a confocal laser scanning microscope. DAPI staining was performed to detect the nuclei, and differential interference contrast images were also obtained. As shown in Fig. 2b, fluorescence was predominantly observed in the nucleus of onion epidermal cells expressing 35S-GhWRKY39::GFP, while fluorescence was present throughout the cytoplasm and nucleus of cells expressing the control 35S-GFP construct. These results demonstrate that the GhWRKY39 protein is located in the nucleus, and this localization might indicate the proposed function of the protein.
Subcellular localization of the GhWRKY39::GFP fusion protein in onion epidermal cells. a Schematic diagram of the 35S-GhWRKY39::GFP fusion construct and 35S-GFP construct. b Onion epidermal cells transiently expressing either the 35S-GhWRKY39::GFP or 35S-GFP construct were observed using a confocal laser scanning microscope. Onion cell nuclei were visualized by DAPI staining
GhWRKY39 promoter analyses
To clarify the mechanism underlying the expression patterns of GhWRKY39 in response to multiple stresses, a 1,377-bp fragment of the GhWRKY39 promoter (Gen-Bank: KF220643) was obtained using high-efficiency TAIL-PCR (hiTAIL-PCR) (Liu and Chen 2007). Computational predictions using the PlantCARE databases revealed that different putative cis-acting elements were present in this region, which suggests that GhWRKY39 plays a role in the response to environmental stresses. Specifically, we identified elicitor-related elements, such as ARE, the CGTCA-motif and the GARE-motif, and development-related elements including an O2-site, TGA and circadian elements. All of the identified cis-elements are listed in Table 1.
To test the activity of the GhWRKY39 promoter, the isolated fragment was fused to the GUS reporter gene, and transgenic Arabidopsis plants with the ProGhWRKY39::GUS fusion construct were generated. Four independent transgenic lines were used for histochemical GUS staining. As shown in Supplementary Fig. 2, GUS staining was mainly detected in the root apex and shoot apical meristem (SAM) at the germination stage. In 2-week-old transgenic seedlings, GUS expression was confined to the root apex and leaf, and weak GUS staining was observed in neonatal leaf. When transgenic lines were at the reproductive stage, GUS activity was observed at a plurality of locations in the plant, such as the root, rosette leaves and flower. These results indicate that GhWRKY39 is developmentally regulated.
In addition, GUS expression was induced by various treatments in transgenic seedings. GUS activity was only detected in the shoot apical meristem in the absence of stress in all four lines (Fig. 3a). However, significant induction of GUS expression was detected in the transgenic lines after various treatments were applied (i.e., R. solanacearum, R. solani and NaCl). Upon application of R. solanacearum, R. solani and NaCl, GUS expression was was strongly induced in the vascular region, root and leaf; moreover, the GUS-stained region varied at different time points (Fig. 3b–j). These results suggest that GhWRKY39 is a stress-inducible gene, and its expression is regulated spatially and temporally.
Histochemical GUS assays in ProGhWRKY39::GUS transgenic Arabidopsis plants. GUS activity in response to various stresses was examined. a Control seedling (non-stressed). b–g Histochemical analysis of GUS activity in ProGhWRKY39::GUS plants following treatment with R. solanacearum or R. solani for 1, 3 and 16 h. h–j Histochemical analysis of GUS activity in ProGhWRKY39::GUS plants treated with 200 mM NaCl for 1, 3 and 16 h
Differential regulation of GhWRKY39 in response to various biotic and abiotic stresses
To explore the effect of biotic stresses on the expression pattern of GhWRKY39, qPCR was performed with cotton seedlings that were inoculated with the bacterial pathogen R. solanacearum and fungal pathogen R. solani. As shown in Fig. 4a and b, both pathogens elevated the transcriptional level of GhWRKY39, although the induction time points were different. The maximum GhWRKY39 transcript level was observed 4 days after treatment with R. solanacearum or R. solani. These data suggest that GhWRKY39 may be involved in the plant pathogen defense response.
Expression profiles of GhWRKY39 under diverse stress conditions. qPCR was performed on total RNA that was extracted from leaves at the indicated time and treated with a R. solanacearum, b R. solani, c SA, d MeJA, e NaCl or f MV. The polyubiquitin gene (UBI) was utilized as an internal control, and the experiment was repeated at least twice. The data are presented as the mean ± SE of three independent experiments (n = 6). Different letters above the columns indicate significant differences (P < 0.05) according to Duncan’s multiple range test using SAS software (version 9.1)
To explore the molecular mechanism underlying the response to various biotic stresses, the expression of GhWRKY39 was examined following treatment with exogenously applied MeJA and SA, which are signaling molecules involved in plant defense signaling pathways. As shown in Fig. 4c and d, the expression of GhWRKY39 was induced by SA and MeJA, and the maximum transcript level occurred at 8 h. Additionally, after NaCl and MV treatments, the accumulation of GhWRKY39 mRNA was markedly increased at 8 and 6 h, respectively. These results indicate that GhWRKY39 is responsive to multiple defense-related signaling molecules and suggested a role for GhWRKY39 in multiple plant defense pathways.
Overexpression of GhWRKY39 conferred enhanced pathogen resistance and affected the expression of defense-related genes
To investigate the function of GhWRKY39 in plant defense, full-length GhWRKY39 was cloned into the plant binary vector pBI121 under the control of the CaMV 35S promoter and transformed into N. benthamiana. Ten independent transgenic lines were obtained by selection with kanamycin and confirmed by genomic PCR detection. Three representative lines (OE1, OE2 and OE3), which exhibited different levels of target gene expression (data not shown), were used for further functional analyses.
To analyze disease resistance in the transgenic plants, 8-week-old WT and transgenic plants were inoculated with R. solanacearum or R. solani. Major disease symptoms were observed in the WT plants 7 days after inoculation with R. solanacearum, with evident chlorosis and enlarged water-soaked lesions; however, slight disease symptoms appeared in all three transgenic lines (Fig. 5a). The detached leaves of WT plants exhibited less resistance to R. solani infection than the transgenic plants. The leaves of all plants were crinkled, distorted and curled; however, the WT plants exhibited significantly more severe disease symptoms compared with the transgenic lines (Fig. 5c).
Overexpression of GhWRKY39 in N. benthamiana conferred enhanced resistance to bacterial and fungal infection. a, c Leaf symptoms of N. benthamiana infected with R. solanacearum and R. solani. b, d The expression of pathogenesis-related (PR) genes in transgenic plants was analyzed by qPCR. WT wild-type, OE overexpression
Some WRKY transcription factors have been shown to play significant roles in activating the transcription of defense genes, especially pathogenesis-related (PR) genes (Yu et al. 2001). To elucidate the possible mechanisms of enhanced pathogen resistance in transgenic plants, the expression levels of defense-related genes, including PR1a, PR1c, PR2 (β-1,3-glucanase), PR4 and NPR1, were determined using qPCR after the plants were infected with R. solanacearum or R. solani. As shown in Fig. 5b, the transcript levels of PR1a, PR1c and PR2 were significantly increased in the transgenic plants. Moreover, the expression of the SA signaling-related gene NPR1 (Yu et al. 2001; Menke et al. 2005) was also slightly enhanced. Similar results were obtained following infection with R. solani, except that the transcription of NPR1 was decreased in transgenic plants compared with WT plants (Fig. 5d). Therefore, we speculate that GhWRKY39-dependent activation of PR genes plays a pivotal role in enhanced disease resistance in transgenic plants and that this resistance might be related to SA-dependent signaling pathways.
Overexpression of GhWRKY39 decreased the accumulation of H2O2 following pathogen infection
Generally, different defense pathways could be activated when plants are attacked by pathogens. Pathogen invasion is often followed by the production of reactive oxygen species (ROS), which plays a critical role in defense responses (Alvarez et al. 1998; Yoshioka et al. 2003). Of the various types of ROS, only H2O2 can cross plant membranes; thus, it plays a direct role in cell-to-cell signaling. To examine whether the enhanced resistance of the transgenic plants was related to ROS accumulation, we measured the accumulation of H2O2 by histochemical analysis via DAB staining after 7-day inoculation with R. solanacearum or R. solani. Based on the visible accumulation of a brown precipitate, as shown in Fig. 6a and b, the leaves of the transgenic plants accumulated lower levels of H2O2 compared with WT leaves. Furthermore, microscopic analysis of the plants following pathogen infection also revealed less H2O2 accumulation in the leaves of transgenic plants compared with WT plants, which is in accordance with the above-mentioned results. These results suggest that the overexpression of GhWRKY39 could enhance defense resistance either by inhibiting the production of pathogen-induced ROS (mainly H2O2) or effectively scavenging excessive H2O2, and the resistance mechanisms mediated by plant may be overlapping.
Overexpression of GhWRKY39 in N. benthamiana decreased the accumulation of H2O2 following R. solanacearum and R. solani infections. a, b Expression of GhWRKY39 in N. benthamiana decreased the accumulation of H2O2 after R. solanacearum and R. solani treatments. The level of H2O2 in leaves was determined using 1 mg/ml DAB as a substrate. The top figures show H2O2 accumulation, and the bottom figures illustrate the microscopic observations of the brown precipitate. WT wild-type, OE overexpression
Effect of GhWRKY39 on salt stress tolerance in transgenic plants
The developmental stage of plants is related to their level of salt stress tolerance (Gao and Xiang 2008). To test the response of GhWRKY39-overexpressing plants to salt stress, the seeds of WT and transgenic plants were surface sterilized and germinated on MS agar medium that was supplemented with different concentrations of NaCl (0, 50, 100 and 200 mM). As shown in Fig. 7a, there was no significant difference between WT and transgenic lines in the presence of 0 and 50 mM NaCl. When the NaCl concentration was increased to 100 and 200 mM, decreased germination rates of WT and OE seeds were observed; however, the germination of the WT seeds was more severely inhibited than that of the OE lines.
Enhanced salt tolerance in N. benthamiana overexpressing GhWRKY39. a Germination rates of WT and OE plants on MS medium containing different concentrations of NaCl. Germination was scored daily, and the results at 9 days after germination are presented. b The post-germination seedling development of WT and transgenic lines on MS medium supplemented with different concentrations of NaCl. The seeds were sown on MS medium for 3 days until radicle emergence was observed. The seedlings were then transferred onto MS medium with different concentrations of NaCl. The plates were oriented vertically to orient the roots of the seedlings in an upright position, and a photograph was taken 2 weeks after transfer. c Root lengths of the seedlings exposed to different concentrations of NaCl were measured 2 weeks after germination. d Photograph of representative 8-week-old WT and OE plants grown in soil containing 200 mM NaCl for 2 weeks. e, f Symptoms of the leaves and roots of the plants after being grown in soil containing 200 mM NaCl for 2 weeks. g Survival rates of 8-week-old plants that were treated with 200 mM NaCl for 2 weeks. h Stomatal changes observed via microscopy after salt treatment, and the stomatal aperture is shown in i. The data are presented as the mean ± SE of three independent experiments (n = 6). Different letters above the columns indicate significant differences (P < 0.05) according to Duncan’s multiple range test using SAS software (version 9.1). WT wild-type, OE overexpression
Next, to examine whether GhWRKY39 affects the growth of seedlings under salt stress conditions, seeds of WT and transgenic lines sown on MS medium for 3 days showing radicle emergence were transferred to medium containing different NaCl concentrations, ranging from 0 to 200 mM NaCl. As shown in Fig. 7b and c, the transgenic lines exhibited longer roots than the WT plants in the presence of 100 and 200 mM NaCl.
To further confirm the salt tolerance conferred by overexpression of GhWRKY39 at the vegetative growth stage, WT and T3 transgenic plants were grown under the same conditions and irrigated with salt water (200 mM) for 2 weeks. Severe growth inhibition was observed in the WT and transgenic plants; however, the growth of transgenic plants was less growth inhibited, which was evidenced at the whole plant, leaf and root levels (Fig. 7d–f). Salinity is often associated with drought, and plants control water loss during drought stress by regulating stomatal closure. Therefore, the stomatal response was examined under normal and salt stress conditions. As shown in Fig. 7h and i, a similar stomatal response was observed in WT and OE plants under normal conditions, while salt treatment resulted in less stomatal opening in transgenic lines compared with WT plants. Taken together, these data indicate that GhWRKY39 overexpression can confer increased salt stress tolerance in transgenic plants during seed germination and vegetative growth.
Overexpression of GhWRKY39 decreased the accumulation of ROS under salt stress and resulted in increased tolerance to oxidative stress
High salinity, which is one of the major environmental stresses, is well known to induce the production of cellular ROS (Borsani et al. 2005). To investigate whether the overexpression of GhWRKY39 in transgenic plants could confer elevated tolerance to salt stress, leaves were detached from WT and transgenic plants, submitted to 200 mM NaCl treatment and collected after 0, 0.5, 1, 1.5 and 2 h. The accumulation of H2O2 and O2 − was then detected by the 3,3′-diaminobenzidine (DAB) and nitro blue tetrazolium (NBT) staining methods, respectively. As shown in Fig. 8a and b, the leaves of WT and transgenic plants accumulated different levels of ROS after the salt stress treatment. Specifically, H2O2 and O2 − accumulation in transgenic lines was remarkably slower than that in WT plants. These results indicate that the overexpression of GhWRKY39 may result in decreased production of ROS or more effectively scavenging of excess ROS after salt stress.
Analysis of ROS accumulation in WT and OE N. benthamianas in response to NaCl and MV treatment. a, b NaCl-induced H2O2 and O2 − accumulation were detected by DAB and NBT staining, respectively. c Leaves from WT and OE plants were incubated in different concentrations of MV (0, 10 and 20 μM) under greenhouse conditions. d Relative chlorophyll contents were determined in the leaves of WT and OE plants after MV treatments. Leaves that were floated in water were utilized as the control. The data are presented as the mean ± SE of three independent experiments (n = 6). Letters above the columns indicate significant differences (P < 0.05) according to Duncan’s multiple range test using SAS software (version 9.1). WT wild-type, OE overexpression
Methyl viologen (MV), which is a potential redox mediator, was used to examine whether GhWRKY39 was responsive to oxidative stress. As shown in Fig. 8c, no abnormalities appeared in the leaves incubated in water without MV. After incubation in different concentrations of MV for 72 h, WT and transgenic plants exhibited symptoms of bleaching or chlorosis. However, MV treatment led to more severe damage in WT plants, and this result was further confirmed by measuring the chlorophyll content of the leaves after MV treatment (Fig. 8d). These results suggest that overexpression of GhWRKY39 can elevate plant tolerance to oxidative stress during the vegetative stage.
The expression and activity of antioxidant enzymes were regulated by GhWRKY39 overexpression
To explore the possible mechanism underlying the reduced ROS levels in transgenic plants during the response to salt stress, qPCR was performed to detect the expression of genes encoding ROS-scavenging enzymes, i.e., APX, CAT, GST and SOD in addition to the ROS producer, respiratory burst oxidase homolog (RbohA and RbohB). The transcript levels of CAT and APX and especially SOD and GST were much higher in the transgenic plants, and no obvious differences in the transcript levels of RbohA and RbohB were observed between transgenic and WT plants under normal conditions (Fig. 9a). However, when treated with 200 mM NaCl for 2 weeks, the expression patterns of the four genes encoding ROS-scaveng enzymes were only slightly altered, whereas the expression of RbohA and RbohB was decreased (Fig. 9b). These results indicate that the overexpression of GhWRKY39 influences the antioxidant system; thus, the regulatory roles of GhWRKY39 in the ROS scavenging pathway should be further explored. Next, the total activities of the antioxidant enzymes, i.e., (SOD, POD and CAT), were examined after pathogen infection and salt stress treatment (Fig. 10). Compared with its activities in WT plants, the activity of SOD was significantly increased in the transgenic lines, which is in accordance with the expression of antioxidant-related genes. Interestingly, the activities of POD and CAT were slightly increased upon salt treatment, while pathogen infection resulted in relatively higher POD and CAT activities. These results suggest that GhWRKY39 may be involved in the regulation of ROS pathways and that the role of GhWRKY39 in the ROS scavenging pathway is complex.
Expression of antioxidant enzyme genes in WT and transgenic N. benthamianas. a The expression of antioxidant enzymes under non-stress conditions. b The expression of antioxidant enzymes under salt stress conditions for 2 weeks. The data are presented as the mean ± SE of three independent experiments. The values indicated by different letters are considered significantly different at P < 0.05 as determined by the Duncan’s multiple range tests. WT wild-type, OE overexpression
Effects of pathogen infection and salt stress treatment on the activities of SOD, POD and CAT in WT and transgenic N. benthamianas. a, b SOD, POD and CAT activities following a 7-day inoculation with R. solanacearum and R. solani. c The activities of the antioxidant enzymes SOD, POD and CAT in plants after irrigation with salt water (200 mM NaCl) for 2 weeks. The data are presented as the mean ± SE of three independent experiments, and values indicated by different letters are significantly different at P < 0.05 as determined using Duncan’s multiple range tests. FW fresh weight, WT wild-type, OE overexpression
Discussion
WRKY transcription factors are members of a large superfamily of regulatory proteins in plants (Rushton et al. 2010). Recently, increased attention has been focused on WRKY transcription factors and their involvement in the regulation of plant responses to various biotic and abiotic stresses (Pandey et al. 2009; Li et al. 2011; Hu et al. 2012; Wang et al. 2013a, b). However, functional analyses of WRKY transcription factors have been mostly extensively focused on model plants, such as A. thaliana and O. sativa, and little progress has been made toward understanding the function of WRKY proteins in cotton. To explore the function of the WRKY transcription factors in cotton, a IId WRKY gene named GhWRKY39 was isolated from G. hirsutum, which is an important, widely distributed fiber and oil crop species.
Due to the high similarity between the sequence of the WRKY gene from cotton and those of several WRKYs from A. thaliana, B. napus and Z. mays, we confirmed that the gene isolated from cotton was a WRKY transcription factor gene. The phylogenetic tree and conserved primary motif (C-region: VSSFK[K/R]VISLL) further indicated that the gene was a member of subgroup IId (Park et al. 2005). Moreover, the conserved primary motif (C-region), which is one of the distinct characteristics of WRKY Group IId, has been reported to potentially function as a nuclear localization sequence (Park et al. 2005). In accordance with the above-mentioned characteristic of members of WRKY subgroup IId, a C-region sequence comprising a nuclear targeting sequence was identified in the N-terminus of the protein, and subcellular analysis indicated that GFP-tagged GhWRKY39 was indeed localized in the nucleus. These results indicate that, similar to GhWRKY15, GhWRKY39 may function in the nucleus (Yu et al. 2012).
The expression pattern of a gene is usually an indicator of its function, and the inducibility of GhWRKY39 expression by pathogen infection and high-salt stress suggested that GhWRKY39 might play roles in defense responses and be involved in the regulation of stress pathways. As shown in Fig. 5, plants overexpressing GhWRKY39 were more resistant to R. solanacearum and R. solani infection than WT plants, and transgenic plants also exhibited increased expression of PR genes. This finding is consistent with a previous report that overexpression of GhWRKY15 in tobacco caused increased resistance to virus and pathogenic fungi in addition to enhancing the expression of PR1, PR2 and PR4, which contributed to the disease resistance of the transgenic plants (Yu et al. 2012). Previous studies have demonstrated that WRKY DNA binding-proteins can specifically recognize W-box elements that are conserved in the promoters of many PR genes (PR1, PR2, PR3 and PR5) and therefore induce the expression of these defense-related genes (Yu et al. 2001; Zheng et al. 2007). In addition, it has been reported that plant innate immunity is composed of two interconnected branches, known as pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI) (Jones and Dangl 2006). Therefore, we deduced that the defense mechanism mediated by GhWRKY39 may be involved in PTI. PTI and ETI activate local as well as systemic defense responses (referred to as systemic acquired resistance [SAR]), which are modulated by phytohormones, especially jasmonic acid (JA) and SA (Durrant and Dong 2004; Bostock 2005). JA-dependent plant defenses are generally activated by necrotrophic pathogens and chewing insects, whereas SA-dependent defenses are often triggered by biotrophic pathogens. JA and SA signaling usually act antagonistically; however, synergism between these two phytohormones has also been observed (Mur et al. 2006). The results of our expression analysis also indicated that GhWRKY39 may be involved in SA- and JA- dependent signaling pathways. In conclusion, we suggest that the molecular mechanism mediated by GhWRKY39 is fairly complex during the defense response and that GhWRKY39 may act as an integrator of multiple signaling pathways.
Accumulating evidence indicates that the ROS network is important for the induction of disease resistance (Kotchoni and Gachomo 2006). Evidence for the role of ROS in triggering or executing the hypersensitive response (HR), usually caused by ETI, has been demonstrated by pharmacological studies showing that a blockade of ROS accumulation inhibited cell death (Levine et al. 1994; Hammond-Kosack and Jones 1997). The effect of ROS in defense responses and activation of the HR have been mainly associated with NADPH oxidase, which catalyzes the reduction of O2 to O2 −. Further dismutation of O2 − by SOD generates the most stable ROS, H2O2 (Lamb and Dixon 1997). Our results demonstrated that GhWRKY39-overexpressing plants showed enhanced antioxidase activity following pathogen infection. Therefore, the GhWRKY39-mediated defense response may be related to PTI and ETI, and GhWRKY39 may regulate antioxidase to reduce the accumulation of ROS and inhibit cell death.
In addition to the vital roles of WRKYs in disease resistance, abiotic stresses, such as salt and drought, have a close relationship with WRKY proteins. For example, Zheng et al. (2013) suggested that ThWRKY4, a WRKY gene from Tamarix hispida, could positively mediate abiotic stress tolerance by modulating ROS and the expression of stress-responsive genes. TaWRKY10-overexpressing plants exhibited decreased H2O2 and O2 − accumulation than WT plants under drought and salt stress conditions (Wang et al. 2013a). Overexpression of a chrysanthemum transcription factor gene, DgWRKY3, in tobacco enhanced tolerance to salt stress, as reduced accumulation of malondialdehyde (MDA) and hydrogen peroxide (H2O2) as well as higher antioxidant enzyme activity were observed in transgenic plants compared with WT plants (Liu et al. 2013). Transgenic Arabidopsis overexpressing TaWRKY2 exhibited enhanced salt and drought tolerance compared with controls, while overexpression of TaWRKY19 conferred tolerance to salt, drought and freezing stresses in transgenic plants (Niu et al. 2012). Our results showed that overexpression of GhWRKY39 in N. benthamiana improved plants’ tolerance to salt stress. In contrast, Li et al. (2010) reported that heat stress-induced AtWRKY39 positively regulated the cooperation between the SA- and JA-activated signaling pathways that mediated the response to heat stress. Therefore, we conclude that the function of WRKY39 is diverse, and it may be involved in multiple pathways.
Based on these results, we propose that GhWRKY39 is involved in at least two regulatory pathways: one that is related to pathogen infection and one that is involved in the response to salt stress. GhWRKY39 may be involved in crosstalk between the complicated biotic and abiotic stress response pathways. Although the overexpression of GhWRKY39 has been explored in N. benthamiana, the mechanism underlying the function of GhWRKY39 should ideally be elucidated in transgenic cotton. Moreover, additional research concerning GhWRKY39 is needed, including examinations of its interactions with other WRKY proteins or MAPK cascade members.
Abbreviations
- APX:
-
Ascorbate peroxidase
- CAT:
-
Catalase
- Ca:
-
Capsicum annuum
- CTAB:
-
Cetyltrimethyl ammonium bromide
- DAB:
-
3,3′-Diaminobenzidine
- GFP:
-
Green fluorescent protein
- Gh:
-
Gossypium hirsutum
- GST:
-
Glutathione-S-transferase
- hiTAIL-PCR:
-
High-efficiency TAIL-PCR
- MeJA:
-
Methyl jasmonate
- MS medium:
-
Murashige and Skoog medium
- MV:
-
Methyl viologen
- Nb:
-
Nicotiana benthamiana
- NPR1:
-
Non-expression of PR1
- OE:
-
Overexpression
- ORF:
-
Open reading frame
- PR:
-
Pathogenesis-related
- RACE:
-
Rapid amplification of cDNA ends
- ROS:
-
Reactive oxygen species
- R. solanacearum :
-
Ralstonia solanacearum
- R. solani :
-
Rhizoctonia solani
- qPCR:
-
Quantitative PCR
- SA:
-
Salicylic acid
- SOD:
-
Superoxide dismutase
- UTR:
-
Untranslated region
- WT:
-
Wild-type
References
Alvarez ME, Pennell RI, Meijer PJ, Ishikawa A, Dixon RA, Lamb C (1998) Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92:773–784
Baumann K, De Paolis A, Costantino P, Gualberti G (1999) The DNA binding site of the Dof protein NtBBF1 is essential for tissue-specific and auxin-regulated expression of the rolB oncogene in plants. Plant Cell 11:323–334
Boland JF, Linthorst HJM, Cornelissen BJC (1990) Plant pathogenesis-related protein induced by virus infection. Annu Rev Phytopathol 28:113–138
Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK (2005) Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123:1279–1291
Bostock RM (2005) Signal crosstalk and induced resistance: straddling the line between cost and benefit. Annu Rev Phytopathol 43:545–580
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Dang FF, Wang YN, Yu L, Eulgem T, Lai Y, Liu ZQ, Wang X, Qiu AL, Zhang TX, Lin J, Chen YS, Guan DY, Cai HY, Mou SL, He SL (2013) CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant, Cell Environ 36:757–774
Durrant WE, Dong X (2004) Systemic acquired resistance. Annu Rev Phytopathol 42:185–209
Eulgem T, Somssich IE (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10:366–371
Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5:199–206
Gao L, Xiang CB (2008) The genetic locus At1g73660 encodes a putative MAPKKK and negatively regulates salt tolerance in Arabidopsis. Plant Mol Biol 67:125–134
Hammond-Kosack KE, Jones JD (1997) Plant disease resistance genes. Annu Rev Plant Physiol Plant Mol Biol 48:575–607
Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51:463–499
Higashi K, Ishiga Y, Inagaki Y, Toyoda K, Shiraishi T, Ichinose Y (2008) Modulation of defense signal transduction by flagellin-induced WRKY41 transcription factor in Arabidopsis thaliana. Mol Genet Genomics 279:303–312
Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT (1985) A simple and general method for transferring genes into plants. Science 227:1229–1231
Hu J, Barlet X, Deslandes L, Hirsch J, Feng DX, Somssich I, Marco Y (2008) Transcriptional responses of Arabidopsis thaliana during wilt disease caused by the soil-borne phytopathogenic bacterium, Ralstonia solanacearum. PLoS One 3:e2589
Hu YR, Dong QY, Yu DQ (2012) Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci 185–186:288–297
Jabs T, Dietrich RA, Dangl JL (1996) Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science 273:1853–1856
Jiang YQ, Deyholos MK (2006) Comprehensive transcriptional profiling of NaCl-stressed Arabidopsis roots reveals novel classes of responsive genes. BMC Plant Biol 6:25
Jones JD, Dangl JL (2006) The plant immune system. Nature 444:323–329
Kotchoni SO, Gachomo EW (2006) The reactive oxygen species network pathways: an essential prerequisite for perception of pathogen attack and the acquired disease resistance in plants. J Biosci 31:389–404
Lamb C, Dixon RA (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48:251–275
Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Peer VY, Rouze P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Levine A, Tenhaken R, Dixon R, Lamb C (1994) H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79:583–593
Li SJ, Fu QT, Chen LG, Huang WD, Yu DQ (2011) Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermo tolerance. Planta 233:1237–1252
Li SJ, Zhou X, Chen LG, Huang WD, Yu DQ (2010) Functional characterization of Arabidopsis thaliana WRKY39 in heat stress. Mol Cells 29:475–483
Liu YG, Chen YL (2007) High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 43:649–656
Liu QL, Zhong M, Li S, Pan YZ, Jiang BB, Jia Y, Zhong HQ (2013) Overexpression of a chrysanthemum transcription factor gene, DgWRKY3, in tobacco enhances tolerance to salt stress. Plant Physiol Biochem 69:27–33
Lu WJ, Chu XQ, Li YZ, Wang C, Guo XQ (2013) Cotton GhMKK1 induces the tolerance of salt and drought stress, and mediates defense responses to pathogen infection in transgenic Nicotiana benthamiana. PLoS One 8:e68503
Mason G, Provero P, Vaira AM, Accotto GP (2002) Estimating the number of integrations in transformed plants by quantitative real-time PCR. BMC Biotechnol 2:20
Menke FL, Kang HG, Chen Z, Park JM, Kumar D, Klessig DF (2005) Tobacco transcription factor WRKY1 is phosphorylated by the MAP kinase SIPK and mediates HR-like cell death in tobacco. Mol Plant Microbe Interact 18:1027–1034
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681
Mur LAJ, Kenton P, Atzorn R, Miersch O, Wasternack C (2006) The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol 140:249–262
Murray SL, Ingle RA, Petersen LN, Denby KJ (2007) Basal resistance against Pseudomonas syringae in Arabidopsis involves WRKY53 and a protein with homology to a nematode resistance protein. Mol Plant Microbe Interact 20:1431–1438
Niu CF, Wei W, Zhou QY, Tian AG, Hao YJ, Zhang WK, Ma B, Lin Q, Zhang ZB, Zhang JS, Chen SY (2012) Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant, Cell Environ 35:1156–1170
Pandey SP, Somssich IE (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol 150:1648–1655
Park CY, Lee JH, Yoo JH, Moon BC, Choi MS, Kang YH, Lee SM, Kim HS, Kang KY, Chung WS, Lim CO, Cho MJ (2005) WRKY group IId transcription factors interact with calmodulin. FEBS Lett 579:1545–1550
Ross CA, Liu Y, Shen QJ (2007) The WRKY gene family in rice (Oryza sativa). J Integr Plant Biol 49:827–842
Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15:247–258
Rushton DL, Tripathi P, Rabara RC, Lin J, Ringler P, Boken AK, Langum TJ, Smidt L, Boomsma DD, Emme NJ, Chen X, Finer JJ, Shen QJ, Rushton PJ (2012) WRKY transcription factors: key components in abscisic acid signaling. Plant Biotechnol J 10:2–11
Shah J, Kachroo P, Nandi A, Klessig DF (2001) A recessive mutation in the Arabidopsis SSI2 gene confers SA- and NPR1-independent expression of PR genes and resistance against bacterial and oomycete pathogens. Plant J 25:563–574
Shi J, Zhang L, An HL, Wu CA, Guo XQ (2011) GhMPK16, a novel stress-responsive group D MAPK gene from cotton, is involved in disease resistance and drought sensitivity. BMC Mol Biol 12:22
Shinozaki K (1999) Plant response to drought and salt stress: overview. Tanpakushitsu Kakusan Koso 44:2186–2187
Sun C, Palmqvist S, Olsson H, Boren M, Ahlandsberg S, Jansson C (2003) A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell 15:2076–2092
Tausz M, Sircelj H, Grill D (2004) The glutathione system as a stress marker in plant ecophysiology: is a stress-response concept valid? J Exp Bot 55:1955–1962
Thordal-Christensen H, Zhang ZG, Wei YD, Collinge DB (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 11:1187–1194
Tuteja N (2007) Abscisic acid and abiotic stress signaling. Plant Signal Behav 2:135–138
Varagona MJ, Schmidt RJ, Raikhel NV (1992) Nuclear localization signal(s) required for nuclear targeting of the maize regulatory protein Opaque-2. Plant Cell 4:1213–1227
Wang H, Miyazaki S, Kawai K, Deyholos M, Galbraith DW, Bohnert HJ (2003a) Temporal progression of gene expression responses to salt shock in maize roots. Plant Mol Biol 52:873–891
Wang W, Vinocur B, Altman A (2003b) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218:1–14
Wang HH, Hao JJ, Chen XJ, Hao ZN, Wang X, Lou Y, Peng Y, Guo Z (2007) Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol Biol 65:799–815
Wang M, Li SW, Yang HF, Gao Z, Wu CA, Guo XQ (2012) Characterization and functional analysis of GhRDR6, a novel RDR6gene from cotton (Gossypium hirsutum L.). Biosci Rep 32:139–151
Wang C, Deng PY, Chen LL, Wang XT, Ma H, Hu W, Yao NC, Feng Y, Chai RH, Yang GX, He GY (2013a) A wheat WRKY transcript ion factor TaWRKY10 confers tolerance to multiple abiotic stresses in transgenic tobacco. PLoS One 8:e65120
Wang Y, Dang FF, Liu ZQ, Wang X, Eulgem T, Lai Y, Yu L, She JJ, Shi YL, Lin JH, Chen CC, Guan DY, Qiu AL, He SL (2013b) CaWRKY58, encoding a group I WRKY transcription factor of Capsicum annuum, negatively regulates resistance to Ralstonia solanacearum infection. Mol Plant Pathol 14:131–144
Xing DH, Lai ZB, Zheng ZY, Vinod KM, Fan BF, Chen ZX (2008) Stress- and pathogen-induced Arabidopsis WRKY48 is a transcriptional activator that represses plant basal defense. Mol Plant 1:459–470
Yang YO, Shah J, Klessig DF (1997) Signal perception and transduction in defense responses. Genes Dev 11:1621–1639
Yang PZ, Chen CN, Wang ZP, Fan BF, Chen ZX (1999) A pathogen- and salicylic acid-induced WRKY DNA-binding activity recognizes the elicitor response element of the tobacco class I chitinase gene promoter. Plant J 18:141–149
Yang L, Tang R, Zhu J, Liu H, Mueller-Roeber B, Xia H, Zhang H (2008) Enhancement of stress tolerance in transgenic tobacco plants constitutively expressing AtIpk2β, an inositol polyphosphate 6-/3-kinase from Arabidopsis thaliana. Plant Mol Biol 66:329–343
Yoshioka H, Numata N, Nakajima K, Katou S, Kawakita K, Rowland O, Jones JD, Doke N (2003) Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 15:706–718
Yu D, Chen C, Chen Z (2001) Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell 13:1527–1540
Yu FF, Huaxia YF, Lu WJ, Wu C, Guo XQ (2012) GhWRKY15, a member of the WRKY transcription factor family identified from cotton (Gossypium hirsutum L.), is involved in disease resistance and plant development. BMC Plant Biol 12:144
Zheng Z, Mosher SL, Fan B, Klessig DF, Chen Z (2007) Functional analysis of Arabidopsis WRKY25 transcription factor in plant defense against Pseudomonas syringae. BMC Plant Biol 7:2–15
Zheng L, Liu GF, Meng XN, Liu YJ, Ji XY, Li YB, Nie XG, Wang YC (2013) A WRKY gene from Tamarix hispida, ThWRKY4, mediates abiotic stress responses by modulating reactive oxygen species and expression of stress-responsive genes. Plant Mol Biol 82:303–320
Acknowledgments
This work was financially supported by the Genetically Modified Organisms Breeding Major Projects of China [Grant Numbers 2009ZX08009-092B] and the National Natural Science Foundation of China [Grant Number 31171837].
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shi, W., Liu, D., Hao, L. et al. GhWRKY39, a member of the WRKY transcription factor family in cotton, has a positive role in disease resistance and salt stress tolerance. Plant Cell Tiss Organ Cult 118, 17–32 (2014). https://doi.org/10.1007/s11240-014-0458-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-014-0458-8