Abstract
An orthologue of the vacuolar H+-pyrophosphatase (H+-PPase) gene, AmVP1, was isolated from a desert plant, Ammopiptanthus mongolicus (Leguminosae), by RACE-PCR. AmVP1 has a total length of 2,875 bp, with an open reading frame of 2,316 bp, which encodes a predicted polypeptide of 771 amino acids. Sequence analysis revealed that it has high similarity with the VP1 proteins from other plants. AmVP1 was strongly induced by drought stress, but only responded initially to a salt stress. In addition, a 1.8 kb upstream sequence of AmVP1 was isolated from the genomic DNA of A. mongolicus by TAIL-PCR. Cis-element as well as promoter prediction analysis indicated that it contained three promoter sequences and more than 50 cis-elements. Heterologous expression of AmVP1 in the yeast mutant ena1 could partially suppress its hypersensitivity to NaCl. Over-expressing AmVP1 resulted in enhanced tolerances to both drought and salt stresses in transgenic Arabidopsis plants. The transgenic plants accumulated more sodium and potassium in their leaves after salt stress, and retained more water while producing less malondialdehyde during drought stress. A comparative study of salt tolerance between AtVP1 (an H+-PPase from Arabidopsis) and AmVP1 transgenic Arabidopsis suggested that the efficiency of AmVP1 is more than threefold higher than AtVP1. Our work suggested that AmVP1 functioned as a typical VP1 gene, but might be a more efficient orthologue than AtVP1 and therefore a valuable gene for improving plant salt and drought tolerances.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The vacuolar H+-pyrophosphatase (H+-PPase) is involved in various cellular processes, including abiotic stress tolerances in plants (Fukuda et al. 2004; Mitsuda et al. 2001; Queiros et al. 2009; Wisniewski and Rogowsky 2004; Zandonadi et al. 2007). In the Na+ compartmentali-zation, the H+-PPase, together with the vacuolar ATPase, forces the electrochemical gradient of protons (Gao et al. 2006), which subsequently drives the vacuolar Na+/H+ antiporters to detoxify the Na+ in cytoplasm (Gao et al. 2006; Taiz 1992). During the adjustment of osmosis, H+-PPase also drives the proton-dependent cation transport proteins, such as Ca2+/H+ antiporter as well as organic acids, sugars, and other compound transporters, to maintain cell turgor (Gao et al. 2006; Taiz 1992). Unlike the vacuolar ATPase, which consists of multiple subunits, the vacuolar H+-PPase is a single unit protein, and therefore an easier target for genetic manipulation (Luttge and Ratajczak 1997; Maeshima 2000). As expected, over- expression of H+-PPase genes results in enhanced tolerances to salt and drought in diversified plant species (Brini et al. 2007; Gao et al. 2006; Guo et al. 2006; Li et al. 2010; Lv et al. 2009).
So far, vacuolar H+-PPase genes have been isolated from different plant species (Bao et al. 2009; Brini et al. 2007; Gao et al. 2006; Guo et al. 2006; Li et al. 2010). However, few vacuolar H+-PPase genes have been isolated from extremely abiotic stress-tolerant plant species (Gao et al. 2006; Guo et al. 2006; Hu et al. 2012). Considering the importance of vacuolar H+-PPase genes, we set out to identify the orthologue from a distinctive desert shrub, Ammopiptanthus mongolicus. It is distributed naturally in the northwestern part of China, an area marked by seasonally extreme drought and temperatures (over 40 °C in the summer and under −30 °C in the winter), poor soil quality with high salinity, and extraordinarily high ultraviolet- irradiation (Wang et al. 2007). A new H+-PPase gene orthologue, AmVP1, was identified and functionally characterized in yeast and Arabidopsis. AmVP1 functioned as a typical VP1 gene, and seemed to confer more salt stress tolerance than AtVP1 to transgenic Arabidopsis plants.
Materials and methods
Isolation of AmVP1
A pair of degenerate PCR primers, AmVP1CRF(forward: 5′-CTTGGTGGGTCTT CTATGGCT-3′, and reverse: 5′-AGGCAAGGCCAAATATAACATT-3′) was designed for amplifying the cDNA fragment of H+-PPase orthologue from A. mongolicus, based on the multi-alignment of the full-length mRNA sequences from different plant species. Two primers (forward: 5′-CTGGGCTGGACTTATTATTGGGTTTGTG-3′, and reverse: 5′-GATGTTGCTGATTCCTGCCG-3′) were designed to perform 3′-RACE cDNA synthesis using the Smart™ RACE cDNA Amplification Kit (Clontech, Palo Alto, CA, USA). Five primers [AmVP15RRT (5′-ACAGACAAGAATGCC-3′), AmVP15RA1 (5′-TGGGGTCTGATCTTTTTGGT-3′), AmVP15RS1 (5′-CAGATCAGCACCGACATCAG-3′), AmVP15RA2 (5′-TCAACCATGATTTCACTGCC-3′), and AmVP15RS2 (5′-GATACCACCACCGACTCTCC-3′)] were designed to perform 5′-RACE cDNA synthesis using 5′- RACE Amplification Kit (Takara Japan). The clones obtained were sequenced and the overlapping region with the first clone was confirmed. After re-construction of the open reading frame, a fragment containing the open reading frame was re-obtained by PCR with primer AmVP1EL (forward: 5′-TTTTTTTCTGGCGAGGATGG-3′, and reverse: 5′-GCAAACCTATTCCGCTCCAA-3′) from the root cDNA library and sequenced for further confirmation.
Amplification of AmVP1 promoter
Genomic DNA was extracted from the roots of A. mongolicus, and used as templates for Tail-PCR amplification. Those primers were designed to perform Tail-PCR according to Liu and Chen (2007): AmVPPro-1-0 (5′-AGAACACAATCCCCACCACAGCACACACCG-3′), AmVPPro-1-1(5′-ACGATGGACTCCAGTCCC ACTATCTCCGTCGCAAGC TCCGA-3′), AmVPPro-2-0 (5′-GCTTGTAGGGAGT GCTGGTGTTCAATGTG-3′), AmVPPro-2-1(5′-ACGATGGACTCCAGTCCGGCC AGTTGCTAAGCGATTGATCGGAATG-3′), LAD1-1(5′-ACGATGGACTCCAGA GCGGCCGC(G/C/A)N(G/C/A)NNNGGAA-3′), and AC1(5′-ACGATGGACTCCA GAG-3′). PCR products were cloned into pMD19-T vector and sequenced by Invitrogen Inc., USA.
Real-time PCR
Determination of AmVP1 expression patterns under salt and drought stresses
Seeds of A. mongolicus were surface-sterilized with HgCl2, and grown on MS (Murashige and Skoog 1962) medium plate in a 16-h light/8-h dark cycle at 24 °C.
Ten days after sowing, seedlings were carefully pulled out and moved to the liquid MS medium containing 200 mM NaCl to incubate for various time periods (0 h, 1, 2, 4, 8, 12, and 24 h) and then sampled for RNA extraction. For drought treatment, seedlings were moved to a 13-cm-filter paper, and kept in a chamber for 1 and 1.5 h respectively under the light condition of 120 μmol m−2 s−1 at 24 °C. Total RNAs were extracted from the root of salt and drought treated seedlings and non-treated seedlings using TRIzol reagent (Invitrogen). First-strand cDNA synthesis was performed with 5 μg of total RNA using a first-strand cDNA synthesis kit (Shenergy Biocolor). The products were subsequently used as templates for Real-time PCR analysis. The Real-time PCR was performed using SYBR Green I PCR kit (Toyobo) on an iCycler (Bio-Rad) according to the manufacturer’s suggestions with AmACTIN2 as a reference.
Partial cDNA fragment of internal control AmACTIN2 was obtained by PCR with degenerate primers: (forward: 5′-GAAGCACAATCCAAAAGAGGTAT-3′, and reverse: 5′-GAGCCTCCGATCCAGACACT-3′). The resulting fragment was cloned to pMD19-T and sequenced by Invitrogen Inc. (USA).
Specific primers for real-time PCR to respective genes were as follows: AmVP1 (forward: 5′-GAGATAGTGGTGCCGGTGTGT-3′, and reverse: 5′-AGTCCCCGTAACCATTCTTGCT-3′), AmNHX2 (Wei et al. 2011) (forward: 5′CCATCCAGCATT CGTGCCTTAC3′, and reverse: 5′-GACCATTGCGTTCAGATGGTGAG-3′), AmACT2 (forward: 5′-CCATCCAGGCTGTGCTTTCT-3′, and reverse: 5′-AGATCA CGCCCTGCAAGGT-3′).
Determination of AmVP1 and AtVP1 expressions
The primers for the real-time PCR analysis of respective genes were as follows: AmVP1 (forward: 5′-GAGATAGTGGTGCCGGTGTGT-3′, and reverse: 5′-AGTCCC CGTAACCATTCTTGCT-3′), AtVP1 (forward: 5′-CTCTTGATGCCGCTGGAAAC A-3′, and reverse: 5′-GCCCAATGATAACTTTAGGGGTCA-3′) and AtACTIN2 (forward: 5′-CCGAGGCTCCTCTTAACCCA 3′, and reverse: 5′-ACCAGAATCCAG CACAATACCG-3′).
Yeast expression vectors construction
The AmVP1 ORF was amplified with primer AmVP1EL (forward: 5′-TTTTTTTCTGGCGAGGATGG-3′, and reverse: 5′-GCAAACCTATTCCGCTCCAA-3′) using KOD-Plus polymerase (TOYOBO Inc., Japan). The resulting fragment was cloned into pMD19-T vector and sequenced by Invitrogen Inc (USA). The DNA fragments of AmVP1 was then digested from the pMD19-T vector by BamHI/SalI and subcloned into the BamHI/SalI site of the p426ADH vector between the ADH promoter and the CYC1 terminator.
Functional assays using the yeast mutant
The ena1 yeast mutant (kindly presented by Professor Yang Ai-Fang) was transformed with p426ADH-AmVP1, and p426ADH as a control using LiAc/polyethylene glycol method. All yeast strains including the wild type were grown at 28 °C for 16–18 h to reach OD600 1.5–2.0 in selective yeast nitrogen base (YNB) medium lacking uracil (Difco) or YPD (1 % (w/v) yeast extract, 2 % (w/v) peptone, and 2 % (w/v) dextrose; Sigma) liquid medium (wild type). After adjusting OD600 to 1.0, aliquots (2.5 μl) of adjusted cultures and two fivefold serial dilution of the cultures were spotted onto YPD medium supplemented with or without 0.5 mol l−1 NaCl. Growth status was compared after culturing the strains at 28 °C for 3 days.
Generation of transgenic Arabidopsis plants
The ORF of AmVP1 was digested from the AmVPI/pMD19-T vector with BamHI/SalI and subcloned into the BamH1/Sal1 site of pPZPY122 plasmid between the 35 s promoter and Nos-polyA terminator. Primers AtVP1F (5′-TATGGTACCAT GGTGGCGC-3′; the underlined section is an engineered KpnI site) and AtVP1R (5′-ATGGTCGACTTAGAAGTACTTGAAA-3′; the underlined section is an engineered SalI site) were used to PCR amplify the ORF of AtVP1. The PCR product was cloned into pPZPY122 plasmid and the resultant vector was named AtVP1/pPZPY122. The resulting constructs were introduced into Agrobacterium tumefaciens LBA4404 by the freeze and thaw method (Holsters et al. 1978). Arabidopsis plants (Columbia-0) were transformed using the floral-dipping method (Clough and Bent 1998). Putative transgenic plants were selected on the plates supplemented with 90 mg l−1 gentamicin in a 16-h light/8-h dark cycle at 24 °C, and were further verified by PCR.
Semi-quantitative PCR
Total RNA extraction and First-strand cDNA synthesis were carried out as described previously. Using AtACTIN2 as an internal control, RT-PCR was performed with following primers, AmVP1RT (forward: 5′-ATGGAGGTTTGTCAGTGATAGCC-3′, and reverse: 5′-AATGCTGGATGGACGAGGAA-3′) and AtACT2 (forward: 5′-TGAAAGTTGCCACCTATGCC-3′, and reverse: 5′-CCATCCCAGCAATGTCCC-3′).
Salt and drought stress treatment
The seeds of T4 homologous transgenic lines and transgenic plants with the empty vector were surface-sterilized with 0.01 % HgCl2(w/v), and germinated on the MS plate containing 90 mg l−1 gentamicin in a 10-h light/14-h dark cycle (salt tolerance test) or 16-h light/8-h dark cycle(drought tolerance test) at 24 °C. Seven days after sowing, well-rooted seedlings were moved to 10-cm-side square pots with soil (peat soil:vermiculite:pearlite [v/v/v] 3:9:0.5 purchased from Shanghai Institute of Landscape Science) presoaked with plant nutrient medium, and grown in a 10-h light/14-h dark cycle(salt tolerance test) or 16-h light/8-h dark cycle(drought tolerance test) at 24 °C. The pots were flooded for 2 h every 5 days.
Eight plants and eight of each of the two AmVP1-overexpressing transgenic lines (M-10, M-11) were selected for salt stress treatment at day of 35; the water was supplemented with 200 mM NaCl. Eight plants and eight of each of the two AmVP1-overexpressing transgenic lines (M-10, M-11) were selected for drought stress treatment at day of 20. Plants were flooded for 2 h and then kept for 15 days for water deprivation.
Measurements of relative water content (RWC)
Five plants from each line were taken to determine the relative water content. RWC measurements were carried out as described in the Gaxiola et al. (2001). Means and standard deviation will be calculated for further analysis.
Determinations of Na+ and K+ contents
Eight rosette leaves from each of the transgenic lines were taken to determine the Na+ and K+ contents. After 24 h at 75 °C, the dry weight was measured. Na+ and K+ were extracted with 0.001 N HAc at 90 °C for 3 h. The supernatants were analyzed by atomic absorption. Means and standard deviation will be caculated for further compare analysis.
Determination of lipid peroxidation
Six plants from each line were taken to determine the MDA (malondialdehyde) content. MDA measurement was carried out as described in the Cao et al. (2009b). Means and standard deviation will be further caculated for compare analysis.
Determination of efficiencies of AmVP1 and AtVP1 in conferring salt stress tolerance
One AmVP1 transgenic line (M-10) and ten AtVP1 transgenic lines were grown under 10-h-light/14-h-dark photoperiod for 30 days before determining the efficiencies of AmVP1 and AtVP1 in conferring salt stress tolerance. Plants were watered with 180 mM NaCl with an interval of 5 days. After 30-day salt treatment, line M-10 and two AtVP1 transgenic lines (A-3, A-7) exhibiting the similar phenotype with M-10, together with another AtVP1 transgenic line (A-2) exhibiting poorer salt tolerance, were selected to measure malondialdehyde content and examine the AmVP1 and AtVP1 expression levels using real-time PCR with AtACTIN2 as a reference.
Results
Isolation and molecular characterization of AmVP1
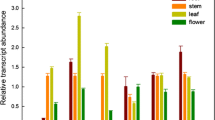
To facilitate the isolation of salt stress responsive genes, a cDNA library was made from the mRNA extracted from the salt treated radical of A. mongolicus. A 2,875 bp cDNA, with an open reading frame of 2,316 bp, was then obtained by RACE -PCR from the cDNA library. It encodes a polypeptide of 771 aa, with 12 putative transmembrane helixes, predicted by the online program SOSUI (http://bp.nuap.nagoya-u.ac.jp/sosui) (Zhang et al. 2008). Sequence analysis indicated that AmVP1 had the five conserved domains of H+-PPase (Drozdowicz and Rea 2001) as well as eight unique amino acids which are different from the six glycophytes H+-PPase (Fig. 1). It was therefore tentatively named as AmVP1. AmVP1was strongly induced by drought stress, but only responded to salt stress at the initial stage (Fig. 2). After 1 h exposure to the air, its transcript level in roots increased about 3.5-fold but decreased to about 2.6-fold at the time point of 1.5 h, correlating well with a drought responsive gene, AmNHX2 (Wei et al. 2011) (Fig. 2b). Its transcript level increased to about 2.2-fold after 1 h salt treatment, but declined dramatically shortly afterwards and remained relatively stable at a slightly higher level between 2 and 4 h (about 1.25-fold). It began to decrease dramatically again 8 h after salt treatment (Fig. 2a).
AmVP1 sequence analysis by Genedoc software. a Five conserved boxes of AmVP1 and six other H+-PPases from glycophytes. b Eight different amino acid sites between AmVP1 and six other H+-PPases from glycophytes. Identical amino acids are highlighted in black, and residues with conservative substitutions are shaded in gray. Genebank accession numbers are: AtVP1 (NP_173021), NtVP1 (CAA58701), OsVP1 (BAA31523), ZmVP1 (CAG29370), HvVP1 (Q06572), and TaVP1 (ABX100 14). Star means the different amino acids
Expression patterns of AmVP1 under stresses. a Expression patterns of AmVP1 under salt stress. For salt treatment, 10-day-old seedlings growing on MS plate were carefully pulled out and moved into the liquid MS medium containing 200 mM NaCl to incubate for different periods (0, 1, 2, 4, 8, 12, and 24 h). b Expression patterns of AmVP1 under drought stress. For drought treatment, the pulled out seedlings were kept in a chamber on the 13 cm filter papers for 1 and 1.5 h. Total RNA extracted from the root of treated and untreated seedlings (as a control) was used for the real time RT-PCR analysis, with AmACTIN2 as an internal control. AmNHX2, a salt and drought responsive gene, was employed as a positive control. The entire experiments were repeated at least three times
Isolation and analysis of AmVP1 promoter
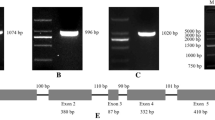
An 1,866 bp promoter sequence was isolated from the genomic DNA of A. mongolicus. Using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/), we analyzed the sequence of the AmVP1 and AtVP1 promoter, and predicted their key cis-acting elements and the location of these elements. AmVP1 promoter harbored multiple cis-acting elements, such as light responsive elements, phytohormone responsive elements and environmental stress signal responsive elements etc. (Table 1). Three predicted promoter sequences (Fig. 3) were detected in AmVP1 promoter using Neural Network Promoter Prediction software (http://www.fruitfly.org/seq_tools/promoter.html).
Promoter sequence of AmVP1. Capital character: part coding region of AmVP1; ARE: cis-acting regulatory element essential for the anaerobic induction; CGTCA-motif: cis-acting regulatory element involved in the MeJA-responsiveness; MBS: MYB binding site involved in drought-inducibility; TATC-Box: cis-acting element involved in gibberellin-responsiveness; TCA-element: cis-acting element involved in salicylic acid responsiveness; AuxRR-core: cis-acting regulatory element involved in auxin responsiveness; Number 1, 2 and 3: predicted promoter sequence, transcription start shown in larger font. Cis-elements constitutes was analyzed by website of PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/), and promoter sequence was predicted by Neural Network Promoter Prediction (http://www.fruitfly.org/seq_tools/promoter.html)
Functional characterization of AmVP1 using yeast mutant ena1
Previous work showed that hetero-expressions of vacuolar H+-pyrophosphatase genes in yeast mutant ena1 could partly suppress its hypersensitivity to NaCl (Gaxiola et al. 1999). The similar method was exploited to initially characterize the function of AmVP1. As shown in Fig. 4a, AmVP1 could partly restore the NaCl tolerance of ena1.
Functional characterization of AmVP1 using yeast mutant and identification as well as phenotypic analysis of AmVP1 transgenic Arabidopsis plants. a Functional characterization of AmVP1 using ena1 yeast mutant. The photograph corresponds to yeast at third day after growth on YPD or YPD + 0.5 M l−1 NaCl medium. b RT-PCR analysis of AmVP1 expression in representative transgenic lines, M-10, M-11 and M-16; (−), H2O; Vc, vector control. c Phenotypes of transgenic plants under salt stress. The photograph corresponds to plants 25 days after treatment with 200 mM NaCl. d Phenotypes of three plants of each transgenic line after water-deprivation for 15 days
Ectopic over-expression of AmVP1 in Arabidopsis
To characterize AmVP1 functionally in planta, its open reading frame (ORF), driven by 35S promoter, was introduced into Arabidopsis using the floral-dipping method (Clough and Bent 1998). Twenty one putative transgenic lines were obtained and eight leaves from each of the lines were taken to determine the transcript levels of AmVP1 by reverse transcription RT-PCR. A 400 bp AmVP1 specific fragment was detected in all the putative transgenic lines, but not in the control plants (transgenic plants with the empty vector) (Fig. 4b). A higher AmVP1 transcript level was accumulated in M-10 than in M-11 and M-16, with AtACTIN2 as an internal control (Fig. 4b). M-10 and M-11 were representatively used in the following analyses.
AmVP1 over-expression resulted in enhanced tolerance to salt stress
The transgenic plants, grown under 10 h-light/14 h-dark photoperiod for 35 days, were subject to salt stress treatment. The growth of all the plants was inhibited by the 200 mM NaCl solution. However, M-10 and M-11 grew better than the control plants. After 25 days in the solution, the control plants exhibited inhibitory growth state and severe chlorosis, whereas the transgenic plants remained healthier and greener (Fig. 4c). Eight rosette leaves from each of the transgenic lines were taken to determine the Na+ and K+ contents. As shown in Fig. 5a, b, more Na+ (38–48 %) and slightly higher K+ content were accumulated in the leaves of transgenic plants than in those of the control plants after salt stress, while no significant differences in the contents of Na+ and K+ were detected between the transgenic plants and the control plants under the normal conditions (Fig. 5a, b).
Physiological variations of transgenic plants under stresses. a and b Na+ and K+ contents in empty-vector transgenic plants (Vc) and AmVP1 transgenic plants under normal conditions (control) or salt stress (NaCl). Values are means ± SD (n = 8 for each). c MDA contents in the empty-vector transgenic plants (Vc) and AmVP1 transgenic plants under a water-deficit stress. Nw normal water; 15dD, 15-day water deprivation; Wr, 16 h after re-watering. Values are means ± SD (n = 6 for each). d Relative water content in plants under a water deficit stress. Values are means ± SD (n = 5 for each)
AmVP1 transgenic plants are also drought tolerant
Both transgenic and control plants, grown under 16-h-light/8-h-dark photoperiod for 20 days, were water-deprived for 15 days at 24 °C before being re-watered. As shown in Fig. 4d, after 15-day water deprivation, the control plants were severely wilted, while the two transgenic lines were lightly wilted. The drought tolerance of three transgenic lines correlated with the expression level of AmVP1. Line M-10, exhibiting the higher expression level, showed more drought tolerance than M-11.
The MDA content in both the transgenic and control plants significantly increased after 15-day water deprivation. However, it increased much more dramatically in the control plants, 12-fold compared to 2.2- and 3.5-fold increases in M-10 and M-11 respectively (Fig. 5c). After re-watering, MDA content in the control plant decreased to about tenfold, still higher than those in M-10 and M-11 (about two and threefold respectively) (Fig. 5c).
Five plants from each line were taken to determine the relative water content. Both the control and transgenic plants showed a significant reduction in their RWC during the drought stress. Nine days after water deprivation, the control plants began to lose water (the value was 0.87), and a sharp reduction of RWC was observed at days 11 and 13 (the respective RWC were 0.50 and 0.21). However, no significant water loss was observed in the two transgenic lines until day 13 (RWC of M-10 and M-11 dropped to value of 0.73 and 0.56 respectively) (Fig. 5d).
AmVP1 was likely a more efficient orthologue than AtVP1
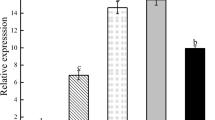
The functional difference between AmVP1 and AtVP1 was briefly analyzed by examining the rough ratio of their transcript levels to the tolerances to a salt stress in the respective transgenic plants. Two AtVP1 transgenic plants (A-3, A-7), with AtVP1 transcript level being about threefold of AmVP1 (about 8.1-fold of AtActin-2), exhibited the similar salt tolerant phenotype with M-10 (in which AmVP1 transcript level being 2.7-fold of AtActin-2), when being watered with 180 mM NaCl solution at a 5-day interval (Fig. 6). Another AtVP1 transgenic plant A-2, with AtVP1 transcript level being about 1.5-fold of AtActin-2, exhibited a poor salt tolerance.
A comparison of functional efficiencies between AmVP1 and AtVP1. a Phenotypes of different plants grown under normal conditions (upper) or after a salt stress (below). b MDA contents measured before and after a salt stress in the respective plants. Values are means ± SD (n = 4 for each). c Relative transcript levels of AmVP1 and AtVP1 in the respective plants. A-3, A-7 and A-2, three AtVP1 transgenic plant lines
Discussion
Ammopiptanthus mongolicus is a relic of the Tertiary Period. It was mainly distributed in the coast of the Ancient Mediterranean in the early period of the Tertiary Period, indicating that it was adapted to wet and warm climates. Therefore, the subsequent evolution of its extreme tolerances to a combination of abiotic stresses could be logically attributed to the gradual climate change (e.g. from warm and wet to extremely hot/cold and dry/salty), incurred by the geological change (Wei et al. 2012). The distinctive character makes A. mongolicus a valuable system for exploiting the mechanistic evolvement of abiotic stress tolerances in trees. In literature, there are several reports attempting to address its antifreezing, drought and salt tolerance mechanism (Cao et al. 2009a; Liu et al. 2010; Wei et al. 2011, 2012). In this work, we set out to identify the H+-PPase gene orthologue, which has been reported responsible for regulating the osmotic stress tolerance, from the radical cDNA library of A. mongolicus by RACE-PCR.
Previous studies revealed that the transcript level of the vacuolar H+-PPase genes was responsive to salt stress, cold, hypoxia and the lack of nutrient elements (Carystinos et al. 1995; Colombo and Cerana 1993; Darley et al. 1995; Fukuda et al. 2004; Kasai et al. 1998; Queiros et al. 2009). In this study, it was also shown that AmVP1 could be strongly induced by drought stress. This indicated that AmVP1 might play a role in the osmotic adjustment. The transcript level of AmVP1 also responded to the salt stress at the initial stage, which was in contrast to the observation made on the AtVP1 transcript level, indicating that AmVP1 might be involved in the early responsive pathway. This is partially in agreement with the observation on the responsive pattern of ThVP1 (an H+-PPase gene orthologue from Thellungiella halophila), which strongly responds to salt stress (Gao et al. 2006). Besides, we isolated the AmVP1 promoter sequence to further investigate the possible expression pattern of AmVP1. As seen in the Table 1, it harbored multiple phytohormone and environmental stress signal responsive cis-elements as well as tissue specific expression related elements. It indicated that AmVP1 may have various expression patterns. It was surprising that AmVP1 promoter differed mainly with AtVP1 promoter not only in the types of cis-elements but also in the number of cis-elements (Table 1), indicating the different regulation mechanism between AmVP1 and AtVP1.
It has been repeatedly reported that over-expression of vacuolar H+-PPase genes increases both salt and drought tolerances in plants (Bao et al. 2009; Brini et al. 2007; Gao et al. 2006; Li et al. 2010; Park et al. 2005). In this study, ectopic over-expression of AmVP1 also enhanced salt tolerance, with more sodium and potassium accumulated in leaves, and drought tolerance, with much lower MDA detected after drought stress, as well as stronger water-maintaining capability during drought stress. Those results are consistent with the observations reported previously (Bao et al. 2009; Brini et al. 2007; Gao et al. 2006; Gaxiola et al. 2001; Li et al. 2010). Besides, heteroexpressing AmVP1 could partly restore the NaCl tolerance of ena1 which was also used to demonstrate the function of Arabidopsis AVP1 by Gaxiola et al. (1999). Those results collectively implicate that AmVP1 functions as AtVP1 to drive the antiporters, such as Na+/H+ antiporter, to detoxify the Na+ in cytoplasm under high salt stress, or to drive other compound transporters, such as organic acids, sugars, to maintain cell turgor under water deficit stress (Gao et al. 2006; Taiz 1992).
As far as stress tolerant genes are concerned, it is still an open question if there exist more efficient orthologues in extremely stress-tolerant resource plants. In 2006, Gao reported a study in attempting to elucidate a possible difference in the functional efficiency between ThVP1 and AtVP1 in transgenic tobacco plants. However, it was found that the ectopic over-expression of ThVP1 conferred a similar level of salt tolerance as AtVP1, although ThVP1 strongly responded to the salt stress in Thellungiella halophila while the expression of AtVP1 in Arabidopsis showed no distinct change under salt stress (Gao et al. 2006). In this study, according to the ratio of their transcript levels and their tolerance to a salt stress, the functional efficiency of AmVP1 is about threefold higher than AtVP1. The differential efficiency of AmVP1 and AtVP1 might be due to one or more of the eight different amino acids in the key sites of H+-PPase between AmVP1 and those from the other six typical glycophytes (Fig. 1b). In other words, the differences in salt and drought sensitivity between A. thaliana and A.mongolicus may be partly attributed to the difference in functional efficiency between AmVP1 and AtVP1. AmVP1 is therefore a valuable gene for improving plant salt and drought tolerances.
References
Bao AK, Wang SM, Wu GQ, Xi JJ, Zhang JL, Wang CM (2009) Overexpression of the Arabidopsis H+-PPase enhanced resistance to salt and drought stress in transgenic alfalfa (Medicago sativa L.). Plant Sci 176:232–240
Brini F, Hanin M, Mezghani I, Berkowitz GA, Masmoudi K (2007) Overexpression of wheat Na+/H+ antiporter TNHX1 and H+-pyrophosphatase TVP1 improve salt- and drought-stress tolerance in Arabidopsis thaliana plants. J Exp Bot 58:301–308
Cao PX, Song J, Zhou CJ, Weng ML, Liu J, Wang FX, Zhao F, Feng DQ, Wang B (2009a) Characterization of multiple cold induced genes from Ammopiptanthus mongolicus and functional analyses of gene AmEBP1. Plant Mol Biol 69:529–539
Cao YJ, Wei Q, Liao Y, Song HL, Li X, Xiang CB, Kuai BK (2009b) Ectopic overexpression of AtHDG11 in tall fescue resulted in enhanced tolerance to drought and salt stress. Plant Cell Rep 28:579–588
Carystinos GD, Macdonald HR, Monroy AF, Dhindsa RS, Poole RJ (1995) Vacuolar H+-translocating pyrophosphatase is induced by anoxia or chilling in seedlings of rice. Plant Physiol 108:641–649
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Colombo R, Cerana R (1993) Enhanced activity of tonoplast pyrophosphatase in Nacl-grown cells of Daucus-carota. J Plant Physiol 142:226–229
Darley CP, Davies JM, Sanders D (1995) Chill-Induced changes in the activity and abundance of the vacuolar proton-pumping pyrophosphatase from mung bean hypocotyls. Plant Physiol 109:659–665
Drozdowicz YM, Rea PA (2001) Vacuolar H+ pyrophosphatases: from the evolutionary backwaters into the mainstream. Trends Plant Sci 6:206–211
Fukuda A, Chiba K, Maeda M, Nakamura A, Maeshima M, Tanaka Y (2004) Effect of salt and osmotic stresses on the expression of genes for the vacuolar H+-pyrophosphatase, H+-ATPase subunit A, and Na+/H+ antiporter from barley. J Exp Bot 55:585–594
Gao F, Gao Q, Duan XG, Yue G, Yang AF, Zhang JR (2006) Cloning of an H+-PPase gene from Thellungiella halophila and its heterologous expression to improve tobacco salt tolerance. J Exp Bot 57:3259–3270
Gaxiola RA, Rao R, Sherman A, Grisafi P, Alper SL, Fink GR (1999) The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc Natl Acad Sci USA 96:1480–1485
Gaxiola RA, Li JS, Undurraga S, Dang LM, Allen GJ, Alper SL, Fink GR (2001) Drought- and salt-tolerant plants result from overexpression of the AVP1 H+-pump. Proc Natl Acad Sci USA 98:11444–11449
Guo SL, Yin HB, Zhang X, Zhao FY, Li PH, Chen SH, Zhao YX, Zhang H (2006) Molecular cloning and characterization of a vacuolar H+-pyrophosphatase gene, SsVP, from the halophyte Suaeda salsa and its overexpression increases salt and drought tolerance of Arabidopsis. Plant Mol Biol 60:41–50
Holsters M, de Waele D, Depicker A, Messens E, van Montagu M, Schell J (1978) Transfection and transformation of Agrobacterium tumefaciens. Mol Gen Genet 163:181–187
Hu Y, Zeng Y, Guan B, Zhang F (2012) Overexpression of a vacuolar H+-pyrophosphatase and a B subunit of H+-ATPase cloned from the halophyte Halostachys caspica improves salt tolerance in Arabidopsis thaliana. Plant Cell Tissue Organ Cult 108:63–71
Kasai M, Nakamura T, Kudo N, Sato H, Maeshima M, Sawada S (1998) The activity of the root vacuolar H+-pyrophosphatase in rye plants grown under conditions deficient in mineral nutrients. Plant Cell Physiol 39:890–894
Li ZG, Baldwin CM, Hu Q, Liu H, Luo H (2010) Heterologous expression of Arabidopsis H plus -pyrophosphatase enhances salt tolerance in transgenic creeping bentgrass (Agrostis stolonifera L.). Plant Cell Environ 33:272–289
Liu RL, Liu MQ, Liu J, Chen YZ, Chen YY, Lu CF (2010) Heterologous expression of a Ammopiptanthus mongolicus late embryogenesis abundant protein gene (AmLEA) enhances Escherichia coli viability under cold and heat stress. Plant Growth Regul 60:163–168
Luttge U, Ratajczak R (1997) The physiology, biochemistry and molecular biology of the plant vacuolar ATPase. Adv Bot Res 25:253–296
Lv SL, Lian LJ, Tao PL, Li ZX, Zhang KW, Zhang JR (2009) Overexpression of Thellungiella halophila H+-PPase (TsVP) in cotton enhances drought stress resistance of plants. Planta 229:899–910
Maeshima M (2000) Vacuolar H+-pyrophosphatase. Bba-Biomembranes 1465:37–51
Mitsuda N, Takeyasu K, Sato MH (2001) Pollen-specific regulation of vacuolar H+-PPase expression by multiple cis-acting elements. Plant Mol Biol 46:185–192
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plantarum 15:473–497
Park S, Li JS, Pittman JK, Berkowitz GA, Yang HB, Undurraga S, Morris J, Hirschi KD, Gaxiola RA (2005) Up-regulation of a H+-pyrophosphatase (H+-PPase) as a strategy to engineer drought-resistant crop plants. Proc Natl Acad Sci USA 102:18830–18835
Queiros F, Fontes N, Silva P, Almeida D, Maeshima M, Geros H, Fidalgo F (2009) Activity of tonoplast proton pumps and Na+/H+ exchange in potato cell cultures is modulated by salt. J Exp Bot 60:1363–1374
Taiz L (1992) The plant vacuole. J Exp Biol 172:113–122
Wang W, Chen JJ, Li JN, Zhang YH, Shao ZY, Kuai B (2007) Extraordinary accumulations of antioxidants in Ammopiptanthus mongolicus (Leguminosae) and Tetraena mongolica (Zygophyllaceae) distributed in extremely stressful environments. Bot Stud 48:55–61
Wei Q, Guo YJ, Cao HM, Kuai BK (2011) Cloning and characterization of an AtNHX2-like Na+/H+ antiporter gene from Ammopiptanthus mongolicus (Leguminosae) and its ectopic expression enhanced drought and salt tolerance in Arabidopsis thaliana. Plant Cell Tissue Organ 105:309–316
Wei Q, Kuai B, Hu P, Ding Y (2012) Ectopic-overexpression of an HD-ZipIV transcription factor from Ammopiptanthus mongolicus (Leguminosae) promoted upward leaf curvature and non-dehiscent anthers in Arabidopsis thaliana. Plant Cell Tissue Organ. doi:10.1007/s11240-012-0151-8
Wisniewski JP, Rogowsky PM (2004) Vacuolar H+-translocating inorganic pyrophosphatase (Vpp 1) marks partial aleurone cell fate in cereal endosperm development. Plant Mol Biol 56:325–337
Zandonadi DB, Canellas LP, Facanha AR (2007) Indolacetic and humic acids induce lateral root development through a concerted plasmalemma and tonoplast H+ pumps activation. Planta 225:1583–1595
Zhang GH, Su Q, An LJ, Wu S (2008) Characterization and expression of a vacuolar Na+/H+ antiporter gene from the monocot halophyte Aeluropus littoralis. Plant Physiol Bioch 46:117–126
Acknowledgments
We are grateful to Qiang Gao and Dr. Aifang Yang of the Shandong University, who generously offered the yeast mutant ena1. We thank Wei Wang and Dr. Hongxuan Lin of the Institute of Plant Physiology & Ecology for their help in the measurement of Na+/K+ contents. This work was supported by grants (2008ZX08009-004, 2009ZX08009-061B) from the National Program of Transgenic Variety Development of China and a grant from the Science and Technology Committee of the Shanghai Municipal Government (Grant no. 08DZ2270800).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Wei, Q., Hu, P. & Kuai, B.K. Ectopic expression of an Ammopiptanthus mongolicus H+-pyrophosphatase gene enhances drought and salt tolerance in Arabidopsis . Plant Cell Tiss Organ Cult 110, 359–369 (2012). https://doi.org/10.1007/s11240-012-0157-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-012-0157-2