Abstract
Based on both unique morphological characteristics of the gamont, distinct changes caused to the host erythrocyte and analysis of partial 18S rRNA gene sequences, a new parasite of the genus Hepatozoon Miller, 1908 is described from the snake Philodryas nattereri Steindachner (Squamata: Dipsadidae) in northeastern Brazil. The new species, Hepatozoon musa n. sp., is characterized by large and curved mature gamonts (18.9 ± 0.9 μm in length and 3.8 ± 0.3 μm in width) that considerably engorge infected host erythrocytes and displace the nucleus laterally, which become longer and thinner. Phylogenetic estimates indicate the new species is more closely related to the recently described Hepatozoon cuestensis O’Dwyer, Moço, Paduan, Spenassatto, Silva & Ribolla, 2013, from Brazilian rattlesnakes. These recent findings highlight the need for further studies of Hepatozoon to better determine the biodiversity of this common but poorly-studied parasite group.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Members of the genus Hepatozoon Miller, 1908 are the most frequently reported blood parasites of reptiles (Telford, 2009). Surprisingly, the systematics and diversity of these parasites remains poorly understood. They are part of the Apicomplexa, a phylum in which it has been estimated that over 99% of species remain undescribed (Morrison, 2009). Recent estimates of relationships based on 18S rRNA gene sequences have also indicated inconsistencies between predicted phylogeny and current taxonomy. Hepatozoon have traditionally been placed in the family Hepatozoidae Wenyon, 1926, which along with members of the Karyolysidae Wenyon, 1926 and Haemogregarinidae Léger, 1911 form the haemogregarines. However, phylogenetic trees indicate that the genus Karyolysus Labbe, 1894 is part of a lineage within a paraphyletic Hepatozoon (see Maia et al., 2012, Haklová-Kočíková et al., 2014). Recent molecular analyses have also indicated that many different genetic lineages of parasites can occur in reptile species, in particular within geckos (Tomé et al., 2016) and snakes (Tomé et al., 2014). A proposed taxonomic revision erected a new genus, Bartazoon Karadjian, Chavatte & Landau, 2015, to include most species previously included in Hepatozoon, except for those from mammalian carnivores (Karadjian et al., 2015). However, this new genus is not monophyletic in several analyses while new genetic lineages continue to be identified, e.g. Tomé et al. (2016) and Maia et al. (2016). Therefore, we conservatively continue to refer these species to Hepatozoon. These issues highlight that there is a clear need for further assessment of these parasites, employing both morphological and genetic tools, to better delimit diversity and to place this within an evolutionary framework.
Many species of Hepatozoon have been identified in snakes from Brazil, but mainly based only on morphological characteristics of the gamont in blood cells (reviewed in Moço et al., 2012). A recent study employing molecular techniques identified three new species in the rattlesnake Crotalus durissus Linnaeus from Brazil, highlighting the value of this methodology (O’Dwyer et al., 2013). At the same time, estimates of phylogeny of Hepatozoon and related parasites in geckos from Brazil indicated a possible scenario of diversification in South America followed by later spread to other regions for a large clade of parasites, although also suggesting that more data would be needed to confirm this (Harris et al., 2015). Assessment of Hepatozoon in other snakes in South America therefore can be used to shed further light on this possibility and improve knowledge of this poorly known but common group of parasites. Here we report the finding of a novel Hepatozoon species from the snake Philodryas nattereri Steindachner from Upanema, Rio Grande do Norte, in northeastern Brazil. Morphological characteristics of the gamont are very different from those previously identified in this genus of snake, Hepatozoon philodryasi Carini, 1910 from the host species Philodryas patagoniensis (Girard) (see Carini, 1910). Phylogenetic estimates based on 18S rRNA sequences also indicate the distinctiveness of these parasites, and therefore we propose that they represent a new species, Hepatozoon musa n. sp., which is described here.
Materials and methods
Sample collection
Six P. nattereri from three municipalities in the State of Ceará (Fortaleza 3°19′0″S, 41°25′0″W; Itapipoca 3°29′38″S, 39°34′44″W; and Flecheiras 03°16′39″S, 39°16′11″W) and one sample from the State of Rio Grande do Norte (Upanema, 05°38′32″S, 37°15′26″W) were studied. All the specimens were taken to NUROF-UFC, Universidade Federal do Ceará, where blood was taken by tail venipuncture. Blood samples were stored in 96% ethanol for molecular assessment, and blood smears were immediately prepared. Smears were air-dried, fixed with methanol and stained with Giemsa (1:9 distilled water).
Morphological analysis
The slides were examined for parasites using an Olympus CX41 microscope with an in-built digital camera (SC30) (Olympus, Hamburg, Germany). Intensity of infection was estimated as the number of parasites per 5,000 erythrocytes. Intraerythrocytic mature gamont stages and infected host erythrocytes were measured at 1,000× magnification using cell ^B software (basic image acquisition and archiving software; Olympus, Münster, Germany). Length, width and area were taken using segmented and freehand selection tools after setting the scale, in ImageJ v1.50 (Abramoff et al., 2004). Fifty infected erythrocytes, 50 uninfected erythrocytes and 50 mature gamonts were measured (Table 1); data are given in micrometres as the range followed by the mean and standard deviation in parentheses.
Genetic characterisation
Molecular characterisation of Hepatozoon parasites was performed via the polymerase chain reaction (PCR) amplification using both the HEMO primers (Perkins & Keller, 2001) and the HEP primers (Ujvari et al., 2004) so that a longer consensus sequence of a 1,400 bp fragment of the 18S rRNA gene could be obtained. Briefly the PCR protocol involved 20 µl reaction mixture containing 4.0 U of Blend MasterMix, 0.6 μM of each primer, 0.4 mg/µl of albumin (BSA), 2.0 μl of DNA and remaining volume was filled with distilled water. The reaction mix for HEMO primers was amplified using the following cycles: 94°C (3 min), 35 × [94°C (30 s), 48°C (30 s) and 72°C (1 min)], and 72°C (10 min). The reaction mix for HEP primers was amplified by: 94°C (3 min), 35× [94°C (30 s), 60°C (30 s) and 72°C (1 min)], and 72°C (10 min) (Harris et al., 2011). Positive (a known infected sample) and negative controls (distilled water substituting the DNA extraction) were run with each reaction. All positive products were sent to a commercial company for sequencing (Beckman Coulter Genomics). The new sequences were submitted to the GenBank database under accession number KX880079.
Phylogenetic analysis
The sequences generated in this study were aligned with related sequences retrieved from GenBank. Although a longer fragment was generated in this study, only a short fragment was used in the phylogenetic analysis, so that more comparative sequences from GenBank could be utilised. The final dataset contained 55 taxa and was 540 bp in length. Maximum likelihood (ML) analysis with random sequence addition (100 replicate heuristic searches) was used to estimate evolutionary relationships using the software PhyML (Guindon et al., 2010). Support for nodes was estimated using the bootstrap technique, with 1,000 replicates. The model of evolution employed was chosen using the AIC criteria carried out in jModeltest 2.1.9 (Posada, 2008). Bayesian inference (BI) was implemented using MrBayes v.3.2 (Huelsenbeck & Ronquist, 2001; Ronquist et al., 2012). The analysis was run for 1 × 107 generations, saving one tree every 1,000 generations. All trees prior to reaching stationary were discarded as ‘burn-in’ samples (25%), and remaining trees were combined in a 50% majority consensus tree. Following Barta et al. (2012), Babesiosoma stableri Schmittner & McGhee, 1961 and Dactylosoma ranarum Labbe, 1894 were used as outgroups.
Results
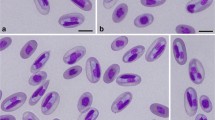
The six samples from Ceará were all negative using microscopy, and also gave negative results with PCR. However, the sample from Upanema was heavily infected, with 114 gamonts identified in 5,000 erythrocytes, or 2.28% of erythrocytes infected (Fig. 1). Measurements showed that infected cells were greatly enlarged (area of 198 μm2 compared to an average 157 μm2 for uninfected cells; Welch Two Sample t-test, t = 13.002, df = 88.277, P < 0.001). This is not surprising given that gamonts were 18.94 μm long, longer than a typical uninfected erythrocyte.
Molecular analysis
The single sample that was positive for microscopy gave successful PCR results for both HEP and HEMO primer pairs. The resulting sequences were unambiguous, and matched published Hepatozoon sequences when compared to data from GenBank using BLAST similarity searches (Altschul et al., 1990). We could therefore be confident that we had amplified DNA from the same parasite that was identified through microscopy.
Both ML and BI produced similar estimates of relationships (Fig. 2) that indicated the same major clades as other recent phylogenetic analyses (Harris et al., 2015; Tomé et al., 2016). The new sequence generated in this study was indicated to be the sister taxon to the recently described Hepatozoon cuestensis O’Dwyer, Moço, Paduan, Spenassatto, da Silva & Ribolla, 2013 from the rattlesnake Crotalus durissus from southern Brazil (O’Dwyer et al., 2013).
Based on these observations, of a morphologically distinct gamont, a genetic characterisation based on the 18S rRNA gene sequences, and a new host species, we propose that the parasite identified should be considered a new species, which we here describe as Hepatozoon musa n. sp.
Family Hepatozoidae Wenyon, 1926
Genus Hepatozoon Miller, 1908
Hepatozoon musa n. sp.
Type-host: Philodryas nattereri Steindachner (Colubridae), Paraguay green racer.
Type-locality: Upanema (5°38′32″S, 37°15′27″W), Rio Grande de Norte, Brazil.
Type-specimens: Hapantotype deposited in the collection of CIBIO, University of Porto, under accession number Co1031.
Prevalence and parasitaemia levels: No P. nattereri from the State of Ceará (6 individuals) were infected. A single specimen from Upanema, Rio Grande do Norte, was infected with 2.28% infected erythrocytes (114 mature gamonts in 5,000 erythrocytes).
Etymology: The term refers to the form of the gamonts of the new species, which shows similarity to the fruit called “banana”, common to plants of the genus Musa.
Description (Fig. 1)
Gamont. Mature gamonts (Fig. 1) 16.6–20.9 (18.9 ± 0.9) in length, 3.1–4.4 (3.8 ± 0.3) in width; area 59.7–94.9 (77.6 ± 8.5) μm2 (Table 1). Nuclei situated typically in the central portion of gamont. Host cell nuclei typically displaced laterally; infected erythrocytes larger than average uninfected cells (198.1 vs 157.5 μm2). Gamonts with considerable curvature at both ends. The nucleus of infected cells was reduced in size and changed in shape, becoming longer and thinner.
Discussion
Only one species of Hepatozoon has previously been identified in snakes of the genus Philodryas, H. philodryasi from Philodryas patagoniensis (see Carini, 1910). However, our report of Hepatozoon is the first of this parasite genus in P. nattereri. Philodryas patagoniensis is a species that occurs in forested environments, with a distribution from northern Brazil to Uruguay and Argentina. It also occurs in the Cerrado and Atlantic Forest, from Bahia to southern Brazil (Uetz & Hošek, 2015). This is quite different from P. nattereri, a species related to open environments, occurring in the Caatinga (from Piauí to Minas Gerais), Cerrado (Goiás and Tocantins) and Chaco (Paraguay) (Vitt et al., 2002; Uetz & Hošek, 2015), although there are records for the Atlantic Forest localities (Morato et al., 2011; Hamdan & Lira-da-Silva, 2012). Thus although the two species can be found in sympatry in some locations (e.g. Distrito Federal-Goiás and South Bahia) (França et al., 2008; Hamdan & Lira-da-Silva, 2012; Freitas, 2014), in general they occupy different ecological niches, and in the region where the specimens from this study were collected P. patagoniensis does not occur. Moreover, morphologically the gamonts identified in this study are very different from those of H. philodryasi. Carini (1910) indicated that this parasite was between 11–13 μm long, 2.0–3.5 μm wide, and that parasitised erythrocytes were not altered by the parasite. The Hepatozoon found in P. nattereri is much larger than this, averaging 18.9 μm in length and 3.8 μm in width, and parasitised erythrocytes are strongly altered, being considerably swollen by the presence of the parasite. We can therefore discard the possibility that the species identified in this study is H. philodryasi. Based on our analysis of 18S rRNA sequence data, the newly identified parasite is most closely related to Hepatozoon cuestensis, from C. durissus from southern Brazil (Fig. 2). The genetic distance is around 1%, which is considerable for the slow evolving 18S rRNA marker, and is higher than the divergence between H. cevapii O’Dwyer, Moço, Paduan, Spenassatto, da Silva & Ribolla, 2013 and H. massardii O’Dwyer, Moço, Paduan, Spenassatto, da Silva & Ribolla, 2013, two species from the host Crotalus durissus (Fig. 2). At the same time, morphologically the gamonts are again very different. In particular, the area of the gamonts from the parasite from P. nattereri averages 77.8 μm2. Although O’Dwyer et al. (2013) identified considerable variation of gamonts within H. cuestensis, the largest form was only 55.8 μm2. Furthermore in H. cuestensis the nucleus of the gamont is displaced to one end of the parasite. This was not observed in the parasite from P. nattereri.
Our finding of a new species of Hepatozoon in P. nattereri, along with the recent description of three new species from rattlesnakes (O’Dwyer et al., 2013) indicates that haemogregarine parasite diversity in Brazilian snakes is still greatly underestimated. The new species appears to be the sister taxon to H. cuestensis from rattlesnakes, and this pair then related to an undescribed Hepatozoon from geckos, also from Brazil (Harris et al., 2015). There does seem to be a group of essentially South American Hepatozoon, with extensive diversity and with lineages forming basal branches of one of the main clades within the group (Fig. 2). Such geographically cohesive groups do not occur in other well-sampled regions such as North Africa and the Iberian Peninsula (Maia et al., 2012). Greater sampling of Hepatozoon from other regions, particularly Australasia, would be necessary to try to understand these patterns.
Hepatozoon have a heteroxenous life-cycle, in which merogony and gametogony occur in the vertebrate host, with the sexual cycle and sporogony occurring in a definitive invertebrate host (Telford, 2009). Many invertebrates can host Hepatozoon, including ticks, mites and mosquitos. Which invertebrates might host H. musa remain unknown, and clearly further investigation of potential definitive hosts from the same regions in northeastern Brazil is needed. Molecular tools have been used to identify haemogregarine parasites from hosts such as ticks (Herbert et al., 2010; Harris et al., 2013), and similar screening of ectoparasites and mosquitoes in Brazil could help to assess this aspect.
To conclude, we describe a new species of Hepatozoon from Philodryas nattereri from Brazil. This species and other recent descriptions highlight the need for further assessments of these common but poorly known parasites in snakes, not just from South America but also from other geographical regions. In this way the evolutionary history of these parasites can be better assessed, and proposed biogeographic hypotheses tested.
References
Abramoff, M. D., Magalhães, P. J., & Ram, S. J. (2004). Image processing with ImageJ. Biophotonics International, 11, 36–42.
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410.
Barta, J. R., Ogedengbe, J. D., Martin, D. S., & Smith, T. G. (2012). Phylogenetic position of the adeleorinid coccidia (Myzozoa, Apicomplexa, Coccidia, Eucoccidiorida, Adeleorina) inferred using 18S rDNA sequences. Journal of Eukaryotic Microbiology, 59, 171–180.
Carini, A. (1910). Sobre uma hemogregarina de Philodryas schottii. Revista de Medicina (São Paulo), 23, 339–340.
França, F. G. R., Mesquita, D. O., Nogueira, C. C., & Araújo, A. F. B. (2008). Phylogeny and Ecology Determine Morphological Structure in a Snake Assemblage in the Central Brazilian Cerrado. Copeia, 1, 23–28.
Freitas, M. A. (2014). Squamate reptiles of the Atlantic Forest of northern Bahia, Brazil. Check List, 10, 1020–1030.
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., & Gascuel, O. (2010). New algorithms and methods to estimate Maximum-Likelihood phylogenies: Assessing the performance of PhyML 3.0. Systematic Biology, 59, 307–321.
Haklová-Kočíková, B., Hižňanová, A., Majláth, I., Račka, K., Harris, D. J., Földvári, G., et al. (2014). Morphological and molecular characterization of Karyolysus - a neglected but common parasite infecting some European lizards. Parasites & Vectors, 7, 555.
Hamdan, B., & Lira-da-Silva, R. M. (2012). The snakes of Bahia State, northeastern Brazil: species richness, composition and biogeographical notes. Salamandra, 48, 31–50.
Harris, D. J., Borges-Nojosa, D. M., & Maia, J. P. (2015). Prevalence and diversity of Hepatozoon in native and exotic geckos from Brazil. Journal of Parasitology, 101, 80–85.
Harris, D. J., Graciá, E., Jorge, F., Maia, J. P. M. C., Perera, A., Carretero, M. A., & Giménez, A. (2013). Molecular detection of Hemolivia (Apicomplexa: Haemogregarinidae) from ticks of North African Testudo graeca (Testudines: Testudinidae) and an estimation of their phylogenetic relationships using 18S rRNA sequences. Comparative Parasitology, 80, 292–296.
Harris, D. J., Maia, J. P. M. C., & Perera, A. (2011). Molecular characterization of Hepatozoon species in reptiles from the Seychelles. Journal of Parasitology, 97, 106–110.
Herbert, J. D. K., Godfrey, S. S., Bull, C. M., & Menz, R. I. (2010). Developmental stages and molecular phylogeny of Hepatozoon tuatarae, a parasite infecting the New Zealand tuatara, Sphenodon punctatus and the tick, Amblyomma sphenodonti. International Journal for Parasitology, 40, 1311–1315.
Huelsenbeck, J. P., & Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics, 17, 754–755.
Karadjian, G., Chavatte, J.-M., & Landau, I. (2015). Systematic revision of the adeleid haemogregarines, with creation of Bartazoon n. g., reassignment of Hepatozoon argantis Garnham, 1954 to Hemolivia, and molecular data on Hemolivia stellata. Parasite, 22, 31.
Maia, J. P., Harris, D. J., Carranza, S., & Goméz-Díaz, E. (2016). Assessing the diversity, host-specificity and infection patterns of apicomplexan parasites in reptiles from Oman, Arabia. Parasitology. doi:10.1017/S0031182016001372.
Maia, J. P. M. C., Perera, A., & Harris, D. J. (2012). Molecular survey and microscopic examination of Hepatozoon Miller, 1908 (Apicomplexa: Adeleorina) in lacertid lizards from the western Mediterranean. Folia Parasitologica, 59, 241–248.
Moço, T. C., da Silva, R. J., Madeira, N. G., Dos Santos Paduan, K., Rubini, A. S., Leal, D. D. M., & O’Dwyer, L. H. (2012). Morphological, morphometric, and molecular characterization of Hepatozoon spp. (Apicomplexa, Hepatozoidae) from naturally infected Caudisona durissa terrifica (Serpentes, Viperidae). Parasitology Research, 110, 1393–1401.
Morato, S. A. A., Lima, A. M. X., Staut, D. C. P., Faria, R. G., Souza-Alves, J. P., Gouveia, S. F., et al. (2011). Amphibians and reptiles of the Refúgio de Vida Silvestre Mata do Junco, municipality of Capela, state of Sergipe, northeastern Brazil. Check List, 7, 756–762.
Morrison, D. A. (2009). Evolution of the Apicomplexa: Where are we now? Trends in Parasitology, 25, 375–382.
O’Dwyer, L. H., Moço, T. C., Paduan, K. D. S., Spenassatto, C., da Silva, R. J., & Ribolla, P. E. M. (2013). Description of three new species of Hepatozoon (Apicomplexa, Hepatozoidae) from Rattlesnakes (Crotalus durissus terrificus) based on molecular, morphometric and morphologic characters. Experimental Parasitology, 135, 200–207.
Perkins, S. L., & Keller, A. K. (2001). Phylogeny of nuclear small subunit rRNA genes of hemogregarines amplified with specific primers. Journal of Parasitology, 87, 870–876.
Posada, D. (2008). jModelTest: Phylogenetic Model Averaging. Molecular Biology and Evolution, 25, 1253–1256.
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61, 539–542.
Telford, S. R. (2009). Hemoparasites of the Reptilia (p. 394). Boca Raton, Florida: CRC Press, Taylor and Francis.
Tomé, B., Maia, J. P., Salvi, D., Brito, J. C., Carretero, M. A., Perera, A., et al. (2014). Patterns of genetic diversity in Hepatozoon spp. infecting snakes from North Africa and the Mediterranean Basin. Systematic Parasitology, 87, 249–258.
Tomé, B., Rato, C., Perera, A., & Harris, D. J. (2016). High diversity of Hepatozoon spp. in geckos of the genus Tarentola. Journal of Parasitology, 102, 476–480.
Uetz, P., & Hošek, J. (2015). The Reptile Database. http://www.reptile-database.org. Accessed 19 March 2016.
Ujvari, B., Madsen, T., & Olsson, M. (2004). High prevalence of Hepatozoon spp. (Apicomplexa, Hepatozoidae) infection in water pythons (Liasis fuscus) from tropical Australia. Journal of Parasitology, 90, 670–672.
Vitt, L. J., Caldwell, J. P., Colli, G. R., Garda, A. A., Mesquita, D. O., França, F. G. R., & Balbino, S. F. (2002). Um guia fotográfico dos répteis e anfíbios da região do Jalapão no Cerrado brasileiro. Publications in Herpetology Sam Noble Oklahoma Museum of Natural History, Norman, Oklahoma, USA, 1, 1–17.
Funding
This study was supported by CNPq to Project “Pesquisador Visitante Estrangeiro” (PVE-Process No.401800/2013-1). DMBNojosa was supported by CNPq “Pesquisador Produtividade” (PQ-Process No. 309617/20120). DJH was funded through an IF-FCT contract (IF/01627/2014) under the “Programa Operacional Potencial Humano – Quadro de Referência Estratégico Nacional” funds from the European Social Fund and the Portuguese Ministério da Educação e Ciência.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All applicable institutional, national and international guidelines for the care and use of animals were followed.
Rights and permissions
About this article
Cite this article
Borges-Nojosa, D.M., Borges-Leite, M.J., Maia, J.P. et al. A new species of Hepatozoon Miller, 1908 (Apicomplexa: Adelerina) from the snake Philodryas nattereri Steindachner (Squamata: Dipsadidae) in northeastern Brazil. Syst Parasitol 94, 65–72 (2017). https://doi.org/10.1007/s11230-016-9676-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11230-016-9676-2