Abstract
Nautilus species are the only remaining cephalopods with an external shell. Targeted heavily by the shell trade across their distribution area, these species have a poorly known population structure and genetics. Molecular techniques have been used to assess levels of inter- and intra-population genetic diversity in isolated populations of Nautilus in the northern sections of the Great Barrier Reef (GBR), Australia and in the Coral Sea. Distinct populations, physically separated by depths in excess of 1,000 m were examined. RAPD analysis of genetic differences showed limited differentiation of the “Northern GBR” populations and the “Coral Sea” populations. Discrimination between the two geographic groups was observed from these data. In addition, partial sequencing of the CoxI gene region, yielded 575 bp of sequence, which was aligned for 43 samples and phylogenetic trees constructed to examine genetic relationships. Two distinct clades were resolved in the resulting trees, representing the “Northern GBR” and “Coral Sea” population groups. Inter- and intra-population relationships are presented and discussed. The differentiation of the Nautilus populations from the Northern section of the Great Barrier Reef and those from the Coral Sea were supported by two distinctly different methodologies and the significance of this separation and the potential evolutionary divergence of these two population groups is discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
There are two subclasses of extant cephalopods: the Coleoidea and the Nautiloidea. The Coleoidea includes all known forms of cuttlefish, octopus and squid (>650 species). The Nautiloidea represent a distinctly monophyletic group, containing the only extant cephalopods with an external shell (Bonnaud et al. 2004). The pearly Nautilus (Mollusca, Cephalopoda) is a modern survivor of a previously abundant group of shelled cephalopods and it is a direct descendant of these fossil forms (Woodruff et al. 1987; Wray et al. 1995; Boyle and Rodhouse 2005). Although often cited as a living fossil (Wells 1986; Davis 1987; Basil et al. 2000) genetic studies indicate that current Nautilus species may only have originated between 1 and 5 million years ago (Woodruff et al. 1987).
The biology (Ward 1987; Suzuki et al. 2000; Springer et al. 2004; Boyle and Rodhouse 2005), sensory chemotaxis (Basil et al. 2000, 2005; Ruth et al. 2002; Westermann and Beuerlein 2005) and functional morphology (Wani 2004; Westermann et al. 2004, 2005) of Nautilus pompilius have been well described. It is known that Nautilus are generally found among the coral reefs of the Indo-Pacific, where they are not only predators (like their remote ancestors), but they also scavenge on detrital matter settling on the reef bottom. During daylight hours, Nautilus are found at depths of 200–400 m, ascending to shallower depths during the nocturnal hours to feed and forage (Carlson et al. 1984; Saunders 1984; Ward et al. 1984; Saunders and Landman 1987; Saunders and Ward 1987; Ward 1987; Hanlon and Messenger 1996; Wells et al. 1992).
The number of distinct species of Nautilus which exist is still a subject of debate. Based on shell morphology alone, Ward (1987) identified a total of 11 species and three variants. However, Saunders (1987) recognised five species, not including Allonautilus [N. belauensis Saunders, N. macromphalus Sowerby, N. pompilius Linnaeus, N. scrobiculatus Lightfoot and N. stenomphalus Sowerby] (Saunders 1981a, b, 1987; Ward 1987). Wray et al. (1995) further examined the genetic and geographic divergence of N. pompilius across its Indo-Pacific distribution, suggesting the presence of three geographically distinct clades as opposed to independent genetic lineages. Two species of Nautilus (N. pompilius and N. stenomphalus) were recorded in waters off the Great Barrier Reef, Australia (GBR) in the 1980s (Saunders 1987).
The use of molecular techniques to study population structure and to identify discrete populations is now commonplace across a wide range of marine vertebrate and invertebrate species (Fauvelot et al. 2003; van Herwerden et al. 2003; Yamamoto et al. 2004). In particular, mitochondrial DNA polymorphisms have become increasingly important in determining fish stock structures, because of its maternal mode of inheritance and relatively rapid rate of evolution (Brown et al. 1992; Conseugra et al. 2002) and has become widely used as a population genetics tool in fisheries biology (Ovenden 1990; Perrin and Borsa 2001; Hebert et al. 2003; Aspden et al. 2006; Pegg et al. 2006).
A number of genetic and evolutionary studies have been carried out in a range of cephalopod groups (Bonnaud et al. 1998; Anderson 2000a, b; Lindgren et al. 2005; Söller et al. 2000; Strugnell et al. 2005). Similarly, a number of such approaches have utilised mitochondrial DNA gene sequences, such as CoxI (cytochrome oxidase, subunit I) to investigate and resolve genetic and evolutionary relationships at a range of hierachical levels (Lindgren et al. 2004; Bonnaud et al. 1997; Lindgren et al. 2005; Strugnell et al. 2005; Anderson 2000a, b), however many studies use a combined approach with more than one DNA sequence being compiled to give greater depth and strength to such analyses. Other than the studies by Wray et al. (1995) and Bonnaud et al. (2004), there have been limited studies on the molecular population and evolutionary genetics of Nautilus, although there are other reports where Nautilus DNA sequences have been generated (Merritt et al. 1998; Giribet and Wheeler, 2002; Lindgren et al. 2005). To date, there is no significant work reported on the population genetics of Nautilus in the GBR and there has been no previous attempt to characterise discrete populations of Nautilus from isolated reefs of the Coral Sea. Nautilus are fished for the ornamental shell trade in a number of locations and many of these fisheries are being over-exploited.
Population-structure and -genetics of Nautilus from geographically distinct populations on the Eastern seaboard of Queensland, Australia and in the isolated seamounts of the Coral Sea are presented in this study. These data will provide important new information, which can be incorporated into future conservation plans for these sites. The phylogenetic relationships and the potential evolutionary divergence of these remote and distinct populations are being assessed on the hypothesis that genetic divergence has taken place due to geographic and physical isolation over evolutionary time.
Materials And Methods
Sampling sites
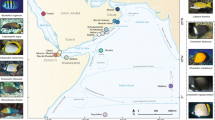
Osprey reef (lat. 13°53.44; long. 146°33.27) is a seamount, 195 km2 in surface area, in the Coral Sea ∼200 km NNE of Cairns (see Fig. 1) and represents the primary sample population for this study. The reef formation at Osprey ranges from low-tide breaking reef to coral gardens at 20 m with steep coral walls dropping to over 1,200 m. Other sampling sites in this “Coral Sea” group included in this study are Shark reef (lat. 14°05.88; long. 146°47.02), ∼15 km South of Osprey and separated by depths in excess of 1,000 m and Bougainville reef (lat. 15°29.00; long. 147°06.50), a further 30 km South of Shark reef and isolated by similar depths. In a separate “Northern GBR” group of reefs (∼400 km North of Cairns and 250 km NNW of Osprey reef) which provide Nautilus samples from the far northern distribution of the species in reefs adjacent to the Great Barrier Reef, samples were obtained from isolated sites around Raine Island: Southern Small Detached reef (lat. 12°34.71; long. 143°51.30), Northern Small Detached reef (lat. 1°39.59; long. 143°58.56), Mantis reef (lat. 12°03.77; long. 143°55.77), and Wishbone reef (lat. 12°05.57; long. 143°57.29). Osprey reef, unlike the GBR sites, has vertical reef walls that are protected from the southeasterly trade winds and is therefore accessible throughout the year. Great Barrier Reef sites were accessed for Nautilus sampling when weather conditions allowed.
Schematic map illustrating the approximate geographic locations of the sites, relative to Queensland, Australia, where Nautilus populations/samples were obtained. Sample animals were collected from between 150–350 m depth in a wire mesh trap with chicken as attractant. Captured animals were stored in fresh sea water at 10°C for 2 h before tentacle biopsy samples were taken and stored in 20% DMSO. Animals were released at dusk at depths of 20–30 m
Capture and release
A barrel shaped trap (90 × 77 cm) constructed from wire mesh (7.5 × 9 cm mesh size) was used. The trap had two entrance funnels of 23 cm diameter on each end of the barrel. Traps were deployed at 200–300 m depth at dusk and retrieved at dawn: traps were lowered on 8 mm rope with an attached float and either set to the seafloor if it is a flat bottom or tethered to the reef if sampling on a vertical wall. Captured Nautilus were kept for a maximum of 15 h in a dedicated, dark refrigerated tank (50 L) at temperatures between 16–19°C, before being released at night in 20–30 m depths.
Tissue sampling
The terminal 4–5 mm section from one labial tentacle from each animal was collected and preserved initially in 20% DMSO (dimethyl-sulphoxide), 100 mM EDTA, saturated NaCl solution stored at 4°C in the field. Subsequently, the tissue was washed in TE and placed into 80% alcohol preservative for storage until required for DNA extraction.
DNA extraction
Total DNA was extracted using a QIAGEN DNeasy Tissue kit, following the manufacturers instructions (Qiagen Pty. Ltd., Poncaster, Vic., Australia).
PCR amplification of RAPDs
Forty commercially available arbitrary 10-mer oligonucleotide primers (Qiagen Pty. Ltd.) were screened with three accessions of Nautilus DNA. Primers used to characterise genetic diversity in this study were selected based on the presence of polymorphic bands between the accessions in this test and are indicated in Table 1 . Accessions for RAPD analysis included 30 Nautilus samples, with duplicate amplification of selected samples run to assess the reproducibility of the procedures. The 25 μl reaction mix consisted of 1 × PCR buyffer, (Amersham, Freiburg, Germany; 100 mM Tris–HCL pH 8.6, 500 mM KCl, 15 mM MgCl2, 1% Triton-X 100), 0.2 mM of each deoxynucleotide triphosphate (dATP, dCTP, dGTP, dTTP; Promega Corporation, Mannheim, Germany), 4 ng bovine serum albumin (Promega), 1 unit DNA polymerase (Amersham), 0.04 mM UBC 10−mer primer and 2 μl (25–50 ng) of the DNA extract. Amplification was carried out in a Perkin-Elmer (Boston, MA, USA) thermocycler (Gene Amp PCR system 2400) for 43 cycles of: 1 min denaturation at 94°C, 1 min annealing at 35°C, 2 min primer extension at 72°C, followed by a final extension cycle at 72°C for 7 min and then held at 4°C until required.
PCR amplification of coxI sequences
PCR was initially undertaken on 3 of the extracted DNA samples using a panel of coxI primers reported from a range of different species. Those utilised by Sparks (2004) from cichlids (Teleosti: Perciformes: Cichlidae), as developed by Folmer et al. (1994), proved to be the most successful in amplifying the corresponding region from Nautilus samples—primers LCO1490 (5′-GGTCAACAAATCATAAAGATATTGG-3′) and HCO2198 (5′-TAAACTTCAGGGTGACCAAAAAATCA-3′). For those samples which did not amplify successfully with these primers (majority of these samples were from the “Northern GBR” group) dedicated primers were designed for the study. The coxI sequences obtained from “Northern GBR” samples were aligned manually using Primer Express® Software Version 2.0 (Applied Biosystems 2001) and then used to determine an optimal primer pair anchored in the conserved regions at each end of this sequence. Two sequencing quality primer sets were prepared by Geneworks (Adelaide, SA, Australia) as: Naut1F 5′-GAACATCTCTTAGTTTATTAATTCGG-3′; Naut1BR 5′-AGAATTGGATCTCCTCCTCC-3′; Naut2F 5′-CCGGGYTTAGTCGGAACATCYCTTAG-3′; Naut2R 5′-KRATTGGRTCYCCTCCTCCAGC-3′ where Y = C/T, R = A/G and K = G/T. PCR was undertaken in 25 μl reactions containing 100 μM each dNTP, 7.5 pmol of each primer, 1.0 mM MgCl2, 1.0 unit Taq polymerase (Promega), 2.5 μl of 10 × PCR buffer (Promega) and ∼25 ng of DNA template. The PCR protocol followed involved an initial 2 min at 94°C followed by 30 cycles of 30 s at 94°C, 30 s at 52°C and 2 min at 72°C, followed finally by 10 min at 72°C. For each sample, following PCR and electrophoresis on a 1% agarose gel, a band was observed at ∼600 bp. This band was excised from the gel and the DNA purified from the agarose with a kit (Promega, Wizard). DNA from the purified band (3 ng) was then placed into a sequencing reaction according to the manufacturer’s instructions for the Big Dye Terminator Version 3.1 Cycle Sequencing Kit (Applied Biosystems). Both forward and reverse strands were sequenced. Sequences were determined using an ABI3130 Genetic Analyser (Applied Biosystems) giving, in each case, a sequence of 575 bases.
Analysis of RAPD data
The RAPD analysis was conducted for the presence and absence of bands. Binary data for the presence or absence (incidence) of each polymorphic band was recorded as a “1” or a “0”, respectively. Bands were only scored if they were reproducible. Faint and ill-defined bands were disregarded. Statistical analyses and calculations of scored data were carried out using Genstat 5, release 6.1 (Payne et al. 2002). Principle coordinates analysis was carried out using a similarity matrix generated using a simple matching coefficient. Allele diversities (expected heterozygosity) were calculated using null marker data, by the method of Zhivotovsky (1999). Heterozygosity values were calculated for all individuals at Osprey reef and for the sample set as a whole. ANOVA calculations (1,023 replicates) for the populations at Osprey reef, Shark reef, Bougainville reef and Southern Small Detached reef were carried out (Payne et al. 2002).
DNA sequence alignment and analysis
The sequences representing the coxI region of DNA from each of the three individuals were aligned manually, using Chromas® Version 1.45 Freeware (Technelysium Pty. Ltd., Tewantin, Qld., Australia) and BioEdit® Version 7.0.4.1 Freeware (Ibis Therapeutics, Carlsbad, CA, USA) using the programme default settings. Alignment of the sequences required no insertion/deletion events (indels). The sections of sequence toward each end of the generated sequences were observed to be fully conserved in these three animals. PCR was then performed on all the remaining accessions of Nautilus in a 25 μl reaction mix as described above. The product obtained from each 25 μl PCR reaction was visualised by agarose gel electrophoresis. To compare the data for consistency, maximum likelihood, neighbour joining and maximum parsimony trees were constructed (Harrison and Langdale 2006) in PAUP* 4.0 Beta 10 Win (Swofford 1998) with a MaxTrees setting of 100 and a Distances value of 500, using the HKY + G model (as indicated by the ModelTest function), with the TBR (tree-bisection-reconnection) algorithm. Statistical support for each analysis was evaluated by the bootstrap procedure (1,000 resampling replicates). A least squares tree was run using these DNA sequence data and generated a γ-α shape parameter, which was used to calculate FSTs (3,024 replicates) in Arlequin® for Pairwise FSTs. Within group genetic distances (between and within the “Northern GBR” and “Coral Sea” groups) were calculated using MEGA Version 3.1, using the programme default settings (Kumar et al. 2004).
Results
DNA was extracted from a number of animals at a range of different sites in both the Coral Sea and the outer edges of the GBR in far north Queensland. A total of 48 samples were extracted and analysed: 23 from animals at Osprey reef (14 from a location known as Entrance and nine from a site called Pavona), nine animals from Shark reef and two animals from Bougainville reef. From the “Northern” section of the GBR; 14 animals were sampled from reefs adjacent to Raine Island (six animals from North Small Detached reef, five from South Small Detached reef, one from Wishbone reef and two from Mantis reef).
RAPD data
For RAPD analysis, a subset of the overall samples were used. These included a total of 16 samples from Osprey reef (ten from Entrance and six from Pavona), three samples from Shark reef, two from Bougainville, one sample from Mantis reef, one from Wishbone reef, two from Northern Small Detached reef and five from Southern Small Detached reef. Together, these represented a collection of 21 samples from the “Coral Sea” distribution and nine from the “Northern GBR” distribution of the accessions.
Within the fingerprints generated by the RAPD primers used for scoring, there were a total of 54 markers scored, three were monomorphic and 51 were polymorphic across all samples. All polymorphic markers amplified across the samples were collated and analysed. Within the data set were a number of primers, which amplified a single polymorphic band. These data were not included in the final analyses of the combined data set, due to their sporadic and inconclusive nature. As has been observed by other authors (e.g. Williams et al. 1990) negative controls occasionally produced bands that are usually faint. However, bands in negative controls were always different from bands in the amplification reactions. Bands observed and included in the analysis here were between 300 and 2,500 bp in size. RAPD profiles included bands with different degrees of sharpness and intensity. Mean number of bands per individual and primer ranged between 8 and 15. Intra- and inter-population variation was examined by the production of a similarity matrix, and from this principle coordinates analyses were conducted. Figure 2a, b shows the PCO ordination of all 30 samples (identified by population) for the first three axes. This represents 43.7% of the variation among all 30 individuals tested (the percentage variance is accounted for, in order: 19.3, 16.4 and 8.0%, respectively). Histograms illustrating the estimated allele diversity of markers obtained from Nautilus samples from both the “Northern GBR” and “Coral Sea” group of populations and allele diversity of Nautilus samples from Osprey reef are shown in Fig. 3a, b, respectively. Using the data for the first three PCO axes, ANOVAs were carried out for the data obtained from populations at Osprey reef, Shark reef, Bougainville reef and South Small Detached reef. ANOVAs indicated differences along the first two axes, Axis 1 F = 5.99, p = 0.003; Axes 2 F = 18.55, p = 2.4−6. Mean points for Mantis reef, Wishbone reef and North Small Detached reef were plotted with the confidence intervals of the four populations above and lie within the 95% confidence interval for the mean of South Small Detached reef (Fig. 4).
The ordination of all 30 Nautilus accessions resulting from principle coordinates analysis of a similarity matrix generated from 51 RAPD markers using the simple matching coefficient. A first and second principle axes, B second and third principle axes. Key to symbols used in both figures is: Osprey reef (◆), Shark reef (■), Bougainville reef (▲); Mantis reef, Wishbone reef (○), North Small Detached reef (△) and Southern Small Detached reef (□)
Data calculated for the first three PCO axes allowed ANOVAs to be carried out for the data obtained from populations Osprey reef, Shark reef, Bougainville reef and South Small Detached reef. Mean points for Mantis reef, Wishbone reef and North Small Detached reef were then plotted with the confidence intervals of the four populations above. Key to symbols used in both Figures is: Osprey reef (◆), Shark reef (■), Bougainville reef (▲); Mantis reef, Wishbone reef (○), North Small Detached reef (△) and Southern Small Detached reef (□)
Partial coxI DNA sequence data
DNA sequencing reactions were carried out on all DNA samples extracted. Partial coxI sequence information was obtained and analysed from a total of 43 samples: in the “Coral Sea” group, 22 animals at Osprey reef (14 from The Entrance and eight from Pavona), nine from Shark reef and two from Bougainville reef. From the “Northern GBR” group; a total of 14 sequences were obtained (five from North Small Detached reef, four from South Small Detached reef and one from Mantis reef). Sequences generated in this study were deposited in GenBank under Accession numbers EF128174–EF128216.
On a Genbank BLAST search this partial sequence aligned (93%) with N. pompilius coxI sequence (Genbank accession: AF120628). Similarly, a specimen identification search on the Barcodes of Life Database (www.barcodinglife.com) (Ward et al. 2005) produced a 99.4% similarity match with voucher sequences from N. pompilius.
Alignment of the coxI partial sequence data (575 bp) obtained from all 43 samples was carried out, with 18 bp (2.96%) variable sites observed. Of these, 11 points of variation (1.91%) were specific to the accessions from the “Northern GBR” populations. The remaining six variable sites were dispersed across samples from the “Northern GBR” group of populations but also accessions from Osprey reef and Shark reef.
Between group population pairwise Fsts were calculated (“Northern GBR” group versus “Coral Sea” group) by the distance method of Tamura and Nei in Arlequin® at 0.936 and Fst p-values were calculated as 0.000, with 3,024 permutations. Standard diversity indices within the “Northern GBR” group for gene diversity was 0.8667 ± 0.0714 and 0.5600 ± 0.0865 for the “Coral Sea” group. Genetic distances within the “Northern GBR” and “Coral Sea” groups were calculated as 0.003 ± 0.001 and 0.001 ± 0.001, respectively, with between group values as 0.023 ± 0.007.
Phylogenetic reconstructions based on the coxI sequence data
Neighbour Joining analysis in PAUP* 4.0 generated a consensus tree (Fig. 5) and was constructed from the alignment of partial coxI sequences from 43 N. pompilius individuals, and rooted against the sequence of Nautilus macromphalus (GenBank Accession DQ472026). Of the 575 bp of partial sequence, 425 characters were constant, 131 variable characters were parsimony uninformative and 18 characters were parsimony informative (with consistency Index values of 0.958 and Retention Index values of 0.987, these are considered to be well supported). The same topology was obtained from Maximum Parsimony (MP) analysis. Bootstrap values, generated from 1,000 resampling replicates, which are over 50% are shown above (NJ) or below (MP) branches.
Consensus tree produced by Neighbour Joining analysis in PAUP* 4.0 constructed from the alignment of partial coxI sequences from 43 Nautilus pompilius individuals. Tree was rooted against the sequence of N. macromphalus (GenBank Accession DQ472026). The same topology was obtained from Maximum Parsimony (MP) reconstructions based on 18 parsimony informative characters, with a consistency index of 0.958 and retention index of 0.987. Bootstrap values (1,000 resampling replicates) higher than 50% are shown above (NJ) or below (MP) branches
Within this tree the presence of two distinct clades were detected. These clades respectively represented the accessions, which comprised the “Northern GBR” group of populations and those in the “Coral Sea” group of populations and were supported with bootstrap values of 100%.
Within each of these clades there were a number of hierarchical levels, but these did not show any population-specific groupings of the samples. The “Northern GBR” clade shows a supported (65 and 56% Bootstrap support) grouping of two samples from Northern small detached reef (NSSD 5ii and NSSD 9ii) with 58 and 61% bootstrap support for their separation from the remainder of the “Northern GBR” group.
In the “Coral Sea” clade, there were a number of unresolved steps (containing accessions from Osprey, Shark and Bougainville reefs). A distinct sub-clade (identified in both analyses) contained five accessions from Osprey reef (O-1593, O-1600P, O-1604E, O-1648E and O-1650E). This has bootstrap support of 61 and 53%.
Discussion
Nautilus are deepwater predators and scavengers, they are also the only living cephalopods with an external shell. Isolated sea mounts provide ideal habitats for these organisms to survive and breed, but they also act as reproductive and biological barriers for the animals. Bounded by water often exceeding 1,500 m in depth, Nautilus shells cannot physically withstand the enormous pressure that travelling to these depths would entail. As such, to leave their habitat and cross ocean expanses to other mounts or reefs would entail swimming in open ocean, a behaviour not readily attributed to Nautilus. This forced retention around a single site will impose limitations on the normal degree of gene flow into and out of a breeding population, possibly resulting in higher levels of in-breeding depression and lower levels of genetic variation.
DNA markers have several advantages over morphometric or protein (isozyme) markers for studying genetic variation and population structure in fish species (Carvhallo and Pitcher 1994; Ferguson and Danzmann 1998). Random Amplified Polymorphic DNA (RAPD) is a hybridisation and amplification-based molecular method for detecting genetic polymorphisms (Welsh and McClelland 1990; Williams et al. 1990). Although the appropriateness of RAPD markers for population genetic inference has been questioned, its relative technical simplicity makes it a cost-effective method for population genetic studies (Jayasankar and Dharmalingam 1997) when used in conjunction with other, more robust, approaches. DNA sequence information is now proving to be a commonly used tool for addressing questions of relationship, population structure and evolutionary history in a range of fish species (Brown et al. 1992; Perrin and Borsa 2001; Conseugra et al. 2002; Fauvelot et al. 2003; van Herwerden et al. 2003; Yamamoto et al. 2004; Aspden et al. 2006; Pegg et al. 2006).
RAPD analysis of genetic differences in this study illustrated weak separation of the “Northern GBR” populations and the “Coral Sea” populations. The RAPD data generated were used to construct a similarity matrix using a simple matching coefficient. The PCO plots generated from this matrix show the first three axes, representing just over 40% of the total genetic variation present in the 30 samples screened.
It is apparent that there are some distinct groupings represented in the data, primarily the separation of the “Northern GBR” group (Mantis, Wishbone, Southern Small Detached and North Small Detached reef) from those individuals in the “Coral Sea” group (Osprey reef, Shark reef and Bougainville reef).
Estimates of allele diversity using null marker data were calculated for this dataset. Figure 3a shows the estimates for the whole data set of individuals from both groups and Fig. 3b the values for the Osprey reef samples alone. The data indicate a fairly high level of heterozygosity at most loci. This interpretation is dependent on treating the markers as traditional alleles (hence the use of the simple matching coefficient above for logical consistency). RAPD data were also used to estimate Heterozygosity (H) values. H-values at each locus for the Osprey reef samples are visibly higher or lower in these samples than in the whole sample, indicative of genetic variation being found between the two groups (Osprey versus remaining sites).
Data calculated for the first three PCO axes allowed ANOVAs to be carried out for the data obtained from populations Osprey reef, Shark reef, Bougainville reef and Southern Small Detached reef. These calculations indicated differences along the first two axes, but not the third, and these suggest that Osprey reef, Shark reef, Bougainville reef and South Small Detached reef are all genetically distinct. The significance of the remaining populations (Mantis reef, Wishbone reef and North Small Detached reef) could not be formally tested due to small sample sizes, but their mean points were plotted with the confidence intervals for these four populations all lie within the 95% confidence interval for the mean of the Southern Small Detached reef population, suggesting that they are fairly closely related, as shown in Fig. 4.
These RAPD are not conclusive evidence of distinct genetic separation in these clusters as RAPD data is not the most robust form of genetic marker. Repeatability varied depending on primer, species and annealing temperature. Since the presentation of RAPD markers by Williams et al. (1990), where dependence of band profiles on parameters such as annealing temperature was reported, a number of studies have shown RAPDs to depend on specific conditions of the reaction such as primer–template concentration, or the transition interval between melting and annealing temperature. Similarly, considerations such as relative time of amplification are more similar than those amplified at different times must also be taken into account with RAPD profiles.
Low population pairwise Fst p-values suggest that the differences indicated here are of significance, but they could be highlighted due to the relatively small numbers of samples in each of the “groups” tested, or that there is limited genetic variation contained within the coxI region sequenced in this study (17 variable position out of 575 bp and 11 discriminatory between the N and S groups).
Within group distance and standard error is lower for within group values, because they each represent, on an evolutionary time-scale, large “super-groups” comprising smaller, distinct geographical populations from each individual sample site, i.e. within the “Coral Sea” and “Northern GBR” locales. This is illustrated by the larger between group value, which is representative of genetic differences between the “Northern GBR” and “Coral Sea” supergroups.
The Neighbour joining and Maximum parsimony consensus tree generated from the coxI sequence data identified the presence of two major clades in the accessions, representing the accessions comprising the “Northern GBR” group of populations and those in the “Coral Sea” group of populations, with bootstrap support of 100% for this distinction. Within these two clades, some minor sister clades are identified and some support is provided for the resolved accessions and the minor groupings, two samples from Northern small detached reef (NSSD 5ii and NSSD 9ii) have bootstrap support of 58 and 61% for their separation from the remainder of the “Northern GBR” group. Maximum likelihood analysis shows no strict separation within the “Northern GBR” clade of any particular set of samples from distinct populations. Similarly, within the “Coral Sea” grouping there were no distinct groupings of samples from specific populations in the tree.
Further divisions of the individual accessions from the three discrete populations which comprise the “Coral Sea” group or the four populations comprising the “Northern GBR” group produce no discernable, population-specific patterns. This was particularly noticeable within the Osprey reef samples (the population with the highest overall representation)—no discrete separation between the samples collected from Pavona (those designated with a P) or those from Entrance (E), although a distinct sub-clade (identified in both analyses) contained five accessions from Osprey reef (O-1593, O-1600P, O-1604E, O-1648E and O-1650E) and has bootstrap support of 61 and 53%.
The relationships between individuals of the different populations, illustrated by the different phylogenetic trees in this study contrast somewhat with that suggested by the RAPD data. This is not unexpected given the conserved nature of the coxI gene sequence compared to the random, potentially whole genome sampling that RAPD data represents. More significance is placed on the sequence information at the longer evolutionary scale, in that divergence of the two groups can be inferred by the phylogenetic information obtained from these data, which is more robust in nature. The RAPD data however is not discounted, for although it may not be as robust as the coxI sequence data, it still suggests the presence of discrete breeding populations, which can, within the limits of this study, be differentiated.
The more basal location of the “Northern GBR” group in the consensus tree may be suggestive of a limited sample size effect (we have not picked up sequence motifs which are more commonly distributed among these animals than our sampling illustrates) or that this represents a more established super-group of populations, with less genetic divergence from the out-group species than those found in the possibly younger “Coral Sea” super-group (this group is potentially ancestral to those in the Coral Sea and gave rise to the individuals which subsequently formed it).
The data presented here illustrate the utility of DNA sequence information to provide useful insights into the genetic and evolutionary history of particular species, however, as indicated earlier, the combined analyses of multiple genetic sequences from each individual (covering both nuclear and mitochondrially encoded sequences) should provide a more though and diagnostic understanding of such relationships and is the avenue which should be pursued in future to resolve such relationships across a number of hierarchical levels.
Most of our understanding on the biology of N. pompilius has come from samples obtained in Palau and the Philippines (Hayasaka et al. 1987; Saunders 1987), although they are distributed across the Indo-Pacific. Nautilus are generally considered to inhabit the reef-slope to around 500 m and, although capable of travelling up to 4.3 km per day, they are not generally perceived to be open water travellers (O’Dor et al. 1993). While ocean-drift dispersal of living animals does occur and has some potential to cause confusion in systematic and population level investigations, it is not considered to be significant overall.
Wray et al. (1995) suggested that systematic and DNA sequence information indicated N. pompilius represented a paraphyletic group of diverging lineages and that these fell into three geographically distinct lineages, one of which was the Australian/Papua-New Guinea clade. This clade had an interesting and unresolved accession, an individual potentially representing a migrant into this sampling area from a distant location, by natural processes of current drift or animal movement.
The results presented in this study are centred on the Australian/Papua-New Guinea lineage that Wray et al. (1995) identified, but at a smaller geographic/population level as opposed to the large-scale geographic scale. Within these parameters, we can determine that there is genetic divergence between the smaller geographic lineages, indicating the presence of distinct groups of populations. We have also identified genetic markers, which can be used to assess discrete populations within these groups.
While these populations are all discrete and isolated (in that they are surrounded by stretches of deep open ocean), it cannot be discounted that there is movement of animals between them, given that all the populations in the Australian/Papua-New Guinea lineage most likely share a common ancestor from their founder populations, most possibly in the Philippines (Roy et al. 2001; Hughes et al. 2002; McRoberts et al. 2002).
Swan and Saunders (1987) suggested that Nautilus will migrate between geographically adjacent populations and O’Dor et al. (1993) suggested that it was possible for Nautilus to survive month-long periods in cooler, deep water, but that long distance transport by sub-surface currents between reefs, while possible, was unlikely. It is unclear as to how such migrations occur, but over geological time-scales, events such as glacial sea movements and large scale-disturbances such as cyclones.
We report here that diversification of N. pompilius is taking place in distinct populations across deep water walls on the Northern Great Barrier reef and in the isolated sea mounts of the Coral Sea. This process has been suggested in a number of studies (Wray et al. 1995 and the references therein), but this is the first report of population-specific genetic differentiation with molecular data, which links specifically to the larger–scale geographic divergence of the species.
References
Anderson FE (2000a) Phylogeny and historical biogeography of the loliginid squids (Mollusca: Cephalopoda) based on mitochondrial DNA sequence data. Mol Phylogenet Evol 15(2):191–214
Anderson FE (2000b) Phylogenetic relationships between loliginid squids (Cephalopoda: Myopsida) based on analysis of multiple data sets. Zool J Linn Soc 130:603–633
Applied Biosystems (2001) Primer express software v2.0 applications-based primer design software user’s manual. Applied Biosystems, Foster City, CA
Aspden W, Pegg G, Briskey L, Sinclair W (2006) Species–specific PCR primers for the mitochondrial genome control region hypervariable region 1 of the reef fish Lutjanus sebae. Mol Ecol Notes 6:499–501
Basil JA, Hanlon RT, Sheikh SI, Atema J (2000) Three-dimensional odour tracking by Nautilus pompilius. J Exp Biol 203:409–1414
Basil JA, Bahctinova I, Kuroiwa K, Lee N, Mims D, Preis M, Soucier C (2005) The function of the rhinophore and the tentacles of Nautilus pompilius L. (Cephalopoda, Nautiloidea) in orientation to odour. Mar Freshw Behav Physiol 38209–38221
Bonnaud L, Boucher-Rodoni R, Monnerot M (1997) Phylogeny of cephalopods inferred from mitochondrial DNA sequences. Mol Phylogenet Evol 7(1):44–54
Bonnaud L, Rodhouse PG, Boucher-Rodoni R (1998) A phylogenetic study of the squid family Onychoteuthidae (Cephalopoda: Oegopsida). Proc R Soc Lond Ser B 265:1761–1770
Bonnaud L, Ozouf-Costaz C, Boucher-Rodoni R (2004) A molecular and karyological approach to the taxonomy of Nautilus. Comptes Rendus Biol 327:133–138
Boyle P, Rodhouse P (2005) Cephalopods: ecology and fisheries. Blackwell Science, Oxford, UK
Brown JR, Beckenbach AT, Smith MJ (1992) Mitochondrial DNA length variation and heteroplasmy in populations of white sturgeon (Acipenser transmotanus). Genetics 132:221–228
Carlson BA, McNibben JN, DeGruy MV (1984) Telemetric investigation of vertical migration of Nautilus belauensis in Palau. Pac Sci 38:183–188
Carvalho G, Pitcher TJ (1994) Molecular genetics in fisheries. Rev Fish Biol Fish 4:269–399
Conseugra S, Garcia de Leaniz C, Serido A, Gonzalez-Morales M, Straus LG, Knox D, Verspoor E (2002) Mitochondrial DNA variation in Pleistocene and modern Atlantic salmon from the Iberian glacial refugium. Mol Ecol 11:2037–2048
Davis RA (1987) Nautilus studies: the first twenty-two centuries. In: Saunders WB, Landman NH (eds) Nautilus. The Biology and Paleobiology of a Living Fossil, New York, pp 3–21
Fauvelot C, Bernardi G, Planes S (2003) Reductions in the mitochondrial DNA diversity of coral reef fish provide evidence of population bottlenecks resulting from Holocene sea-level change. Evolution 57:1571–1583
Ferguson MM, Danzmann RG (1998) Role of genetic markers in fisheries and aquaculture: useful tools or stamp collecting. Can J Fish Aquaculture Sci 55:1553–1563
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome C oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3:294–299
Giribet G, Wheeler W (2002) On bivalve phylogeny: a high-level analysis of the Bivalvia (Mollusca) based on combined morphology and DNA sequence data. Invertebr Biol 121:271–324
Hanlon RT, Messenger JB (1996) Cephalopod behaviour. Cambridge University Press, Cambridge, UK
Harrison CJ, Langdale JA (2006) A step by step guide to phylogeny reconstruction. Plant J 45:561–572
Hayasaka S, Oki K, Tanabe K, Saisho T, Shinomiya A (1987) On the habitat of Nautilus pompilius in Taon Strait (Philippines) and Fiji Islands. In: Saunders WB, Landman NH (eds) Nautilus. The Biology and Paleobiology of a Living Fossil, New York, pp 179–200
Hebert PDN, Cywinska A, Ball SL, de Waard JR (2003) Biological identifications through DNA barcodes. Proc R Soc Lond B 270:313–322
Hughes TP, Bellwood DR, Connolly SR (2002) Biodiversity hotspots, centres of endemicity, and the conservation of coral reefs. Ecol Lett 5:775–784
Jayasankar P, Dharmalingam K (1997) Potential application of RAPD and RAHM markers in genome analysis of scombroid fishes. Curr Sci 72:383–390
Kumar S, Tamura K, Nei M (2004) MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Lindgren AR, Giribet G, Nishiguchi MK (2004) A combined approach to the phylogeny of Cephalopoda (Mollusca). Cladistics 20:454–486
Lindgren AR, Katugin ON, Amezquita E, Nishiguchi MK (2005) Evolutionary relationships among squids of the family Gonatidae (Mollusca: Cephalopoda) inferred from three mitochondrial loci. Mol Phylogenet Evol 36:101–111
McRoberts CM, McClean CJ, Veron JEN, Hawkins JP, Allen GR, McAllister DE, Mittermeir CG, Schueler FW, Spalding M, Wells F, Vynne C, Werner TB (2002) Marine biodiversity hotspots and conservation priorities for tropical reefs. Science 295:1280–1284
Merritt TJS, Shi L, Chase MC, Rex MA, Etter RJ, Quattro JM (1998) Universal cytochrome b primers facilitate intraspecific studies in molluscan taxa. Mar Mol Biol Biotechnol 7:7–11
O’Dor RK, Forsythe J, Webber DM, Wells J, Wells MJ (1993) Activity levels of Nautilus in the wild. Nature 362:626–627
Ovenden J (1990) Mitochondrial DNA and marine stock assessment—a review. Austr J Mar Freshw Res 41:835–853
Payne RW et al (2002) Genstat 5 (release 6.1) reference manual. VSN International, Oxford, UK
Pegg GG, Sinclair B, Briskey L, Aspden W (2006) MtDNA barcode identification of fish larvae in the southern Great Barrier Reef—Australia. Sci Mar 70S2:7–12
Perrin C, Borsa P (2001) Mitochondrial DNA analysis of the geographic structure of Indian scad mackerel in the Indo-Malay archipelago. J Fish Biol 59:1421–1426
Roy K, Balch DP, Helberg ME (2001) Spatial patterns of morphological diversity across the Indo-Pacific: analyses using stombid gastropods. Proc R Soc Lond B 268:1–6
Ruth P, Schmidtberg H, Westermann B, Schipp R (2002) The sensory epithelium of the tentacles and the rhinophore of Nautilus pompilius L. (Cephalopoda, Nautiloidea). J Morphol 251:239–255
Saunders WB (1981a) A new species of Nautilus from Palau. Veliger 24:1–7
Saunders WB (1981b) The species of living Nautilus and their distribution. Veligier 24:8–17
Saunders WB (1984) The role and status of Nautilus in it’s natural habitat: evidence from deep-water remote camera photosequences. Paleobiology 12:469–486
Saunders WB (1987) The species of Nautilus. Nautilus. The biology and paleobiology of a living fossil. In: Saunders WB, Lindman NH (eds) Nautilus. The biology and paleobiology of a living fossil. Plenum Press, New York, pp 35–52
Saunders WB, Landman N (1987) Nautilus. The biology and paleobiology of a living fossil. Plenum Press, New York
Saunders WB, Ward PD (1987) Sympatric occurrence of living Nautilus (N. pompilius and N. stenomphalus) on the Great Barrier Reef, Australia. Nautilus 101:188–193
Söller R, Warnke K, Saint-Paul U, Blohm D (2000) Sequence divergence of mitochondrial DNA indicates cryptic biodiversity in Octopus vulgaris and supports the taxonomic distinctiveness of Octopus mimus (Cephalopoda: Octopodae). Mar Biol 136:29–35
Sparks J (2004) Molecular phylogeny and biogeography of the Malagasy and South Asian cichlids (Teleosti: Perciformes: Cichlidae). Mol Phylogenet Evol 30:599–614
Springer J, Ruth P, Beuerlein B, Westermann B, Schipp R (2004) Immunohistochemical localization of cardio-active neuropeptides in the heart of a living fossil, Nautilus pompilius L. (Cephalopoda, Tetrabranchiata). J Mol Histol 35:21–28
Strugnell J, Norman M, Jackson J, Drummond AJ, Cooper A (2005) Molecular phylogeny of coleoid cephalopods (Mollusca: Cephalopoda) using a multigene approach; the effect of data partitioning on resolving phylogenies in a Bayesian framework. Mol Phylogenet Evol 37(2):426–441
Suzuki T, Fukuta H, Nagato H, Umekawa M (2000) Arginine kinase from Nautilus pompilius, a living fossil. J Biol Chem 275:23884–23890
Swan ARH, Saunders WB (1987) Morphological variation in Nautilus from Papua-New Guinea. In: Saunders WB, Lindman NH (eds) Nautilus. The biology and paleobiology of a living fossil. Plenum Press, New York, pp 85–103
Swofford D (1998) PAUP*V4.0b10win phylogenetic analysis using parsimony. Smithsonian Institution, Sinauer Associates, Sunderland, MA
van Herwerden L, Benzie J, Davies C (2003) Microsatellite variation and population genetic structure of red throat emperor (Lethrinus miniatus) in the Great Barrier Reef, Australia. J Fish Biol 62:987–999
Wani R (2004) Experimental fragmentation patterns of modern Nautilus shells and the implications for fossil cephalopod taphonomy. Lethania 37:113–123
Ward PD (1987) The natural history of Nautilus. Allen and Unwin Press, London
Ward PD, Carlson B, Weekly M, Brumbaugh B (1984) Remote telemetry of daily vertical and horizontal movement of Nautilus in Palau. Nature 309:248–252
Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PD (2005) DNA Barcoding of Australia’s fish species. Philos Trans R Soc B 360:1847–1857
Wells M (1986) Legend of the living fossil. New Sci 10:36–41
Wells MJ, Wells J, O’Dor RK (1992) Life at low oxygen tensions: the behaviour and physiology of Nautilus pompilius and the biology of extinct forms. J Mar Biol Assoc UK 72:313–328
Welsh J, McClelland M (1990) Fingerprinting genomes using PCR with arbitrary primer. Nucleic Acids Res 18:7213–7218
Westermann B, Beuerlein K (2005) Y-maze experiments on the chemotactic behaviour of the tetrabranchiate cephalopod Nautilus pompilius (Mollusca). Mar Biol 147:145–151
Westermann B, Beck-Schildwachter I, Beuerlein K, Kaleta EF, Schipp R (2004) Shell growth and chamber formation of aquarium-reared Nautilus pompilius (Mollusca, Cephalopoda) by X-Ray analysis. J Exp Zool 301A:930–937
Westermann B, Schmidtberg H, Beuerlein K (2005) Functional morphology of the mantle of Nautilus pompilius (Mollusca, Cephalopoda). J Morphol 264:277–285
Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:6531–6535
Woodruff DS, Carpenter MP, Saunders WB, Ward PD (1987) Genetic variation and phylogeny in Nautilus. In: Saunders WB, Landman NH (eds) Nautilus. The biology and paleobiology of a living fossil. Plenum Press, New York, pp 65–83
Wray CG, Landman NH, Saunders WB, Bonacum J (1995) Genetic divergence and geographic diversification in Nautilus. Paleobiology 21:220–228
Yamamoto S, Morita K, Kitano S, Wanatabe K, Koizumi I, Maekawa K, Takamur K (2004) Phylogeography of White-Spotted Charr (Salvelinus leucomaenis) inferred from mitochondrial DNA sequences. Zool Sci 21:229–240
Zhivotovsky LA (1999) Estimating population structure in diploids with multilocus dominant DNA markers. Mol Ecol 8:907–913
Acknowledgements
The authors would like to acknowledge the funding for this project, provided by a CQU Seed grant. We are indebted to the biologists and crew of Undersea Explorer—in particular Andy, Jules, Karl and Mick for their help in obtaining and recording all the tissue samples used in this study. We are indebted to our colleagues Drs Christine McPhie, Neil McRoberts and Larelle Fabbro and to three anonymous reviewers for their constructive comments and assistance in the preparation of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sinclair, B., Briskey, L., Aspden, W. et al. Genetic diversity of isolated populations of Nautilus pompilius (Mollusca, Cephalopoda) in the Great Barrier Reef and Coral Sea. Rev Fish Biol Fisheries 17, 223–235 (2007). https://doi.org/10.1007/s11160-006-9030-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11160-006-9030-x