Abstract
Microorganisms are responsible for the conversion and breakdown of organic compounds and contaminants in bioreactors designed for the treatment of different types of waste. Organized in highly complex communities, they are the heart of every wastewater treatment plant and solid residue landfill. The detailed characterization of these communities and their taxonomic classification based on phylogenetic data are of great utility in monitoring the responses of the system to changing operational parameters and the development and optimization of favorable conditions within the bioreactors these communities inhabit. Until recently, only a fraction of the microbial diversity could be assessed, limited by the available sequencing technology, which was not suited for a high-throughput implementation. With the introduction of the recent next-generation sequencing (NGS) methods, an enormous advance has taken place allowing researchers in microbial ecology to generate large amounts of phylogenetic data in a short time and at relatively low costs. In this review, we present and discuss how the increase in available information has influenced recent research and the results available phylogenetic data has produced in the field of wastewater treatment. Furthermore, we introduce the data processing of NGS-based experiments, which has become more complex as the millions of sequences that a single sample can yield require the effective use of computational resources and human bioinformatics skills. To address this part of modern microbial ecology, the most popular sequencing techniques, as well as data analysis workflows, are outlined in this review article.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
For a long time, the biological reactors used for waste treatment (liquids, slurries or solid waste) were considered a “black box”. The insight that microorganisms played a key role in the process led researchers to study the bacteria that inhabited these systems to understand the underlying physiological processes and improve waste degradation. The microbiology of wastewater treatment plants (WWTPs) has since been in the crosshairs of many researchers. In fact, WWTPs were proposed as a model system for microbial ecology (Daims et al. 2006). For decades, the study of the microorganisms present in WWTPs and anaerobic digesters was performed using conventional microbiological techniques. These approaches were usually based on the isolation of pure cultures and their identification by morphological, metabolic and biochemical characteristics of the isolates. Even simpler approaches existed, like the identification of the bacteria involved in filamentous bulking by the microscopic examination of specific staining reactions.
The application of techniques from the area of molecular biology to WWTPs in the 90s was a revolution. These techniques, based on 16S rRNA (18S rRNA for eukaryotic organisms), deepened our knowledge of the microbiota inhabiting the treatment systems to a level not previously expected. Some of these techniques, such as Denaturing Gradient Gel Electrophoresis (DGGE) or Fluorescent in situ Hybridization (FISH), are still used today. DGGE is a simple technique that does not require extensive knowledge of molecular biology. It allows for the rapid and simple monitoring of the spatial–temporal variability of microbial populations, providing an overview of the dominant taxa inside a bioreactor, and is adequate for the analysis of a large number of samples. For these reasons, DGGE is still one of the most popular techniques for the assessment of biodiversity used by environmental engineers.
FISH (or Catalyzed Reporter Deposition-FISH, CARD-FISH) and qPCR (quantitative real-time PCR) are the only quantitative techniques applicable to the analysis of microbial communities. Real time PCR is a sensitive method that even allows for the quantification of gene expression, if a reverse transcription (RT) step is included in the protocol and yields information on the copy numbers of specific DNA sequences in the original sample. This technique is not easy to perform however and, in case of the reverse transcriptase variant, the recovery and conservation of non-degraded RNA may pose a significant challenge. Furthermore, one important drawback of qPCR is the variable number of copies of the rDNA genes, depending on the species considered. Quantitative data (bacterial counts) obtained via qPCR should only be considered reliable in case of the analysis of bacteria in axenic culture, taking into account the species’ rRNA gene copy number. FISH in comparison is an easy and fast to perform method. Moreover, the non-destructive treatment of the samples and subsequent in situ analysis can facilitate knowledge about the spatial distribution of different taxonomic groups in structures like biofilms. Due to its advantages over other techniques, FISH is still routinely used (for a comprehensive review over molecular biology techniques used in wastewater treatment, see Sanz and Köchling 2007; Cabezas et al. 2015; Bailon-Salas et al. 2017). Although FISH, qPCR and DGGE still present valid options, the same does not apply to another and once very widespread technique: the construction and sequencing of genetic libraries of 16S rDNA.
Cloning of the 16S rRNA gene has been extensively employed since the beginning of the 90s until the first decade of the 21st century. The emergence of high-throughput sequencing techniques, also called Next-generation Sequencing (NGS) technologies, has led to the virtual disappearance of the cloning- and sequencing-based approach which employed the Sanger technique. It should be noted that Sanger sequencing however is still in use today for a variety of applications. In microbial ecology for example it is applied to extract genetic information from the DNA contained in bands excised from a DGGE gel. However, for the simultaneous sequencing of large amounts of samples and/or different DNA templates the NGS techniques are unrivaled today.
We are now in the era of the -omics revolution. The different -omics approaches enable us to answer several questions. What is the microbial composition (16S rDNA approaches)? What is their genetic potential (Metagenomics)? Which genes are expressed (Metatranscriptomics)? What is the protein content (Metaproteomics)? Which metabolites are present as the product of the community’s expressed genetic information (Metametabolomics)? (Nyvad et al. 2013) Good reviews on the use of meta-omics and other molecular biology approaches to the study of wastewater treatment systems (with emphasis on anaerobic digestion) were published by Cabezas et al. (2015) and Rodríguez et al. (2015). In this review we are going to focus on approaches using high-throughput sequencing techniques for the analysis of the composition and structure of the microbial communities from wastewater (urban and industrial, aerobic and anaerobic) and solid waste (biodigesters for biogas production from activated sludge, manure, crops, etc.) treatment facilities. The most widely used approach to the study of microorganisms inhabiting these systems today is the analysis of 16S rDNA amplicon libraries generated with one of the NGS techniques.
The most popular commercial platforms for high-throughput DNA sequencing are 454-pyrosequencing (Roche) and Illumina (Illumina Inc.). The 454 Genome Sequencer, introduced in 2005, was the first NGS technology to become commercially available and has been the dominant NGS technology for one decade. Roche’s American competitor Illumina introduced its genome analyzer Illumina MiSeq in 2011. Despite the higher read lengths achievable with the 454 pyrosequencing platform (Table 1), Illumina MiSeq has nowadays replaced pyrosequencing due to the high capacity, lower price and lower error rate of its proprietary technology. Other massive-sequencing technologies (e.g. Ion Torrent’s PGM, SOLiD technology, HeliScope, single-molecule real-time (SMRT) DNA sequencing) are scarcely used in the field of WWT. In fact, all the articles mentioned in this review have used 454-pyrosequencing or Illumina sequencing. For an in-depth description of the fundamentals of the NGS technologies, we recommend the excellent article by Shokralla et al. (2012). However, NGS technologies have several drawbacks. The most significant one is the relatively short achievable read length, which allows for a reliable taxonomic assignment only at the level of genus or even family. Additionally, for paired-end sequencing (using a forward and a reverse primer flanking the sequence of interest), the short read length of approximately 250 bp in case of the Illumina MiSeq technique has to be considered when selecting the primers to allow for the generation of overlapping segments. Despite this, NGS technologies have become very popular in environmental microbiology and nowadays are the de-facto standard to study/analyze the microbiome of WWTs and other waste treatment reactors.
These novel techniques present high-throughput approaches where a large number of reads is generated with each run. Up to millions of sequences can be produced using a single Illumina flow cell (sample carrier). The resulting massive increase in information to process, as compared with the relatively manageable numbers of reads that clone-based Sanger sequencing provides, renders the manual control and editing of individual sequences impossible. Therefore, dedicated computer software is available that provides automated solutions for the preparative data processing steps, such as de-multiplexing, primer/adapter trimming and quality control, as well as for the actual analysis of the sequence reads produced by an experiment.
The most common application of NGS in microbial ecology today is 16S rDNA amplicon sequencing. For this approach, software packages exist that are capable of, e.g. clustering sequences into species-like units and assigning taxonomic rank and identity to these OTUs (operational taxonomic units). Algorithms for these and other data processing tasks are constantly being developed and adapted as new and more powerful computing hardware becomes available. To take full advantage of the newest hard- and software on the market, the programs are often run on distributed systems (computing clusters), while the systems themselves run in Unix-like environments, for example, one of the numerous implementations of the Linux operating system (OS). The use of these systems and the bioinformatics software that runs on them usually requires personnel with bioinformatics skills. Many sequencing analysis programs do not provide a graphical user interface and require that the user interacts with the software via a command line terminal. This holds for the execution of system administration tasks and the installation of computer clusters.
The possibilities and performance of today’s analytic computing pipelines have increased greatly, and so has the complexity of using these systems. Fortunately, several pre-designed workflow pipelines are available for the analysis of NGS experiments related to microbial ecology. The two most popular implementations, Qiime and mothur, will be presented in more detail in Sect. 3.1. While the user still needs to choose from a variety of sub-programs and construct a workflow from these using a command line interface (CLI), the programs are very well documented and supported by the developers and user communities on the internet, facilitating their use among biologists who have not received formal training in programming or computer science.
2 Next-generation sequencing
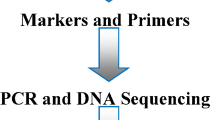
This review is focused on studies of the microbial ecology of waste and wastewater treatment systems where NGS techniques were applied for the identification of the resident microorganisms. These approaches as well as those in use for several decades now, namely cloning-based amplicon sequencing, DGGE and FISH, usually target a marker gene sequence or the gene’s RNA transcript. These genes can be encoding specialized enzymes, which are useful for studying specific metabolic or biodegradation processes. For example, the gene coding for the beta-subunit (dsrB) of the enzyme dissimilatory sulfite reductase is used to analyze sulfate-reducing microbial communities (Zhang et al. 2016b). Methanogenic microorganisms can be specifically targeted by sequencing the gene coding for the alpha subunit of the enzyme methyl-coenzyme M reductase (mcrA), which is involved in the final synthesis step of microbial methane production (Dhillon et al. 2005; Ziganshin et al. 2016). However, the most commonly utilized marker gene in microbial ecology today is the 16S rRNA gene encoding the ribonucleic portion of the small subunit (30S) ribosome in prokaryotic organisms (Bacteria and Archaea). The 16S rRNA gene does not provide any functional information about a microorganism because it is not translated into a protein product. Instead, it acts as an important factor in the initialization of the gene translation process itself. In molecular microbial ecology, 16S rRNA and 16S rDNA serve as a phylogenetic and taxonomic marker due to their universal presence in all prokaryotes and characteristic structure. The gene’s sequence contains strongly conserved regions, which allow for the design of specific PCR primer pairs that flank and amplify a segment of a DNA sequence of higher variability between these conserved stretches. A workflow of an NGS experiment is outlined briefly in Fig. 1 and detailed in the following sections.
Workflow of an NGS experiment. The biomass of bioreactors mainly consists of microorganisms. (1) Samples of biomass are collected, the cells are disrupted, total DNA is extracted and 16S rRNAs are amplified by PCR. (2) A fragment of the 16S rRNA genes is sequenced by a high-throughput sequencing technique (pyrosequencing, Illumina, …), producing a set of reads. (3) Sequences are processed by using different software packages (Qiime, mothur, RDP, …), and the suitable sequences are clustered into Operational Taxonomic Units (OTUs) and compared with databases (SILVA, NCBI,…). (4) Statistical and graphical evaluation is performed with statistical computer programs and dedicated functional libraries (R, Vegan…)
2.1 Sampling/DNA extraction
In most applications, an environmental or bioreactor-derived sample containing microbial biomass is subjected to a suitable whole-DNA extraction protocol. Many extraction methods have been published that yield DNA of sufficient quantity and quality for its subsequent amplification via PCR, achieving the crucial step of cell lysis of microbial cell walls (Yeates et al. 1997; Yeates and Gillings 1998; Lemarchand et al. 2005). The differences in the microorganisms’ resistance against lysis have to be considered when choosing a protocol. A cost-effective alternative to the laboratory methods with self-prepared reagents is the application of ready-to-use commercial kits, which are available for DNA and RNA extraction (Walden et al. 2017). The material costs are higher compared with employing home-made methods, and recipes are often compensated quickly by the substantial saving of time for the preparation and maintenance of the reactants as well as the overall shorter execution time of the commercial extraction procedures. In some cases the microbial target cells have to be separated from either their eukaryotic host cells or surface particles. Soil particles, for example, are known to contain humic and fulvic acids, which are capable of inhibiting the activity of the enzymes used in the PCR amplification or other downstream applications when dragged into the reaction mix with the DNA template solution (Matheson et al. 2010; Nair et al. 2014). In this case additional steps to separate the biomass from the organic matter it adheres to may be necessary. Another scenario in which the bacterial cells need to be separated from the sample matrix prior to DNA extraction is that of environments with a very low cell density. In this case the separation step serves the purpose of increasing the concentration of the sampled biomass. Especially commercial methods for environmental DNA extraction often accept only small volumes of raw sample matter, whereas in the case of the home-brew methods the generous use of reactants can create unnecessarily large amounts of often toxic waste, for example, phenol or chloroform. Examples of methods that are apt for the separation of microbial cells from different surfaces are the centrifugation through a particulate gradient (Bakken 1985), flow cytometry or ultrasound sonication (dos Furtado and Casper 2000).
Whenever possible, direct lysis is the preferable method though, as in the separation step of the cells from the carrier material the loss of a possibly important portion of the microbial cells has to be expected given that the chosen method may not be equally efficient for different microorganisms. This would affect the recovered abundances of different microorganisms, systematically introducing further bias into the downstream experiments.
In the case of unavoidably low-template DNA concentrations, additional amplification steps of the sample DNA might be necessary in the form of Multiple Displacement Amplification (MDA, non-PCR based) or by applying a nested amplification protocol using a PCR product, obtained in low quantity from spurious amounts of DNA, as a template again in a second amplification round. In general, though, additional amplification steps should be avoided whenever possible because they are known to introduce significant bias into the resulting abundance distribution of the generated products, again resulting in a distorted representation of the structure of the studied microbial communities. Furthermore, with each PCR round, the formation of amplification artifacts such as chimaeras (composite PCR products synthesized from several different template molecules) increase in number. This process leads to PCR products that do not correspond to any single template molecule in the sample, being useless for and detrimental to the analysis of microbial communities.
Wastewater treatment systems usually consist of various stages with distinct microbial cell densities. When studying samples derived from such different sample matrices, choosing a method for harvesting an appropriate amount of biomass for analysis should be considered for each stage. WWTP derived sludge contains many particles and a large surface that microorganisms adhere to, resulting in high cell densities. Samples from such an environment can usually be subjected directly to DNA extraction protocols; further concentration of the biomass might even be detrimental as the capacity of the extraction method could be exceeded. In contrast, when working with aqueous environments of low cell densities, e.g. the effluent of a reactor stage or treatment system, microbial cells need to be accumulated prior to DNA extraction, which can be achieved by filtering them onto a 0.22 μm pore-sized polymer membrane, which retains all but the smallest known microorganisms and can subsequently be used as the template for the application of a variety of standard DNA extraction protocols. Though not being the subject of this review, viruses are part of WWTP ecosystems as well and can be the subject of ecological analyses. Due to their small size and heterogeneity, viral particles present their own challenges regarding isolation, concentration and nucleic acid extraction. Specialized protocols exist for these steps and should be carefully selected and implemented, as commonly used methodology for the analysis of microbial cells/DNA is not adequate (Hjelmsø et al. 2017). Obtaining sufficient amounts of genomic DNA is crucial for the faithful description of the microbial community under survey. Starting the analysis pipeline with too little genetic material can result in an incomplete and distorted representation of the microbial community composition (Bowers et al. 2015).
Given that wastewater and sludge samples can contain very high concentrations of organic compounds, it might not be possible to completely eliminate contaminants throughout the DNA isolation step. The purity of nucleic acids can be determined with a spectrophotometer, an easy and quick to perform analytical step that should not be omitted when working with highly contaminated samples like those derived from wastewater treatment plants. The presence of such contamination will be visible as shoulders or peaks in the absorbance spectrum at wavelengths not corresponding to those of nucleic acids. The genomic DNA extracts should then be subjected to additional purification procedures. Several commercial kits exist, that serve to remove all potentially enzyme-inhibiting compounds remaining at this stage. The purified extracts can then be used for all downstream applications. The next step in a standard amplicon sequencing pipeline is the selective amplification of the genetic information in the DNA extract that is desired to be sequenced subsequently. PCR is the method of choice for this purpose and is used to generate multiple copies of only the desired marker genes for analysis, removing the background noise of the rest of the genetic information in the sample. By using adequate pairs of primers, target-specific oligonucleotides that serve as starter molecules for the amplification cycles, it is ensured that exclusively the 16S rDNA sequences or those coding for specific enzymes of interest of a defined phylogenetic group of microorganisms are amplified. It may still be required to adjust the purified DNA samples to a uniform concentration required by the NGS protocol chosen for amplicon sequencing. In comparison with the laborious protocols of the conventional subcloning-based approach of the past, however, sample preparation today for direct NGS analysis is significantly simpler and faster. As virtually every molecular biology laboratory has the necessary equipment for DNA extraction, amplification and purification at its disposal, access to amplicon sequencing analyses should be easy and ubiquitous in most parts of the world.
At this stage the preparatory laboratory workload is finished and the DNA amplicon solutions/samples can be submitted for sequencing. This step requires specialized personnel as well as expensive equipment and can easily be outsourced to a commercial service provider.
2.2 Next-generation sequencing techniques
Once the standard sequencing technique for all purposes, Sanger sequencing has been replaced by the novel NGS. Sanger sequencing is based on the incorporation of labeled chain-terminators and the subsequent electrophoretic separation of the obtained fragments. In contrast to the additional and time-consuming electrophoresis, the NGS techniques integrate the basecalling step seamlessly into the sequencing protocol. Luckily for microbial ecologists, several of the NGS approaches are well-established and easily available today in this area of research. One astonishing feat all NGS methods share, making them so powerful, is that they allow for the massively parallel sequencing of a large number of DNA templates (up to millions) in one single run, while the equipment performs the signal detection simultaneously for all reactions. This characteristic feature, also called “sequencing by synthesis”, constitutes an important difference in comparison to older techniques, which rely on the electrophoretic separation of the sequencing products as an independent step of the protocol. The NGS methods are capable of using a purified DNA extract directly as the source material for the sequencing step. Cloning and isolating the gene sequences of often thousands of different microorganisms present in environmental samples are not necessary, and principally, the removal of this step from the amplicon sequencing workflow makes high-throughput analyses possible.
NGS consists of a set of relatively young technologies that started to become relevant in metagenomic and 16S-based amplicon sequencing designs in the early 2010s and are constantly being developed and updated while new systems are still introduced, sometimes rendering others obsolete. Therefore, users of these techniques should stay informed about current developments which might be interesting to adopt into their experimental setups. Besides the capability of analyzing thousands and up to millions of DNA template molecules simultaneously in one single run, another common feature of all NGS techniques is the processing of several of these complex samples in parallel in the form of a mixed template. This option further reduces sequencing costs, as a number of samples can be pooled together and treated as one. This approach is called multiplexing and is coupled to the associated de-multiplexing after obtaining the sequence reads, which allows separating the sequences from different original samples again for downstream analysis. Multiplexing is based on the customization of the PCR primers in the preparatory template amplification step. Usually, one of the primers incorporates an additional short and unique nucleotide sequence called a “multiplexing identifier” (MID) and also commonly referred to as a “barcode”. This additional DNA tag allows for the binning of the resulting sequence reads after the reaction and according to their original samples by sorting the sequences using the barcode incorporated in all of the product molecules during the PCR. Bioinformatics software for the treatment and analysis of high-throughput sequencing data can perform this step and subsequently remove the artificial barcode tag as well as any primer sequences prior to the phylogenetic analysis.
Though a wealth of different next-generation sequencing techniques exists and is readily available for the researcher, two methods have been the most popular so far for the application of 16S rDNA amplicon sequencing: Illumina sequencing and Roche 454-Pyrosequencing. While the former is the standard approach today and the latter abandoned as a technology, both methods account for a significant share of the sequencing data within the context of microbial ecology studies of wastewater treatment systems available today. In the following sections these methods are outlined in some detail.
2.2.1 Illumina sequencing
Arguably, the single most widely applied NGS technology used for amplicon and shotgun sequencing in microbial ecology surveys at present, Ilumina sequencing is developed and distributed by the American company Illumina. The firm’s MiSeq and HiSeq sequencing systems, distinguishable mainly by their throughput capacity, account for a great share of the microbial amplicon and metagenomics sequences published in the last years. The Illumina techniques provide extremely high numbers of sequences from mixed and complex templates, which are commonly found in microbial ecology and wastewater treatment systems while also being highly error-resistant (for the exact values and a comparison with competitor systems, see Table 1). With each update of the technology, the average length of the generated reads, which could be considered a problem in the first generation of these sequencers, increases and renders the technique more suitable for the application in monitoring the structure and composition of microbial communities.
The Illumina protocol allows for the bidirectional sequencing of the templates, adding an extra layer of confidence to the analysis while also increasing the net read length by extending the sequence of the template across the overlapping portion beyond both termini. The method applies the “sequencing by synthesis” principle: A DNA polymerase catalyzes the insertion of fluorescence-dye-labeled deoxyribonucleotides (dNTPs) into the growing chain of a DNA strand. The reaction mixture is incubated for up to 300 cycles (Miseq system, see Table 1) sequentially with an equimolar mixture of the four DNA-bases adenine, cytosine, guanine and thymine. The fluorochrome labels are excitable by different specific wavelengths. In the moment of insertion into the growing DNA chain, the fluorochromes emit a light signal, allowing for the recording of the resulting DNA sequence based on the corresponding wavelengths/colors. The entire sequencing process can be divided into four main steps as follows.
-
1.
Preparation of the gene library: The mixed DNA sample from the genomic DNA extraction step is fragmented by mechanical or other means (e.g. sonication or enzymatic digestion) into small parts in a random fashion. The generated DNA molecules are subsequently modified at both ends (5’-terminal and 3’-terminal) by the addition of method-specific adapter-, and optionally, barcode tags. The fragmentation and tagging steps can be performed in combination and are called “tagmentation”.
-
2.
Cluster generation: The prepared DNA fragment library is loaded onto the lane of a flow cell, a device/tray containing the sequencing reaction compartments. The template DNA fragments are immobilized by the hybridization of one of their termini with a complementary oligonucleotide bound to the cell’s surface (adapter A). Serving as a template for amplification, the surface-bound fragments are then copied by a DNA polymerase. The resulting double-stranded DNA product is subsequently denatured and the template molecule is released again into the reaction mix, while the free end of the amplification product hybridizes with one of the complementary oligonucleotides (adapter B) immobilized in close vicinity on the flow-cell surface. The DNA fragments, now forming a bridge-like structure, are then serving as the template sequence in a new round of amplification. The resulting products are subsequently denatured, which now gives rise to two complete template molecules, each covalently bound to the reaction cell surface. Accordingly, this process is called bridge amplification and is executed repeatedly until spatially separated clonal clusters of all the fragments in the original sample DNA are generated. The following removal of the reverse strands in the flow cell is the final preparative step and leaves the sample ready for the actual sequencing protocol.
-
3.
Sequencing: The Illumina method uses fluorescently labeled dNTPs that are bound to a chain-terminating molecule, ensuring that the synthesis of the DNA strand is only advanced by a single base per detection cycle. Only one of the four competing DNA bases can be incorporated into the growing DNA strand, which then emits a light signal of a specific wavelength as a response to the excitation wavelength it is exposed to in the reaction channel. According to the response wavelength and intensity, one of the four bases is detected or called (hence the term “base call”). Subsequently, the terminator portion of the last incorporated dNTP is removed from the growing copy strands. This procedure is repeated over and over up to 300 times in the current implementation of the technology, extracting the sequence information of the template DNA molecule progressively cycle by cycle. The data output of the sequencing step consists of the recorded DNA sequences in the commonly used fastq format. Files of this type include the actual DNA sequence along with quality-related information, indicating the probability of each registered base call being erroneous (PHRED score).
2.2.2 Roche 454-pyrosequencing
The 454 pyrosequencing method developed by 454 Life Sciences and established by Roche as the first of the commercially available and successful NGS systems was the technology of choice until recently. In 2013 though, Roche announced that the method would be discontinued and support for existing systems stopped in 2016. Although this technology is now obsolete and disappearing from laboratories, a large number of published studies have obtained their results using pyrosequencing. Therefore, the technical principle is outlined briefly here. The main preparation steps for the template DNA of a sample are similar to the process performed in the Illumina protocol. Accordingly, in shotgun sequencing applications, as for metagenomic studies, the DNA is fragmented, for example by nebulization or sonication, followed by extension with the necessary adapter sequences. For 16S rDNA amplicon sequencing, these steps are omitted both in the pyrosequencing and the Illumina protocols, as the amplicons that serve as the template for sequencing already possess an appropriate length and contain the exact region of interest. In this case, the fragments are tailored according to the requirements of the experiment, before sequencing by the use of specific primer pairs for the PCR step. The adapter and barcoding sequence tags are incorporated into the template amplicons as preparatory steps in the laboratory as well.
Subsequently, for both applications, the DNA fragments are clonally amplified with a similar objective as the bridge amplification in Illumina sequencing, namely to generate densely packed clusters of identical DNA molecules that can then be subjected to the sequencing reaction. In 454-Roche pyrosequencing, the clonal multiplication of the fragments is performed by a PCR protocol applied in an oil–water emulsion. Droplets in the emulsion contain microscopically small capture beads bearing complementary adapter sequences that act as anchors for the template DNA. These DNA fragments are denatured to single-stranded molecules and added to the beads together with all the necessary reagents for the PCR. The aqueous droplets form a complete and autonomous reactor space and produce highly concentrated template beads.
The emulsion is broken up and the beads are loaded onto a picotiter plate containing over a million microscopic wells. Ideally, one bead containing a high concentration of identical DNA fragments fits into one well on the picotiter plate. Again, the principle of the sequencing reaction is “sequencing by synthesis”. Therefore, the plate is incubated with one of the four DNA bases sequentially. In case the complementary nucleotide is present, it is incorporated into the growing DNA strand, causing the release of pyrophosphate, which sets in motion a chain of enzymatic reactions. This process results in the emission of light that is recorded by a camera and quantified. The detection of two identical nucleotides in succession yields approximately twice the signal strength of the detection of a single nucleotide. Herein lies one important problem of the technique though, which certainly was a contributing factor in its discontinuation. With the incorporation of longer stretches of identical nucleotides into the growing DNA molecule, the recorded light intensities become less predictable and proportional to the length of these so-called homopolymers, causing incorrect sequence readouts. Repetitive stretches of DNA frequently occur in nature which, especially in the cases treated in this review -the use of relatively short fragments of phylogenetic information for taxonomic assignment of microorganisms- introduces a significant bias into the results. In this regard, the overestimation of the overall biological diversity of a community due to the microdiversity contributed by these erroneous homopolymers has been described in the literature (Quince et al. 2009). To counter this effect, several software solutions have been developed that try to correct or “denoise” the reads and restore the original sequences. Some of the denoising algorithms can require high amounts of computing power (e.g. Denoiser, Reeder and Knight 2010), while newer solutions can be applied on a common personal computer (e.g. Acacia, Bragg et al. 2012). In any case, homopolymeric stretches in recorded DNA sequences do introduce a factor of uncertainty into the experiment. In contrast, the alternative NGS techniques, like Illumina sequencing, do not present this problematic trait.
The final step in 454 pyrosequencing consists in exporting the raw data registered by the sequencer in the form of flowgrams, machine-only readable data files, which contain all the data recorded during the run. After converting the flowgrams into the common fastq format for example, using proprietary (Roche SFF Tools) or open-source software (http://maasha.github.io/biopieces/), the sequence reads are ready for analysis.
From a practical standpoint, Illumina sequencing is the method to recommend today for high-throughput applications like deep amplicon sequencing or metagenomics. The discontinuation of the pyrosequencing technique in 2016 would not allow for sensible investment in the technology, already carrying the status of a “zombie” platform (The Molecular Ecologist 2016). The high accuracy of the Illumina methods and their steadily increasing read-length, coupled with the possibility of paired-end sequencing and the immense data output at comparatively low prices, additionally offset 454-pyrosequencing’s greatest advantage, which has always been the higher achievable read-lengths. Furthermore, for 16S rDNA amplicon sequencing, a larger number of short reads is considered to yield more robust results than the analysis of fewer but longer sequences (Liu et al. 2007).
3 Data analysis
3.1 Software programs
The raw sequence reads of cloning-based amplicon sequencing experiments were usually generated via the Sanger technique and displayed in the form of chromatograms showing peaks labeled with the corresponding base called at each position. These chromatograms (or electropherograms) were examined and, if necessary, corrected manually, an often tedious task including the trimming of the primer sequences. Sequence files in the standard FASTA format were then exported and compared to a 16S rDNA database like SILVA (https://www.arb-silva.de/) (Quast et al. 2013), the National Center for Biotechnology Information’s (NCBI) GenBank (https://www.ncbi.nlm.nih.gov/) (Clark et al. 2016) or the Ribosomal Database Project (RDP, https://rdp.cme.msu.edu/) (Cole et al. 2014). Because of the sheer amount of sequence data generated by the NGS methods, a manual approach is not feasible anymore and choosing the fitting data analysis pipeline presents an important step in amplicon sequencing today.
Several software programs are available that can either be used offline on the experimenter’s personal computer or online in an internet browser via a graphical user interface. Here we outline the most commonly used software solutions, namely QIIME, mothur (both offline/local installations) and the RDP amplicon sequence pipeline (web-based user interface), though further, less popular programs for the same purpose do exist, for example, CLOTU (Kumar et al. 2011) or W.A.T.E.R.S. (Hartman et al. 2010), the latter offering a graphical user interface for the assembly of custom workflows. QIIME (Quantitative Insights Into Microbial Ecology, Caporaso et al. 2010a) is a popular and complete analysis pipeline that can be easily installed as a virtual machine and run under different operating systems, such as Linux, MacOS and Windows. Dedicated versions of the software are available for the analysis of Illumina as well as Roche-454-Pyrosequencing data. When performing high-throughput amplicon sequencing, many steps are required to transform the original bulk of machine data into phylogenetic and ecological information. These steps are performed by the QIIME software interactively with the user providing the overall workflow structure (the selection and order of the processing steps) and configuration for each step. QIIME works as a framework that connects and runs different third-party programs and implementations of algorithms (e.g. UPARSE, Edgar 2013; UCHIME, Edgar et al. 2011; pyNAST, Caporaso et al. 2010b), which can be conveniently integrated into the personalized workflow. In this regard, QIIME allows the experimenter a high degree of freedom when it comes to tailoring a customized analytical pipeline adapted to the particular study at hand. Often there are several tools available for a job, e.g. for chimera detection, taxonomic assignment, etc. This possibility of personalization is facilitated by the modularity of the software, which consists of a large set of scripts written in the Python programming language. The individual programs are called from the command line with the inclusion of a series of launch parameters provided by the user, defining a set of obligatory and optional constraints and conditions.
A typical application workflow for Illumina sequencing data would begin with demultiplexing the reads that are generated by the sequencer in the FASTQ format. Sequences originating from different samples that were sequenced together are separated again by the identifier barcodes incorporated at the ends of the reads. Quality control of the raw sequence reads presents another important preparatory step. The reads that do not comply with the quality cutoff set by the user are filtered out and removed from further analysis. The quality metric is usually defined as the average Phred score, which represents the probability of a base call being erroneous (Bokulich et al. 2013). Subsequently, and to allow for the extraction of meaningful phylogenetic and diversity-related information from the dataset, the sequence reads are grouped into operational taxonomic units (OTUs), which are used as a substitute for microbial species. Each OTU is assigned one representative sequence, which facilitates the fast computation of the following steps in the pipeline as opposed to computing the analysis for each of the often numerous and highly similar sequence reads. OTU assignment is performed by one of several methods that the user can choose from, utilizing reference-based or de novo approaches. OTU picking furthermore includes the comparative analysis of the set of representative OTUs against a reference 16S rDNA library of choice, for example, the Greengenes database (DeSantis et al. 2006) or one of the alternatives listed above, resulting in the taxonomic assignment of the OTUs. At this point all the data for the ecological analysis of the microbial communities present in the samples is generated. The taxonomic OTU assignments are then used for the study of the communities’ composition and the corresponding numbers of reads are employed as an approximation of the true species counts or abundance values. A series of meaningful diversity metrics can be calculated from these numbers (see Sect. 3.2, diversity indices) to describe the alpha-diversity of the sampled microbial consortia. The analytical and statistical procedures for these steps can be performed e.g. by the QIIME software. Alternatively, the user can use the OTU abundance distributions and taxonomic metadata files and use the computer software of choice to do exploratory data analysis and generate diagrams and graphs to visualize the results. The open-source statistical software environment R (R Development Core Team 2008), for example, provides some specialized libraries that serve to analyze and present ecological data. The vegan package contains numerous functions for the calculation of diversity indices, rarefaction analysis and more complex statistical methods for ecological studies (Oksanen et al. 2018). Further libraries offering a similar functionality are ade4 (Dray and Dufour 2007) and BiodiversityR (Kindt and Coe 2005). QIIME presents the easier-to-use “out of the box” solution and offers built-in functions, for example, to generate phylogenetic trees and bar charts that depict the composition of the analyzed samples by their corresponding proportions and abundance-based heatmaps for selected taxa. The program’s functions are called from a command line interface and are, therefore, easy to store and reproduce. An entire workflow can be saved as a script in a text file and adapted and re-used for future analyses or the re-analysis of existing sequence and abundance data.
The mothur software presents a viable alternative to QIIME. Developed by Patrick Schloss and his group at the University of Michigan, mothur provides methods that work with data generated by a variety of NGS technologies, such as the Illumina HiSeq and MiSeq methods, 454 pyrosequencing, PacBio, IonTorrent and even Sanger sequencing (Schloss et al. 2009). Mothur offers similar functionality as the QIIME package, providing the necessary tools and methods for the processing, analysis and visualization of complex microbial NGS data as QIIME does. Its underlying philosophy is different though: The mothur software consists of a series of re-implementations of the individual algorithms specific for sequence analysis developed as a standalone software application. Due to this design choice, the software is easier to use even though the individual steps are called from a terminal similar to the QIIME interface. This makes mothur simpler to interact with than the looser but more flexible framework of analytical methods provided by the QIIME software. Mothur can also be natively installed on most of the common operating systems without the need for a virtual machine or knowledge about the usage of the Linux command line. This overall more approachable nature of mothur is emphasized by the availability of Standard Operational Procedures (SOPs) for certain common workflows, which further helps to orient the user in creating a working pipeline. QIIME would be recommended for the experienced experimenter with some knowledge of bioinformatics and the desire for a highly customizable set of features to include in a workflow. It should be mentioned that the functionality regarding the presentation of results is richer in QIIME. Mothur users should probably familiarize themselves with additional software like R to create the final steps of the data analysis and visualization, for example, the creation of charts and heatmaps.
An even more accessible solution is available in the form of the web-based RDP amplicon sequence pipeline (RDPipeline), which can be used as an online service without the necessity of installing any software on the user’s personal computer (Cole et al. 2014). Especially in the case of relatively small amounts of sequence reads, the source files can be uploaded in common formats like FASTA or FASTQ to the RDP Project’s server and then be processed server-side via a graphical point-and-click user interface. For vast datasets, the tools of the RDPipeline, most of which are open-source, can be downloaded via the project’s GitHub code repository and then run locally on the experimenter’s own computer, similar to QIIME. QIIME, mothur and the RDPipeline are three of the most popular tools nowadays for the analysis of amplicon sequencing datasets. Additional tools exist and the comprehensive comparison published by Nilakanta and coworkers (Nilakanta et al. 2014) should be of help in the decision process on which program to use in any particular case.
The relative ease with which environmental 16S rDNA sequences can be generated today, owing to the high-throughput capacity of the readily available NGS methods, are now causing a flood of new entries in the public sequence databases. The 16S rDNA sequence collection of the Ribosomal Data Project (RDP) contained 30,000 entries in the year 2000 and increased six-fold to 180,000 until the year 2006 at the time cloning and Sanger-sequencing were the gold standard for amplicon sequencing (Chai et al. 2006). At the time of the last update in September 2016, the database was accommodating a total of over 3,300,000 16S rDNA sequences (Center for Microbial Ecology 2016: http://rdp.cme.msu.edu/). The authors of the corresponding paper state that about 85% of the bacterial sequences in the RDP database are derived directly from environmental samples and not from microorganisms in culture (Cole et al. 2014).
The phylogenetic information gained from the analysis of a microbial community via 16S rDNA-based deep-sequencing is robust, especially when large numbers of sequence reads are generated (Liu et al. 2007). The abundance distribution of the gathered reads can be used for the determination of the completeness of a sampling effort, which should always be part of any study in microbial ecology. Rarefaction analysis serves this purpose and is of the first steps to perform with OTU-clustered abundance values. A rarefaction plot shows the number of different obtained OTUs as a function of the number of reads sampled. The shape of the plot depends on the completeness of the sampling effort and can, therefore, be used to evaluate if further sampling is necessary or if the community under study has been exhaustively sampled. If the rarefaction curve reaches a plateau, it can be concluded that, even if more samples are taken, not many new species or OTUs would be encountered. In contrast, the presence of a steadily rising slope indicates that there are still more OTUs to be uncovered in case more reads are sampled.
3.2 Ecological statistics/diversity
The number of OTUs at this point, i.e. the number of reads falling into the corresponding bin as a result of a clustering process, can be used as a substitute for the species counts that are applied in classic community ecology. This substitution is commonly employed in microbial community ecology. Exhaustive sampling efforts allow for the calculation of further indices of alpha diversity. This approach can only be an estimate, however, as the number of copies of 16S gene copies in a bacterial genome is not uniform among different taxa (Vetrovsky and Baldrian 2013), a factor that is usually not considered in amplicon sequencing experiments. Efforts have been described to eliminate the numerical bias arising from this variability by accounting for individual differences between microorganisms via the introduction of additional data processing steps (Kembel et al. 2012). These techniques are still in need of improvement, however, and their use is even being advised against by some authors (Louca et al. 2018). The cautious estimation of relative abundance using sequence counts remains viable though for the time being, but the results should not be considered to be 100% correct.
The calculation and comparison of the relative OTU/species abundances open up a plethora of interesting methods for the evaluation of community composition and structure. Using a simple abundance distribution table alpha- and beta-biodiversity can be characterized, where alpha diversity describes the structure within a population while beta-diversity entails a comparative study of such multiple communities.
Richness estimators present easy-to-calculate and useful basic descriptive metrics. This type of indicator complements the rarefaction analysis by predicting the true number of different species (or OTUs) in a sample. The richness is estimated based on an extrapolation using certain key abundance values from the dataset, such as singletons (very rare OTUs that only occur once in the data set), doubletons (species that are represented by two sequences/entries) and the total number of OTUs found. Examples for richness estimators are Chao1 (Chao 1984), the abundance-based coverage estimator ACE (Good 1953) and the jackknife estimator (Efron and Stein 1981). These simple estimators present a quick way to extrapolate the richness of species and to evaluate the validity of a taxonomical survey in terms of sampling completeness.
The structure of the community under analysis can be further assessed using the classical diversity indices known from community ecology. Again, species counts are substituted by relative abundance values. The researcher can choose from a series of metrics that differ mainly in their weighting of specific groups of OTUs. Absolute OTU richness can be considered the simplest measure of biodiversity because it reflects the total number of different organisms in a community. Species that are very rare in the sample (e.g. singletons and doubletons) contribute in the same degree to the richness as OTUs that are highly abundant, perhaps encountered thousands of times in the form of high copy number reads. This shows that species richness alone is a very sensitive metric towards rare species, not considering the concept of evenness, which itself is an important part of the concept of biological diversity. As an example, a microbial community with one extremely abundant species and a thousand singletons is obviously less diverse than a community with a thousand different species that are all relatively abundant. Extremely rare members of a community are unlikely to possess important ecological functions and may be present in a dormant state as dead cells or acting as a pool of seeder organisms, which may increase in number once the physicochemical environmental conditions change in their favor. Accounting for this important part of diversity, evenness (or the degree of equal distribution of the abundances of each OTU in an ecosystem) can be measured as an individual metric. Furthermore, several descriptors of biodiversity have been developed that take into account the richness and evenness of species.
One of the most frequently used/popular indices of biodiversity is the Shannon–Wiener index, which was originally developed as a measure of entropy in information theory (Shannon 1948). As an ecological metric, the Shannon index takes into account the number of different species/OTUs (i.e. pure richness) as well as their abundance. It is still the diversity index most sensitive to rare species excluding the raw basic species richness. Based on the Shannon index, the Pielou index of evenness can easily be derived as the quotient of the Shannon–Wiener index and its maximum possible value, which is calculated with the assumption that each species encountered is equally abundant. The Simpson index of diversity (Simpson 1949) and the Berger–Parker index (Berger and Parker 1970) are further metrics that describe ecological diversity with a simple value. These measures are more balanced (Simpson) or less sensitive (Berger–Parker) to the presence of very rare taxa in the analyzed communities. The indices mentioned are grouped under the term Hill numbers developed by Hill (1973). Comparing these measures of diversity and equity across different studies and even between different samples from a single survey becomes imprecise, as the metrics do not scale linearly with increasing values for species richness (Jost 2006). The same author recommends converting the popular diversity indices into effective numbers of species or values of “true diversity”. The effective numbers of OTUs represent the diversity based on a particular index and are linearly comparable between microbial communities, providing more useful information when analyzing and comparing the microbial diversity, for example, in different ecosystems. Rarefaction analysis and the calculation of the diversity descriptors mentioned are available in the amplicon sequencing analysis frameworks, Qiime and mothur, for example.
Studies of microbial communities in wastewater systems often address questions going further than the description of the microorganisms in terms of abundance and by simple descriptors such as the diversity indexes mentioned above. Wastewater treatment systems serve a practical purpose, the conversion and elimination of organic matter and other pollutants from a variety of effluents. The research dedicated to these systems is mostly concerned with objectives such as the elucidation of patterns in the behavior of the microorganisms, changes in their compositional or structural setup as a response to changing physico-chemical parameters or over periods of time. Multivariate statistical methods allow for example analyzing the influence of an abiotic factor or the change in a certain operational parameter on the resident microbial community’s composition or diversity. Both, exploratory multivariate methods (“how can the variation in a data set/microbial community composition be best explained?”) like Cluster Analysis and Principal Component Analysis (PCR), as well as hypothesis-driven techniques (“can the null-hypothesis rejected that the changes in the community under study are random or unrelated to certain physico-chemical or other parameters?”) like Canonical Correspondence Analysis (CCA) can be applied to data sets generated with amplicon sequencing or fingerprinting techniques if additional information, like physico-chemical conditions, geographic or temporal parameters are integrated in the analysis. The main objective of cluster analysis lies in partitioning a data set by grouping the most similar species/OTUs together and maximizing the distance to the other groups, possibly leading to insights into the relations between and functional networks formed by species and the underlying drivers in the form of the known or controlled variables. PCA is a popular multivariate method that uses linear transformations of the existing and measured variables to generate new ones, the so-called principal components. These transformations result in a reduction of the number of dimensions needed to explain the variance/variability in a data set and thus make compositional or structural characteristics of a microbial community more discernible. This is reflected by the common depiction of the results of a PCA analysis in form of a biplot, a two-dimensional representation of the original data, the abundance values of the different OTUs or taxonomic groups and the variables, like e.g. organic loading rates, hydraulic retention times, influent composition, temperature, etc. As an example, a study investigating the microbiota of a multi-stage gibberellin-treating bioreactor made use of both PCA and CCA to observe correlation between the structure of the bacterial population of the system and and operational parameters like sulfate concentration, temperature and dissolved oxygen (Ouyang et al. 2017). Another instance of successful application of multivariate analyses in wastewater treatment microbial ecology is the survey of nitrifying bacteria in a full-scale activated sludge treatment system, where the authors used various biomolecular analytical methods (FISH, qPCR, amplicon-NGS) and were able to explain changes in community structure by the first three components/variables calculated via PCA, combining operational parameters and environmental factors (Awolusi et al. 2018).
An in-depth review article from the time of the advent of high-throughput sequencing techniques exists as an excellent resource for researchers desiring to familiarize themselves with the the application of multivariate statistical methods in microbial ecology (Ramette 2007). The same author later participated in creating a web application (GUSTA ME) which interactively guides a user through the multiple steps of a multivariate analysis in microbial ecology and provides walkthroughs of multivariate data analyses of existing data sets (Buttigieg and Ramette 2014).
The simpler statistical methods like the above mentioned Rarefaction analysis, diversity and evenness metrics, abundance predictors, as well as all common multivariate methods can be performed on abundance and variable/operational data from wastewater ecology experiments using the R statistical computing environment, in connection with dedicated libraries like the vegan package for community ecology (Oksanen et al. 2018). A basic skill level in bioinformatics or computer programming is recommended for the R approach. R, as well as readymade pipelines like mothur and Qiime, are capable of providing graphical output in a format and resolution apt for printed publications. The use of spreadsheet software can be problematic when dealing with large tables, being the norm these days, as the sequencing methods are becoming more and more powerful and abundance tables with thousands of different OTUs are common. Additionally, pipeline or R scripts are easier to reproduce and adapt to new datasets than difficult-to-document sequences of point and click actions.
In summary, 16S rDNA amplicon sequencing in conjunction with NGS technology represents a powerful tool at the disposal of many laboratories today, with the ability to provide insight into microbial communities, revealing their compositional and structural characteristics. Functional aspects of the members of the studied communities can only be inferred though, via the comparison of the gathered sequence reads with highly similar and metabolically characterized entries in the 16S rDNA databases. Owing to the restriction of the 16S rRNA gene’s role of a phylogenetic marker, a more direct assessment of metabolic and ecological microbial functions is not possible via 16S rDNA amplicon sequencing. Either a function-related gene can be selected for amplicon generation or, if a more complete picture of the metabolic possibilities of the communities under study is desired, using the omics disciplines should be considered. A detailed presentation and discussion of these complex and diverse technologies would be beyond the focus of this review though. Excellent reviews exist that give a detailed view and examples for application in the area of environmental and wastewater-related studies (Jünemann et al. 2017; Breitwieser et al. 2017).
4 Anaerobic treatments
4.1 Wastewater reactors
4.1.1 UASB reactors
The Upflow Anaerobic Sludge Bed (UASB) reactor, developed by G. Lettinga and collaborators in the Netherlands during the second half of the 70s, was the first anaerobic reactor widely used throughout the world for the treatment of industrial wastewater (Lettinga 2014). Although the reactors that have evolved from it, i.e. Expanded Granular Sludge Bed reactors (EGSB) and Internal Circulation (IC), are today replacing it at full-scale, it is still the subject of numerous studies and one of the most popular anaerobic reactors for laboratory scale works. In the following section, we will discuss the most common types of industrial wastewater treated by UASB reactors, grouped according to the chemical nature of the pollutants (e.g. organochlorides, LAS, azo dyes) and the microorganisms involved in their removal, as identified by amplicon NGS.Organochlorides make up typical anthropogenic compounds. Because of their broad use, i.e. as solvents for fats, oils, rust, resins, adhesives, paint and varnish; in fumigant, pesticides, paint, ink, perfumes and lacquers formulation; in iron and steel manufacturing, foundries, metal finishing and metal degreasers; in the synthesis of thermoplastics, urethane foam, synthetic rubber, fluorocarbons, vinyl-chloride, etc. (Rodríguez and Sanz 1998), they are released into the environment where they represent a public health concern since they have been shown to be toxic, mutagenic and/or carcinogenic. Although slow biodegradation may occur in groundwater where acclimated populations of microorganisms may exist, organochlorides are usually recalcitrant to biodegradation. This is especially true in the aerobic environment, as poly-chlorinated compounds are more amenable to anaerobic than to aerobic biodegradation by reductive dehalogenation. For these reasons, the anaerobic degradation of both aliphatic and aromatic compounds has been studied at length.
Zhang and co-workers have studied in depth the trichloroethylene (TCE) biodegradation and the bacterial communities in UASB reactors as a function of the pH (from 8 to 6) (Zhang et al. 2015a), temperature (from 20 to 40 °C) (Zhang et al. 2015b) and hydraulic retention time (HRT from 25 to 5 h) (Zhang et al. 2015c). Illumina MiSeq sequencing was used to assess the microbial shifts carried out during the operation of the reactors. The authors found changes in the bacterial communities associated with the parameters studied and their effect on the performance of the reactor. A positive correlation was detected between the relative abundance of the Dehalobacter, a Firmicutes able to carry out the reductive dechlorination of TCE to ethylene, and the TCE removal efficiency. Dehalobacter was not found at pH of 6.0, at which the removal efficiency of TCE was the lowest. TCE removal efficiency increased with temperature from 20 to 35 °C, and it dropped dramatically at 40 °C. The class Dehalococcoidia was detected from 25 to 40 °C but sequences related to the genus Dehalobacter were not retrieved at 40 °C. In addition, TCE removal efficiency decreased when the HRT was lowered from 25 to 5 h. Phylogenetic analyses showed that Bacteroidetes and Firmicutes were the dominant phyla; the class Dehalococcoidia was detected in all samples.
Polychlorinated biphenyls (PCBs) are among the most recalcitrant environmental pollutants. Nonetheless, in batch assays using biomass from a full-scale UASB reactor as inoculum, Gomes et al. (2014) found the genera Sedimentibacter, Tissierela and Fusibacter in the PCB-spiked reactors, while these were absent in the PCB-free reactors. These genera could be implied in the reductive dechlorination of PCBs.
The anaerobic degradation of linear alkylbenzene sulfonate (LAS) in laundry wastewater has been extensively studied by Bernadette Varesche’s research team at São Carlos University (Brazil). Several of their studies have focused on the evaluation of the microbial communities involved in the biodegradation of LAS. Accordingly, they studied the microbial communities of: (1) two UASB reactors treating synthetic wastewater supplemented with LAS and laundry wastewater (Okada et al. 2014); (2) one Fluidized Bed reactor (FBR) for laundry wastewater treatment, with and without sugar added as co-substrate for LAS removal (Braga et al. 2015); and (3) four anaerobic batch reactors inoculated with different sources of biomass and LAS (Motteran et al. 2017). Pyrosequencing (UASB and FBR) and MiSeq-Illumina (batch reactors) were used for high-throughput sequencing. The distinct microbial communities found in the UASB reactors, as a result of the different wastewaters used, were related to the LAS degradation rates obtained (Okada et al. 2014). Thirty-four of the detected genera could be involved in LAS degradation (aromatic compounds degraders, desulforizers, β–oxidizers, ω–oxidizers). The sludge blanket’s microbiota was typical of anaerobic reactor biomass, with a predominance of the phyla Firmicutes, Proteobacteria, Chloroflexi and Synergistetes. The communities from the phase separator compartments resembled biomass from aerobic reactors, with Nitrosomonas genus accounting for 34.6% of the reads. In the FBR, Proteobacteria followed by Bacteroidetes were found to be the dominant phyla in the samples where sugar was added, whereas the Proteobacteria and Gemmatimonadetes predominated in the samples without co-substrate (Braga et al. 2015). Twenty-two genera related to LAS degradation were identified. In the batch assays, Motteran et al. (2017) found that LAS sources influenced the kinetics of methane production. The best inoculum tested was that from a full-scale UASB reactor treating poultry slaughterhouse wastewater. Sequences related to taxa supposedly involved in the degradation of toxic compounds, e.g. VadinCA02, Candidatus Cloacamonas, VadinHB04, PD-UASB-13, were retrieved from this sludge. As a conclusion, it can be said that the treatment process had a relevant effect on the microbial structure.
Also, Delforno et al. (2017a) used three different reactor configurations, UASB, FBR, and EGSB reactors, for laundry wastewater treatment. The microbiota was analyzed by metagenome shotgun sequencing on the Illumina HiSeq platform. The obtained sequences that were affiliated to the Archaea domain, i.e. Methanobrevibacter, Methanothermobacter, Methanosaeta, Methanosarcina and Methanoregula genera, were only retrieved from the EGSB and UASB reactors. The analysis of Shotgun sequences showed genes related to methanogenesis, prevalently involved in the acetoclastic pathway. Unexpectedly, the FBR showed a dominance of aerobic microbiota and pathways for oxygen-dependent aromatic compound degradation. In this reactor, Proteobacteria was clearly the dominant phylum (accounting for 78% of the reads). Sphingopyxis (15%) was the most abundant genus. The presence of oxygen in the reactor was suggested using a color-indicator-based method, which would explain those findings.
UASB reactors are broadly used for the anaerobic treatment of many high-strength industrial wastewaters. In this regard, the decolorization of Alizarin yellow R (AYR), a widely used azo dye, and the microbial community developed in a USAB reactor were studied by Cui et al. (2016). High efficiency was reached at an AYR loading rate of 600 g m−3 * day−1. Sequencing by Illumina MiSeq showed that biodiversity decreased during the treatment, probably due to the reversible inhibition of the anaerobic consortia by the azo dye. Proteobacteria (Enterobacter genus) and Firmicutes (Enterococcus genus) were the most enriched taxa. Both genera are involved in the reduction of azo dyes. The addition of zero-valent iron nanoparticles plus persulfate (NZVI) greatly enhanced the decolorization rate of the dye brilliant red X-3B (Pan et al. 2017). A shift of populations took place in response to the addition of NZVI: Illumina MiSeq revealed that the most abundant genus (Lactococcus) decreased from 33 to 7.9%, while the Akkermansia spp. increased from 1.7 to 20.2%.
Methanogenic archaea were analyzed in a UASB reactor fed with methanol by Cerrillo et al. (2016). Quantitative real-time polymerase chain reaction (qPCR) revealed that the methanogenic population increased twofold concerning the initial inoculum. As could be expected, Illumina sequencing analysis confirmed that the resulting methanogenic population was mainly composed of methylotrophic archaea (Methanomethylovorans and Methanolobus genera). This enriched biomass showed great potential as an inoculum for bioaugmentation reactors that produce biogas from methyl-substrates.
The performance of a modification of the classical UASB, an Up-flow Anaerobic Fixed Bed reactor, for the treatment of PTA (a mixture of terephthalate and benzoic acid) was evaluated by Ma et al. (2015b). The reactor was operated at 33 and 37 °C. Better removal efficiencies were achieved at 30 °C. At the higher temperature, Thauera and Hydrogenophaga (Betaproteobacteria) decreased, whereas the number of Syntrophorhabdus (Deltaproteobacteria) increased. Methanobacterium was the predominant genus at either temperature, indicating the prevalence of the hydrogenotrophic pathway for methanogenesis derived from the degradation of this pollutant.
Anaerobic granular sludge is essential for the efficient performance of UASB reactors. Granules consist of highly settleable microbial aggregates, with a high bacterial density (up to 1011 microorganisms/g, both bacteria and archaea) and high methanogenic specific activity (up to 1 kg-COD (chemical oxygen demand) Kg-VSS−1 day−1 or even more at lab scale). The granulation is a process still not well-understood, so the process of granule formation is being broadly studied at the laboratory and full scale. Some examples of studies related to the formation of UASB granules are presented below. Kim et al. (2016) used a CSTR, inoculated with anaerobic digester sludge, for lactic acid production (pH at 5, temperature 50 °C). After only 5 days of operation, the genus Lactobacillus increased from 0.1 to 91.5%, and Leuconostoc, another lactic acid bacterium, accounted for approximately 7% of total sequences. The mixed liquor in the CSTR was then transferred to a UASB. A gradual decrease of HRT from 8.0 to 0.17 h, corresponding to an increase in the loading rate from 60 to 2880 g-glucose * L−1 day−1, promoted the formation of granules from flocculent sludge. Lactobacillus delbrueckii (80% of the total bacterial sequences) and Leuconostoc sp. (15%) were dominant in the granules. Granulation in a UASB reactor using galactose as the substrate for hydrogen production was evaluated by Sivagurunathan et al. (2016). The authors decreased the HRT from 12 to 2 h and observed the rapid formation of granules at 6 h HRT, which was further enhanced at 3 h HRT. The maximum H2 production rate was obtained at the shorter retention time. Microbial community analysis by pyrosequencing of 16S rRNA genes revealed a shift of the dominant bacteria: whereas Bacilli (Sporolactobacillus and Lactobacillus genera) dominated at 6 h HRT, Clostridium increased during the HRT when the HRT was set to 3 h. The effect of chitosan for the promotion of granulation in UASB reactors in the presence of organic solvents (ethanol, ethyl acetate, and 1-ethoxy-2-propanol) has been recently evaluated by Torres et al. (2018). Chitosan stimulated the production of extracellular polymeric substances (EPS), correlating with the size of the granules. Higher methanogenic activities were evident in the sludge from the chitosan-assisted reactors. Actinobacteria, Bacteroidetes, Chloroflexi, Firmicutes, Proteobacteria, Synergistetes and Cloacimonetes were the dominant bacterial phyla. Geobacter, a bacterium capable of syntrophic growth, and the hydrogenotrophic methanogen Methanocorpusculum were predominant in the granules.
Thermophilic UASB reactors could present an attractive alternative for the treatment of high-temperature wastewater. However, the availability of thermophilic granule seeds is extremely limited, which is the reason for the use of mesophilic granules as the inoculum in practice. Using Illumina MiSeq technology, Zhu et al. (2017a, b) studied the changes in the granules during the transition from mesophilic to thermophilic conditions. Mainly members of the family Anaerolinaceae managed to tolerate the temperature change and contributed to maintaining the physical integrity of the granular structure. In addition, Ruminococcus species showed the most dramatic increase in abundance. In contrast, Syntrophobacter spp. drastically decreased, resulting in the accumulation of volatile fatty acids accompanied by a drop in pH. When the pH was maintained stable by adding bicarbonate, the genus Methanoculleus appeared at values higher than 6.5, most probably being responsible for the production of methane.
Several conclusions can be drawn from the works described above: (1) Though Firmicutes and Proteobacteria are the dominant phyla, the bacterial genera involved depend on the compounds present in the reactor influent. Only members of the genus Clostridium appear to be involved in the degradation of a large number of xenobiotic compounds; (2) From this it can be concluded that it is not possible a priori to infer the detailed composition of the microbiota of an anaerobic reactor treating industrial wastewater, which stresses the necessity of studying each specific case individually; (3) With respect to the archaea, acetoclastic methanogenesis is the predominant metabolic route in many cases: sequences affiliated to the Methanosaeta and Methanosarcina genera were in the majority in a large number of reactors; (4) Short HRT and the addition of EPS-stimulants were capable of promoting granulation. During the formation of the granule, a shift of the populations that served as the inoculum takes place.
4.1.2 Other anaerobic reactors
Anaerobic digestion is the most suitable treatment for industrial wastewater. In addition to UASB reactors, further anaerobic configurations have been applied for the remediation of different types of wastewater. The EGSB reactor can be considered as an evolution of the UASB technology. EGSB reactors are used in many industrial wastewater treatment processes, although comprehensive studies of their microbiota are scarce. Several studies have paid attention to the treatment of wastewater with a high content of sulfate. Higher sulfate concentration was tolerated by an EGSB reactor treating waste brines (Liao et al. 2014). Efficiencies of 80–90% were achieved at an influent sulfate concentration of 3600 mg L−1 and 3% NaCl. Furthermore, synthetic waste brine containing sulfate up to 5.78 kg m−3 day−1 and nitrate up to 6.38 kg m−3 day−1 was degraded with efficiencies of 99.97% and 82.26%, respectively. The bacterial diversity of the sludge was analyzed by 454-pyrosequencing. Proteobacteria (77.7%) was the dominant microbial population, followed by Firmicutes (12.2%) and Chlorobi (2.7%), Bacteroidetes and Synergistetes. Half of the sequences classified at genus level were affiliated to the genus Thauera, which plays an important role in the denitrification of high-strength nitrate wastewater. Wolinella, Arcobacter, Alkaliphilus and Erysipelothix were also present in the granules. Surprisingly, the abundances of Desulfovibrio, Desufuromonas, Desulfococcus and Sulfurovum, typical sulfate-reducing bacteria (SRB), were less than 1%. Wu et al. (2015) used a two-stage EGSB reactor with a high-efficiency removal of COD, subjected to sulfate concentrations up to 2000 mg-SO4 L−1. When the sulfate concentration was increased to 3000 mg L−1, it caused the inhibition of microbial activity. Pyrosequencing showed that Desulfovibrio spp. in the anaerobic granular sludge increased four times during the operation time of the reactor. A three-stage system composed of two anaerobic reactors, CSTR (for hydrolysis and acidogenesis) and EGSB (methanogenesis), as well as one aerobic Sequencing Batch Reactor (SBR), was applied to treat sulfate-rich cellulosic ethanol wastewater (Shan et al. 2017). Stable performance was obtained for a COD removal efficiency of 94.5%, sulfate removal (89.3%), and methane production rate (11.5 L day−1) at an organic loading rate (OLR) of 32.4 kg-COD m−3 day−1. The microbial community composition was evaluated using the Illumina Miseq platform. The acidogenic reactor was rich in acidogenic bacteria (Megasphaera, Parabacteroides, unclassified Ruminococcaceae spp., and Prevotella) and SRB (Butyrivibrio, Megasphaera). The methanogenic reactor was dominated by the candidate phyla Hyd24-12. Members of the Thermotogaceae and Syntrophomonadaceae were also abundant. With respect to the Archaea domain, reads affiliated to Methanosaeta were predominant, indicating that acetoclastic methanogenesis was prevailing in the EGSB reactor. The dominant aerobic bacterium in the SBR reactor was Truepera.
The anaerobic treatment of dye wastewater has been studied at length using a variety of approaches. A study carried out by Zhang et al. (2012) in an SBR showed that Bacteroidetes represented the dominant phylum, while the second and third most abundant phyla were Firmicutes (Enterococcus genus) and Proteobacteria (Novispirillum, Rhodobacter genera). Textile wastewater usually contains, together with the dyes, high levels of sulfate. SBRs were also used for the anaerobic treatment of sulfate-rich synthetic textile wastewater supplemented with lactate, glucose, and ethanol as co-substrates (Rasool et al. 2015). Due to the competition of sulfate and azo dye for electron donors, the co-substrates affected the performance of the reactors and their microbial community. Dye degradation could be improved if lactate and ethanol were used; the reactor fed with lactate showed the highest relative abundance of sulfate-reducing bacteria (SRBs). As in Zhang’s study, Firmicutes and Proteobacteria were the dominant phyla, followed by Chloroflexi, Bacteroidetes, and Actinobacteria. The genus Desulfococcus was found in the highest relative abundance in all the reactors. Decolorization/degradation of the azo dye Procion Red HE-7B under alkaline conditions with sulfate-reducing granular sludge was explored in batch assays by Zeng et al. (2017). The efficiency of HE-7B degradation was improved at a higher COD. Acidifying bacteria (Lactococcus) and complete oxidizers (Desulfobacter and Desulfomicrobium) were identified as key bacteria of the process. Redox mediators have been proven to accelerate the anaerobic biological reduction of azo dyes, significantly increasing the decolorization rate and recovering the inhibited methanogenesis (Dai et al. 2018). The authors found that the family Enterobacteriaceae, within the phylum Proteobacteria, dramatically increased in numbers from the granular sludge used as inoculum to that in the rectors supplied with azo dye and redox mediator. The abundances of the genera Desulfovibrio and Azoarcus, both belonging to the phylum Proteobacteria, as well as Macellibacteroides and Bacteroides, within the phylum Bacteroidetes, decreased significantly. Concerning the methanogens, Methanosaeta and Methanobacterium significantly decreased in the presence of azo dyes although the number of Methanosaeta was partially recovered by the addition of a redox mediator, which promoted the reduction of azo dyes decreasing its toxic effect on the acetoclastic methanogens. In the presence of azo dyes, an unclassified genus inside the Methanobacteriales family was predominantly detected.
The effect of temperature or the addition of redox mediators has also been explored. The treatment of dye-containing wastewater under increasing high-temperature conditions (from 35 to 55 °C) was studied in Moving Bed Biofilm reactors by Li et al. (2015a). The COD removal exhibited performance optima at 40 and 50 °C. Different thermophilic communities were dominant in different temperature ranges: Caldilinea (from 35 to 45 °C) and Rubellimicrobium and Pseudoxanthomonas genera (over 50 °C). This data confirms that the range 45–50 °C is critical for microbial activity. The recent study by Xiao et al. (2018) deserves special mention. The authors have reported a simple method for assaying anaerobic biodegradation of dyes on 96-well microtiter plates. The assay was verified with the anaerobic degradation of methyl red and amaranths dyes by Shewanella oneidensis. This approach could present a useful time-saving and low-cost method for the analysis of anaerobic dye-decolorization.
Several studies have focused on municipal solid waste leachates using different reactor configurations. In a study published by Liu et al. (2015b) with leachate from a municipal solid waste incineration plant, the authors observed that high NH4+ concentrations inhibited the anaerobic treatment of the leachate. The inhibitory effect on anaerobic granular sludge was reversible, however, affecting the bacterial activity but not its survival. Illumina high-throughput sequencing showed that the microbial community compositions of the inoculum (granular sludge from a full-scale UASB reactor used to treat brewery wastewater) and of the sludge after six months of EGSB reactor operation were similar. The dominant phyla of both samples were Euryarchaeota (Methanobacterium), Chloroflexi, Proteobacteria, and Actinobacteria. The results of several studies (Kim et al. 2014; Jang et al. 2015; Xue et al. 2015) show that, in general, the acetoclastic methanogens, i.e. Methanosarcina or Methanosaeta, seem to be the most abundant archaea, while the prevalence of one of these genera over the other depends on the operational and physico-chemical conditions inside the reactor. Furthermore, the hydrogenotrophic Methanobacterium and Methanoculleus are also commonly detected. The predominant bacterial phyla are usually Proteobacteria, Chloroflexi, Bacteroides, Actinobacteria, Synergistetes and Firmicutes.
The anaerobic treatment of low-strength wastewater still presents a methodological challenge in cold and moderate-climate countries. Although the anaerobic reactors have been commonly used for industrial wastewater treatment, some studies focus on showing the feasibility of the different anaerobic reactor types for domestic wastewater treatments. Smith et al. (2014) evaluated the use of an Anaerobic Membrane Bioreactor (AnMBR) with a submerged membrane to improve the psychrophilic (15 °C) treatment of domestic wastewater. The authors suggest an operational strategy to improve treatment performance in low-temperature AnMBR. Illumina sequencing indicated that the membrane biofilm was enriched in highly active methanogens (Methanosaeta and Methanosarcina in the suspended biomass and Methanoregula and Methanospirillum in both suspended and biofilm) and syntrophic bacteria (Syntrophomonas and Smithella, within Syntrophomonadaceae family). A modification of the conventional UASB reactor consisting of downflow sludge circulation was investigated for anaerobic domestic wastewater treatment (Liu et al. 2018). The innovation enhanced granulation and shortened start-up times, resulting in a high COD removal efficiency of 94.8% at a HRT of 6 h. High-throughput sequencing revealed a shift in microbial community composition during the start-up period from Proteobacteria and Firmicutes to Bacteroidetes and Chloroflexi.
Recirculation is a strategy broadly used for anaerobic treatment of low-strength wastewater using UASB-related reactor. Accordingly, Yang et al. have employed a Strengthened Circulation Anaerobic reactor under different HRT and upflow velocities (2017) and an EGSB reactor (2018) for municipal wastewater treatment. In the first of the cited works, the authors proved that acetoclastic and hydrogenotrophic methanogens played an important role in the formation and maintenance of the anaerobic granular sludge under both low and high OLR operation conditions. Due to the difficult granulation in low-strength wastewater, granular activated carbon (GAC) was added to raw flocculent sludge in the second study. The addition of GAC shortened the start-up time, promoted the granulation and increased the size of the granules as well as their specific methanogenic activity (SMA). The EGSB efficiently removed the COD, when the reactor was operated at HRT from 8 to 4 h and the upflow velocity ranged from 1.09 to 2.44 m h−1. Bacteroidetes (36%), Proteobacteria (28%), Firmicutes (14%) and, to a lesser extent, Spirochaetae and Chloroflexi were the main bacterial phyla present. At the lowest HRT, the fermentative genus Aeromonas, and less pronounced Lentimicrobium and Sedimentibacter, increased in relative abundance, accompanied by biogas production, which led to the floating of the granular sludge. Under this condition, a shift of the metanogenic population from the acetoclastic Mehanosaeta to hydrogenotrophic Methanobacterium could be detected.
Other types of wastewater, e.g., heavy oil refinery (Dong et al. 2016b), abattoir (Jabari et al. 2016), poultry slaughterhouse (Delforno et al. 2017b), cassava (Su et al. 2017), starch (Yu et al. 2016a) or swine wastewater (Ducey and Hunt 2013), have also been treated by anaerobic reactors, and their microbial communities have been analyzed by high-throughtput 16S rDNA amplicon sequencing.
Similar to the case of the UASB reactors, wastewater is the main factor in determining the shape of the microbial population in the reactor. The type and configuration of the reactor only contribute to a lesser extent to this process. Therefore, in the anaerobic treatment of azo dyes e.g., sequences affiliated with the Enterococcus genus (Firmicutes) were among the most abundantly observed in both, UASB and SBR reactors. Although Proteobacteria, Firmicutes, Bacteroides and Chloroflexi generally represented the predominant phyla, no clear trends can be established at the level of the lower taxa. With respect to the methanogen-producing archaea, acetoclastic methanogenesis seems to be predominant, although the preeminence of Methanosarcina or Methanosaeta depends on the operational conditions in the reactor.
4.2 Anaerobic waste digestion and biogas production
Biogas production is regarded as one of the most promising renewable energy sources worldwide. It is a technically simple and low-cost technology so that biogas plays an important role in developing countries. In addition, biomethanization can help to manage the huge amount of solid and slurry waste generated in the industrialized countries (e.g. aerobic sludge produced in WWTPs, agricultural waste, food and domestic waste, …), contributing to the energy supply (methane can be flexibly utilized for heat and electricity generation or distributed via the natural gas grid infrastructure) and mitigating greenhouse gas emissions.
The anaerobic digestion (AD) process consists of four consecutive steps: hydrolysis, fermentation/β-oxidation, acetogenesis and methanogenesis. Methanogenic archaea are considered the key players in the AD of the organic matter, which is reflected by the large number of studies of methane-producing communities. However, during the AD of insoluble material, being often the case for the feedstock for biogas production (e.g. manure, crops, municipal solid waste, activated sludge waste,..), the hydrolysis, which is carried out by bacteria, is often the bottleneck of the process. Since high temperatures lower the viscosity of the medium and increase the rate of hydrolysis, positively affecting the substrate degradation and biogas production rates, the thermophilic temperature range is commonly used in AD. Organic wastes from multiple sources have been used as the feedstock for biogas production. In this section we will review the most commonly used types of waste for this purpose.
4.2.1 Waste-activated sludge
Anaerobic digesters have been widely used since the 1920s for the stabilization of primary and secondary sludges from municipal wastewater treatment plants. AD of the large quantities of waste-activated sludge (WAS) generated along with the degradation of organic matter reduces the amount of sludge, destroys pathogens and produces biogas. A pioneering study, applying an NGS-based approach, revealed the high complexity of the communities involved in the anaerobic digestion of sludge (Yang et al. 2014b). The authors identified Proteobacteria (9.52–13.50%), Bacteroidetes (7.18–10.65%) and Firmicutes (7.53–9.46%) as the most abundant phyla. The genera Methanosaeta and Methanosarcina represented the major part of the methanogenic population. One year later, Guo et al. (2015), also using Illumina sequencing, published similar findings: again, the most abundant bacterial populations were Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria. Similarly, Methanosaeta and Methanosarcina were the predominant methane-producing microorganisms detected. Based on cell count data and the identification of specific enzyme-encoding genes, the authors suggest that the acetoclastic methanogenesis is the dominant methanogenesis pathway in the full-scale anaerobic digester.
Similar results have been found in subsequent studies. The phyla Chloroflexi, Bacteroidetes, and Firmicutes were the dominant functional microorganisms in high solids and low solids anaerobic systems, accounting for 84.8% and 90.6% of the total number of sequences, respectively (Lu et al. 2016). Methanosarcina was, again, the predominant methanogen. The abundance of this microorganism, calculated by quantitative reverse transcription PCR (RT-qPCR), was higher in the high solids anaerobic fermentation reactor than in the low solids reactor, resulting in both higher VFA consumption and methane production. Westerholm et al. (2016) reported that the pre-treatment of WAS with microwaves and ultrasounds (increasing its solubility) influenced the microbial community structure over the course of its anaerobic digestion when compared with untreated waste. Bacteroidetes, Proteobacteria and Firmicutes were the dominant phyla in all digesters. The relative abundance of Proteobacteria decreased in association with the substrate availability. On the other hand, the relative abundance and richness of the Firmicutes/Clostridiales positively correlated with substrate availability and biogas generation. Methanogenic communities were highly dominated by Methanosaeta and Methanobrevibacter phylotypes. A gradual decline in Methanobrevibacter and an increase in the abundance of Methanosaeta concilii over time took place in the digester receiving untreated waste, while more diversified archaeal communities were maintained in the pre-treatment digesters. Different approaches have been used to improve the performance of the AD of waste-activated sludge. According to Zhao et al. (2017), the addition of aged refuse, produced massively in landfills, to WAS increased the abundance of the microorganisms responsible for sludge hydrolysis and acidogenesis (e.g. Clostridium, Sporanaerobacter, Proteiniborus, Parabacteroides), enhancing the anaerobic sludge digestion and the methane yield. Yu et al. (2015) studied the effect of three ferric salts on the thermophilic anaerobic digestion of activated sludge. The addition of FeCl3 caused an increase of almost 100% in CH4 production in comparison to the control sample. Pyrosequencing analysis showed a surprising result: Methanosarcina increased from 1.3 to 63.2% in effective reads after the addition of FeCl3.
To conclude this section, an interesting modification of the AD process deserves a brief comment. Dynamic membrane technology, in which dynamic membranes can be formed and re-formed in situ, have several advantages over conventional AD or, even, over anaerobic membrane digesters. Yu and co-workers analyzed the temporal variations in microbial communities during the start-up of an anaerobic dynamic membrane digester (ADMD) (Yu et al. 2014) and compared the performance and underlying microbial community composition of a conventional anaerobic digestion process to that of the ADMD (Yu et al. 2016b). Proteobacteria and Bacteroidetes were the major phyla at the beginning of the start-up phase while Betaproteobacteria followed by Sphingobacteria were detected as the most abundant classes at the end of the start-up. The archaeal community became stable after 38 days of operation. Methanomicrobiales (genus Methanolinea, originally isolated from a municipal sewage sludge digester) and Methanosarcinales (genus Methanosaeta) were found to be responsible for methane production in the ADMD system. Hydrogenotrophic pathways might have prevailed over the acetoclastic route during the start-up. The authors found that during long-term operation, the ADMD enhanced sludge reduction and improved methane production over the conventional process. Pyrosequencing of the 16S rDNA amplicons revealed that Proteobacteria and Bacteroidetes were abundant in the bacterial communities, and Methanosarcina and Methanosaeta in the archaeal communities.
4.2.2 Agricultural and food waste
Billions of tons of agricultural waste (mainly crop straw and manure) are generated yearly throughout the world (e.g. more than 700 million tons of crop straw in China and 500 million tons in the USA). Their inappropriate disposal leads to environmental problems and constitutes a waste of resources. AD is a technically simple and cost-effective technology, for which different types of agricultural waste can be used as feedstock, resulting in the production of biogas.
In a pioneering and frequently cited article on the use of NGS techniques for the analysis of the microbiota of biogas plants, Schlüter et al. (2008) studied the microbial community metagenome of a full-scale biogas plant fed with maize silage (63%) and green rye (35%). The authors detected numerous sequences affiliated to clostridial genomes, including gene-coding for enzymes involved in the hydrolysis of cellulosic material. A significant number of archaeal reads were affiliated to Methanoculleus marisnigri.
The fermentation of maize silage for biogas production was investigated in three full-scale CSTR reactors (Lucas et al. 2015). The microbial communities in the three reactors were highly similar, indicating that identical environmental and process parameters resulted in identical microbial assemblages and dynamics. Firmicutes and Bacteroidetes and the candidate phylum WWE1 dominated the bacterial communities (82–85% of the reads). Sequences affiliated to Sedimentibacter and Streptococcus were the dominant OTUs identified at the genus level. Specialized hydrogenotrophic methanogens (Methanobacterium and Methanoculleus) and the acetoclastic Methanosaeta covered the entire spectrum of methane-producing organisms. However, no Methanosarcina-related generalists, which are considered as essential for efficient biogas process, were detected. A conclusion drawn in the study was that hydrogenotrophic methanogenesis seems to dominate the biogas formation from crops.
Zhou et al. (2017b) used rice straw for biogas production in a CSTR. Four OLRs were analyzed: 1.22, 1.46, 1.70, and 2.00 kg VSsubstrate m−3 day−1, decreasing the biogas at the higher OLR. The biogas production rate was determined at 323 m3 t−1 dry rice straw over the whole process. Bacteroidetes (37–44%), followed by Firmicutes, Chloroflexi, Proteobacteria, Verrucomicrobia, Planctomycetes, Spirochaetes and Fibrobacteres were the most prevalent phyla in all the samples. Although acetoclastic methanogens were identified (Methanosaeta, Methanosarcina), the hydrogenotrophic pathway (Methanobacterium, Methanospirillum, Methanolinea) was the main biochemical route of methanogenesis in the reactor.
Manure from several sources is often stabilized by AD. Despite the differences in manure in terms of substrates (swine, cattle), size (full scale, farm scale), geographical location or operational conditions of the digesters, a core community could be characterized. The bacterial populations in all of the digesters were dominated by the phyla Firmicutes and Bacteroidetes, followed by Proteobacteria and Chloroflexi. Clostridiales and Bacteroidales were detected as the dominant orders, with Clostridium being clearly the most prevalent genus (Li et al. 2015b; Wolters et al. 2016; Cho et al. 2017). The Firmicutes and Clostridia also accounted for the most dominant bacterial taxa in an anaerobic digester treating swine manure under thermophilic conditions (72/64% of the pyrosequencing reads) (Tuan et al. 2014), and a CSTR digester stabilizing poultry litter (76/52%) (Smith et al. 2014). Again, Bacteroidia was the second most abundant class. In both cases, Methanothermobacter (order Methanobacteriales), which has a hydrogenotrophic metabolism, was the predominant methanogen.
Manure, in general, presents a high content in nitrogen so that its co-digestion with other types of waste (especially carbon-rich and nitrogen-poor waste, like straw) has been extensively studied. In an early survey, using 454-pyrosequencing to study the microbial community inhabits a biogas plant fed with agricultural waste (maize silage. green rye and liquid manure). Krober et al. (2009) found that most of the bacterial 16S-rDNA sequences could be assigned to the phylum Firmicutes, followed by Bacteroidetes. The most abundant orders were Clostridiales and Bacteroidales. Most of the archaeal sequences were affiliated to the order Methanomicrobiales, with Methanoculleus (M. bourgensis) being the predominant genus (species). To determine the effect of the feedstock type on the microbial communities involved in AD, Ziganshin et al. (2013) fed biogas reactors with different agricultural waste (maize silage and maize straw, cattle manure, chicken manure, Jatropha press cake and dried distiller grains). The major detected bacterial taxa were the Clostridia and Bacteroidetes, whereas the archaeal community was dominated by the orders Methanomicrobiales and Methanosarcinales. Community composition was mainly influenced by the feedstock type, e.g. the system alimented with chicken manure depended on syntrophic acetate oxidation as the main acetate-consuming process due to the inhibition of aceticlastic methanogenesis; Jatropha led to the enrichment of fiber-degrading specialists belonging to the genera Actinomyces and Fibrobacter. Li et al. (2014) investigated the prokaryotic community compositions during co-fermentation of wheat straw and swine manure. Communities attached to straw particles were overrepresented by the phyla Spirochaetes and Fibrobacteres, while Synergistetes and Euryarchaeota were more abundant in the slurry. The straw-associated genera Fibrobacter, Bacteroides, Acetivibrio, Clostridium III, Papillibacter, Treponema, Sedimentibacter, and Lutispora could be involved in the substrate hydrolysis. The protein-fermenting bacteria Aminobacterium and Cloacibacillus were highly abundant in the slurry. Methanoculleus and Methanosaeta were the most abundant methanogens. Studying the co-digestion of corn straw with cattle manure, Zhang et al. (2016a) showed that the straw was mainly metabolized by acetate-utilizing methanogens, with Methanosaeta dominating the archaeal community.
Other types of livestock manure, such as chicken manure, were co-digested with the microalgae Chlorella sp. to evaluate the impact of different ratios of co-substrates on methane production (Li et al. 2017b). The co-digestion at the ratio manure: algae 8:2 showed a significantly higher methane yield. The number of acetoclastic Methanosaeta significantly decreased, while Methanosarcina, Methanospirillum and Methanobacterium increased in abundance, suggesting that hydrogenotrophic methanogenesis was responsible for the methane production. The co-digestion of the macro-alga Ulva lactuca with dairy slurry also showed an apparent inhibition of the acetoclastic methanogenesis and an increase of the Methanosarcina population (Fitzgerald et al. 2015).
Food waste (FW) is the major component of the organic fraction of municipal solid waste. Its disposal in landfills has become an environmental concern, and many countries have legislated to avoid this usual but polluting environmental practice. AD is a good option for reducing food waste, which, when stabilized, can be used as fertilizer and for soil amendment. The accompanying production of methane adds further value to the process.
Several operational parameters have been investigated for the efficient treatment of FW. One of the key factors to be considered is the total solids (TS) content (wet or dry AD), which can affect the efficiency of the process and the microbial communities involved. Yi et al. (2014) obtained better performance using higher TS concentrations (from 5 to 20%). They found that the phylum Bacteroidetes (Rikenellaceae family, Proteiniphilum genus) increased with increasing TS, while Chloroflexi (all the sequences were affiliated to the Anaerolineaceae family) apparently decreased. The abundance of Firmicutes, the third mayor phylum, remained constant. Methanosarcina dominated the archaeal communities. Its relative abundance correlated positively with increasing TS. A similar study was carried out by Han et al. (2017). The authors analyzed six full-scale anaerobic digesters, three of which operated under “wet” conditions (TS ≤ 10%), and three were run under “semi-dry” conditions (10% ≤ TS ≤ 20%). As in the former study, the removal efficiency of volatile solids (VS) was almost twice as high in the wet digesters as in the semi-dry digesters. The bacterial communities were distinctly characterized by the families Porphyromonadaceae, Sphingobacteriaceae and Syntrophomonadaceae in the wet digesters and by Clostridiaceae, Patulibacteraceae, Pseudonocardiaceae, Lachnospiraceae, Rikenellaceae in the semi-dry digesters. Surprisingly, Methanosarcina (Metanosarcinaceae family) represented the dominant methanogen in the study by Yi, while the Methanobacteriaceae (wet) and Methanomicrobiaceae (semi-dry) families were the dominant taxa under wet and semi-dry conditions, respectively. Another important operational parameter is the OLR, which at high values can lead to the failure of digesters. Li et al. (2016b) studied the microbial community in response to OLR disturbances in a mesophilic CSTR reactor treating FW. Overloading resulted in a proliferation of acidogenic bacteria, which rapidly adapted to the increased OLR, while the abundance of hydrogenotrophic methanogens decreased. As a consequence, the increase of homoacetogens, as alternative hydrogenotrophs for converting excessive H2 to acetate, was induced. Methanothrix. also failed to degrade the excessive acetate. This metabolic imbalance finally led to process deterioration. Interestingly though, the digester gradually returned to its original performance if the OLR was readjusted to a sustainable level.
Food waste has also been co-digested with several classes of manure. Zhang et al. (2017) used a three-stage (high-solids hydrolysis, acidogenesis and wet methanogenesis) anaerobic reactor for the co-digestion with horse manure. As expected, different microbial communities in terms of hydrolyzing bacteria, acidogenic bacteria and methanogenic archaea were selectively enriched in the three separate chambers. The dominant genera, present at abundances from 3 to 10%, were the following: Aminobacterium, Clostridium, Proteiniphilum, Saccharofermentans, Eubacterium, Syntrophomonas, Petrimonas and Thermoflavimicrobium. The individual proportions of these genera accounted for the main differences in the bacterial communities of the three digesters. Moreover, the abundance of the methanogenic archaea (i.e. Methanosarcina, Methanobacterium, Methanosaeta) was increased by 0.8–1.28 times compared with the controls. Dennehy et al. (2017) studied the effect of HRTs in the co-digestion of food waste with pig manure. Digester HRT was progressively decreased from 21 to 15 and finally to 10.5 days, which resulted in a decrease in the specific methane yield, the volatile solids removal, and the accumulation of butyric acid. A shift in the acidogenic bacterial population was observed. The increase in the relative abundance of Clocamonaceae and Spirochatetes bacteria, both syntrophic VFA oxidizers, suggests that the VFA oxidation plays a key role in digester operation at low HRTs.
From the studies discussed above can be concluded that the substrate constitutes the main determining factor for the composition of the microbial populations that inhabit the analyzed digesters. In general, the most abundant bacterial phyla are Bacteroidetes, Firmicutes, Proteobacteria and, to a lesser extent, Chloroflexi and Actinobacteria. The metabolic information obtained at the phylum level is very scarce however, and it would be necessary to increase the phylogenetic resolution down to the genus level to gain practically relevant information. Unfortunately, it does not seem to be possible to establish a common bacterial core or sub-population. Only some of the non-specialized genera, such as Clostridium, whose different species participate in carbohydrate or protein hydrolysis and in the fermentation of sugars and amino acids, are detected in all reactors. In relation to the methanogenic species, a tendency could repeatedly be observed: (i) Methanosaeta and Methanosarcina were the predominant methane-producing archaea during the AD of waste-activated sludge; (ii) many independent studies point out that for agricultural waste (i.e. crop straw or livestock manures) the hydrogenotrophic pathway (carried out by, e.g. Methanoculleus or/and Methanobacterium) is the main route of methanogenesis.
Finally, high temperatures, i.e. from 55 to 65 °C, are more effective than 35–37 °C in producing methane, allow higher organic loading rates and can achieve a higher reduction of pathogens. Because of that, thermophilic conditions have been applied for several feedstocks, either used alone or in co-digestion. (e.g.: Rademacher et al. 2012; Sundberg et al. 2013; Li et al. 2015c; Jang et al. 2016; Chen and Chang 2017). Some general trends can be drawn from the studies carried-out at thermophilic temperatures: (1) The process temperature governs the microbial community composition. In general, when comparing mesophilic and thermophilic digestion, the mesophilic reactors exhibit greater microbial richness and evenness than the thermophilic reactors. (2) The Firmicutes are the most abundant phylum from the domain Bacteria. The Clostridia class and Clostridium genus are the most prevalent taxa from this phylum. (3) Strong positive correlation with operation temperature has been observed for the phylum Thermotogae. Within this phylum, Petrotoga seems to be the most significant genus. (4) Methanothermobacter (family Methanobacteriaceae) is the most prevalent methanogenic archaeal genus. (5) Methanosarcina, but no Methanosaeta, is the only acetoclastic methanogen detected. (6) All the data suggest that the methanogenesis at high temperature is mainly driven by the hydrogenotrophic pathway. Syntrophic acetate oxidation coupled with hydrogenotrophic methanogenesis also occurs under these conditions.
AD is commonly used for the treatment of diverse types of waste, both liquid (e.g. industrial wastewater) and solid (e.g. domestic or agricultural waste for biogas production). Unfortunately, it is not possible to define a common bacterial core population that develops under different substrates and reactor configurations. For example, in a study using 454 Pyrosequencing, Etchebehere et al. (2016) analyzed the microbiota of 20 hydrogen-producing lab-scale bioreactors operated in four Latin-American countries. As expected for hydrogen-producing reactors, the phylum Firmicutes predominated in most of the samples, while the phyla Thermotogae and Proteobacteria were prevalent in a small number of samples. However, the results showed notably uneven communities, heavily influenced by the inocula, reactor configurations, and substrates. For these reasons it is impossible to define a core population down to class level, which would be mandatory to address questions about the metabolic function of the microorganisms in the system. The conclusion is not very optimistic: although it is possible to gain a general idea about the microbial composition of a bioreactor based on the published results, when working with anaerobic systems it is necessary to analyze the biomass of each individual reactor.
5 Aerobic wastewater treatment
5.1 The activated sludge process
The activated sludge (AS) process is the most prominent and representative aerobic suspended growth treatment used for organic matter stabilization. Since the 1920s, AS has been extensively applied for the depletion of organic matter (BOD), nitrogen (nitrification) and phosphorous from domestic and, sometimes industrial, wastewater, establishing the AS process as one of the world’s most widely applied biotechnological processes.
The AS process is an excellent model for microbial ecology studies because its operational conditions are controlled and stable over time. A comprehensive understanding of the microbial ecology of the communities inhabiting AS reactors is critical to improving their performance and for predicting responses to unexpected environmental changes (Daims et al. 2006). For these reasons, many microbiological studies were performed using classical microbial methodologies (isolation and characterization of isolated strains). With the emergence of molecular biology techniques (FISH, DGGE, clone libraries) applied to environmental samples in the 90s, knowledge about the underlying microbiology was significantly increased. However, the low sequencing depth of the PCR-cloning approach only allowed for a superficial view of the dominant members of a microbial community. The application of the high-throughput NGS technologies has allowed researchers to deepen the knowledge of the microbiology of the AS to levels that were barely conceivable less than a decade ago.
Between the years 2011 and 2013, Ye et al., from the University of Hong Kong, published a series of outstanding papers on the microbiology of activated sludge wastewater treatment plants (AS-WWTP) analyzed by 454 high-throughput pyrosequencing. In the first of these frequently cited works, Ye and Zhang (2011) studied the presence of pathogenic bacteria in 14 municipal wastewater treatment plants located in China, the U.S.A., Canada, and Singapore. Overall, the percentage of the sequences closely related to known pathogenic bacteria sequences was about 0.16% of the total sequences. Aeromonas (A. veronii, A. hydrophila), and Clostridium (C. perfringens) were the most abundant potentially pathogenic bacteria. The same treatment plants were then analyzed (Guo and Zhang 2012) to identify and quantify bacteria involved in two problems commonly associated with activated sludge systems, i.e. bulking and foaming. The most abundant and frequent groups of bulking and foaming bacteria (BFB) detected in typical municipal wastewater treatment plants were Nostocoida limicola I and II, Mycobacterium fortuitum, Type 1863, and Microthrix parvicella.
In another outstanding article (Zhang et al. 2012), the authors described in depth the bacterial diversity of these AS-WWTP. The study revealed that multiple samples shared a bacterial core population, which included the two commonly detected genera Zoogloea and Dechloromonas, three genera that are less frequently encountered (i.e., Prosthecobacter, Caldilinea and Tricoccus), and three genera which were not well described at the time (i.e., Gp4 and Gp6 in Acidobacteria and Subdivision3 genera incertae sedis of Verrucomicrobia). In addition, certain unique bacterial populations were detected in each sample, revealing geographical differences among the WWTP from Asia and North America. The next step consisted in the analysis of the bacterial communities inhabiting the sequential stages of a full-scale WWTP: influent, activated sludge reactor, effluent and anaerobic sludge digestor (Ye and Zhang 2013). The dominant classes in all four cases were Alphaproteobacteria, Thermotogae, Deltaproteobacteria, and Gammaproteobacteria. High abundances of sequences affiliated to the genera Mycobacterium and Vibrio, both of which are known to contain pathogenic species, were retrieved from the effluent. Interesting enough, at the order level, the five most dominant taxa in the activated sludge samples were Planctomycetales, Actinomycetales, Rhizobiales, Caldilineales, and Sphingobacteriales, differing significantly from those detected in the influent samples, i.e., Desulfobacterales, Clostridiales, Desulfovibrionales, Lactobacillales, and Bifidobacteriales. About 67% of the sequences retrieved from the sludge digester were affiliated with the Thermotogales order (genus Thermotoga).
A similar study was published by Wang et al. (2012), focusing exclusively on WWTP in China. The authors detected a core population of 60 bacterial genera, which was commonly shared by the 14 analyzed samples, including Ferruginibacter, Prosthecobacter, Zoogloea, Subdivision 3 genera incertae sedis, Gp4, Gp6, etc. Wastewater characteristics (water temperature, conductivity, pH, and dissolved oxygen content) seemed to have a greater effect on the bacterial community variance (25.7%) than operational parameters (23.9%) or geographic location (14.7%). At that time, Ranasinghe et al. (2012) explored the bacterial diversity on 3 small and 17 large scale AS WWTP by 454-pyrosequencing. Proteobacteria and Bacteroidetes were identified as the major phyla, and Betaproteobacteria and Bacteroidia as the major classes regardless of the scale of the plants. In an attempt to test whether the core bacterial populations found in municipal activated sludge in surveys mentioned above (Wang et al. 2012; Zhang et al. 2012) were shared by industrial activated sludge, Ibarbalz et al. (2013) applied 454 pyrosequencing of 16S rRNA genes to analyze the microbiota of seven systems treating wastewater from several industrial plants and one plant that treated domestic wastewater. They observed that each industrial activated sludge system exhibited a unique bacterial community composition, which was distinct from the common profile of bacterial phyla or classes observed in municipal plants. The wastewater characteristics were likely to be the major determinant that drives bacterial composition at high taxonomic ranks. The differences in the bacterial community structure could partly be explained by the differences in the dissolved oxygen and the pH. Another attempt to determine the possible existence of a core community in AS was performed by Saunders et al. (2016). The authors analyzed the microbial communities in 13 Danish wastewater treatment plants with nutrient removal using Illumina sequencing. The plants contained a core community of 63 abundant genus-level OTUs that made up 68% of the total reads. Interesting enough, the genus Nitrotoga (class Betaproteobacteria) was the most abundant putative nitrite oxidizer in a number of activated sludge plants, which challenges the previous assumption that Nitrospira (phylum Nitrospirae) are the primary nitrite-oxidizers in activated sludge systems with nutrient removal. The 16S rRNA gene sequences retrieved from AS systems deposited on databases in 2014 led Ju et al. (2014) to explore the associations between bacterial communities in AS by correlation-based network analysis. Over 760000 sequences from 50 AS samples from globally distributed municipal/industrial WWTPs were analyzed. The results revealed several statistical correlations in the taxonomic associations and a core of functional bacteria (e.g. nitrogen-related bacteria) widely distributed in the different WWTPs.
In a previously cited article (Guo and Zhang 2012), the authors analyzed the presence of bulking and foaming bacteria, which are always present in typical activated sludge and play certain roles in the degradation of organic matter. Jiang et al. (2016) followed the population dynamics of BFB in a full-scale AS-WWTP over 5 years. The population of BFB showed seasonal variations with higher abundance in winter-spring than summer-autumn. Gordonia sp. was positively correlated with NO2-N and negatively correlated with NO3-N, and Nostocoida limicola II Tetraspharea sp. was negatively correlated with temperature and positively correlated with NH3-N in activated sludge. Wang et al. (2016b) compared total and filamentous bacteria in AS under sludge bulking and normal conditions. Sludge bulking resulted in a decrease in total bacterial numbers and bacterial diversity. With the occurrence of sludge bulking, Actinobacteria and Firmicutes increased sharply, whereas Proteobacteria, which was the predominant phylum under non-bulking conditions, decreased markedly. Eleven types of filamentous bacteria were always present under both conditions. Moreover, in addition to the habitually reported filamentous bacteria (e.g. Candidates M. parvicella and Tetrasphaera), novel filamentous species of Trichococcus might be related to the bulking.
One of the biggest challenges of the molecular biology applied to wastewater treatment is to establish relationships between the physical–chemical and operational conditions of the WWT systems and the microorganisms that inhabit them. In this context, three articles published in 2016 are worth highlighting. Using a 454 Pyrosequencing-based approach, Isazadeh et al. (2016) characterized 39 biomass samples from 8 full-scale and 2 pilot-scale AS-WWTPs. To establish possible relationships between the microbiota and a set of environmental parameters, the following variables were considered: influent wastewater characteristics, treatment process (conventional, oxidation ditch, and sequence batch reactor) and reactor configuration (fully aerobic vs. anoxic/aerobic), reactor sizes (pilot-scale vs. full-scale reactors), chemical stresses defined by ozonation of return activated sludge, season (winter vs. summer), interannual variation, and geographical locations. A core set of phyla were present in most samples, i.e. Proteobacteria (average of 38.5% of the total reads), Bacteroidetes (27.6%), Chloroflexi (9.0%), Acidobacteria (8.3%), TM7 (6.1%), Actinobacteria (4.4%) and Firmicutes (3.7%). Some of the most abundant families were: Flavobacteriaceae, Saprospiraceae, Rhodobacteriaceae, Sphingomonadaceae, Comamonadaceae and Anaerolineaceae. Despite the presence of a core set of families, plant-specific abundant families were also identified. The season did not generate specific trends in the composition of the microbial communities, suggesting a slow seasonal community turnover. Reactor scale or ozonation did not significantly affect the alpha diversity of the microbial community in each reactor. Although, based on the statistical analysis, the authors were unable to identify the variables that determine the composition of the communities, it could be concluded that the size of the plant did not influence the stability of the resident microbial population, which has important design and economic implications.
With a similar goal, Gao et al. (2016) examined, by Illumina sequencing analysis, the microbial community composition of 4 full-scale municipal WWTPs with different configurations, i.e. oxidation ditch, anoxic/aerobic, anaerobic/anoxic/aerobic, and two-stage anaerobic/aerobic-anoxic/aerobic processes. Proteobacteria was the most abundant phylum in all sludge samples, accounting for 44.9–56.0% of the total number of bacterial sequences, followed by Bacteroidetes (15.4–37.4%) and Firmicutes (1.7–5.0%). Other phyla detected at lower abundances were Acidobacteria, Chloroflexi, Verrucomicrobia, Planctomycetes, and Nitrospirae. A total of 166 genera were commonly shared, accounting for 95.0 to 96.4% of the classified sequences. Among them, several genera were abundant in, at least, two samples, such as Zoogloea, Dechloromonas, Thauera, Nitrospira, Arcobacter, Ferruginibacter or Sulfuritalea. COD and pH were strongly linked to the microbial community composition, whereas temperature and influent ammonia exhibited less influence. Although fluctuation of anoxic, oxic, and anaerobic conditions was favorable for promoting the growth of diverse microbial populations, it is surprising that there was no significant correlation between bacterial genera and dissolved oxygen.
With the same aim of investigating the link between microbial community structures and functions in activated sludge, Ibarbalz et al. (2016) analyzed the metagenome from six industrial and six municipal WWTPs. They concluded that wastewater influent is the principal factor in shaping the metagenomic composition of activated sludge. For example, petrochemical WWTPs contained a large number of genomes with hydrocarbon and sulfur metabolism potential, whereas metagenomes from the whey processing WWTP were dominated by lactose-using Propionibacterium genomes, and the textile dyeing WWTP was characterized by genomes belonging to the Planctomycetes-Verrucomicrobia-Chlamydiae superphylum. As a general trend, the taxonomic and functional richness of municipal activated sludge was significantly greater than that of industrial activated sludge.
A number of design configurations have evolved since the early conception of the AS process. Membrane reactors are nowadays one of the most prominent systems and will be discussed in the following section. Here we would like to mention the study carried out by Gonzalez-Martinez et al. (2016) on the microbial community structure of ten different wastewater treatment systems and their influent analyzed by pyrosequencing. Seven of these plants were conventional AS systems, while the other three employed the AB (so-called Adsorption-Belebungsverfahren) process. The A-stage is highly loaded and mainly intended for organic matter removal. While the microbial community structures of both influent and the conventional AS were similar, the bacterial communities of the A-stage were case-specific. The authors identified a core group of genera for all the influents (Clostridium, Pseudomonas), A-stage bioreactors (Dechloromonas, Hydrogenophaga, Rhodoferax, Zoogloea), and conventional AS (Acidobacterium, Chloroflexus, Dechloromonas, Flavobacterium, Fluviicola, Rhodocyclus, Rhodoferax, Sterolibacterium), showing that different geographical locations (The Netherlands and Spain) did not affect the functional bacterial communities.
Some generally observed trends should be highlighted: (1) Multiple AS-WWTPs treating domestic wastewater shared a bacterial core population (e.g. Zoogloea, Dechloromonas, Prosthecobacter, and the uncultured Acidobacteria Gp4 and Gp6; (2) however, unique bacterial population form in WWTPs as a function of certain characteristics of the wastewater, operational parameters and geographical location; (3) as commented previously in relation to anaerobic reactor designs, industrial AS-WWTPs harbor bacterial communities of unique composition, which are different from those observed in municipal plants. The characteristics of the wastewater influent appear to be the principal factor in shaping the bacterial composition of AS; (4) Nostocoida limicola, Mycobacterium fortuitum and candidatus Microthrix parvicella are the most abundant and frequently found bacteria during the bulking and foaming processes. Generally, Actinobacteria and Firmicutes increase and Proteobacteria decrease in abundance during the occurrence of bulking.
In concluding this section, it should be s tated that, because AS is considered as the archetype of aerobic WWTPs, the archaeal communities that inhabit these systems have been scarcely studied. A study of 20 full-scale AS WWTPs in China (Niu et al. 2017) showed that the archaeal communities were dominated by Methanosarcinales (84.6%). A core archaeal population (94.5%) composed of Methanosaeta, Methanosarcina, Methanogenium and Methanobrevibacter was shared among WWTPs. Surprisingly, the archaeal community’s richness and structure were related to the elevation above sea level of the WWTP.
For further information, interested readers can see the mini-review published by Cydzik-Kwiatkowska and Zielinska (2016) on bacterial communities in full-scale aerobic WWTP. The authors discuss the influence of deterministic and stochastic factors in the dynamics of bacterial communities, with special emphasis on the key role of extracellular polymeric substances and microorganisms involved in nutrient removal.
5.2 Improvement of the activated sludge process (MBR, AGS)
Multiple configurations that are based on AS systems have been developed. One of the latest and most promising is the membrane bioreactor (MBR). MBRs combine the membrane separation process (e.g. microfiltration or ultrafiltration) with the AS treatment and exhibit several advantages over the conventional AS process: effluent of high quality, small footprint, higher suspended solids concentration allowing high loading rates, or the easy retrofit and upgrade of old wastewater treatment plants. MBRs are now widely exploited for the treatment of municipal and industrial wastewaters.
Biofouling is one of the major and persistent operational problems in AS-MBR causing the clogging of the filtration membranes. Therefore, it is not surprising that, in one of the pioneering works in applying the NGS techniques to these reactors, Lim et al. (2012) studied the structure of the bacterial community of an MBR and its relationship with biofouling. The authors observed that the microbial composition of the biofilm on the bio-membrane (biocake) was very different from that in the mixed liquor. The genera Enterobacter and Dyella were found to be closely associated with the initial and late biofouling stages, respectively. Moreover, Enterobacter cancerogenus could play an important role in the formation of biofilms by quorum sensing. Fouling-related biofilms have also been studied by Inaba et al. (2017), in this case by the NGS Illumina and confocal microscopy techniques. According to these authors, organic loading rate is the main factor that determines the architecture, as well as the chemical and microbiological composition of the fouled membranes. At low OLR polysaccharides and microbial cells were the main components of the film. In contrast, high OLR resulted in thick biofilms mainly composed of extracellular lipids. Members of the class Gammaproteobacteria constituted the majority (i.e., 57–82%) of the microbiomes for all fouled membrane samples. Under low OLR conditions, the Flavobacteriia and Betaproteobacteria classes were frequently detected. Several dominant OTUs, e.g. Fluviicola, Aquimonas, Rhenheimera, Brevundimonas, seem to be involved in the biofilm formation, with Alishewanella playing a pivotal role in the development of the polysaccharide-rich biofilms. Under high OLR conditions, the class δ-Proteobacteria emerged as the subsequent major group. The microbiota was then dominated by Pseudomonas, but other predominant OTUs, e.g. Pelobacter, Naumannella or Alkaligenes, all of them potentially being capable of forming biofilms, were also identified.
The influence of several parameters, such as the aeration rates (Ma et al. 2013); the COD/N ratio (Han et al. 2015); as well as the COD, NH4-N, and NaHCO3 concentrations (Tian et al. 2015a) on the microbial community structure and composition of MBR, have been addressed by 454 high-throughput pyrosequencing. In a highly cited article, Ma et al. (2013) analyzed the microbial community under low and high aeration rates. Microbial diversity was decreased under high aeration condition. The relative abundances of Betaproteobacteria and Gammaproteobacteria decreased by 41.5% and 66.6%, consistent with the observed membrane fouling mitigation during a reactor operation. The nitrifyers Nitrospira and Nitrosomonas were the dominant phylogenetic groups at the genus level in both reactors. Han et al. (2015) observed severe membrane fouling when a high COD/N ratio was applied compared with the MBR fed with low COD/N ratio wastewater. Higher COD/N ratios favored the enrichment of the phylum Bacteroidetes, which is considered as potentially EPS-producing, and the denitrifying genera Azospira, Thauera and Zoogloea. Betaproteobacteria, Gammaproteobacteria, Alphaproteobacteria, Bacteroidetes, and Actinobacteria formed the major group found in a comparable study by Tian et al. (2015a). A large number of unclassified bacterial sequences were also detected in the biofilm, suggesting a wide variety of uncharacterized species in MB reactors. The COD and NaHCO3 concentrations in the influent promoted the growth of denitrification-related species, such as Dokdonella, Azospira, Hydrogenophaga, Rhodocyclaceae (sic), and Thauera. The presence of aerobic denitrifiers, like Comamonas, Enterobacter, and Aeromonas show that MBR could simultaneously perform aerobic as well as anoxic denitrification, particularly during the treatment of sewage with low ammonia content.
MBR systems provide a clean effluent compared with conventional activated sludge processes, so MBR effluents are used for irrigation. However, one of the primary concerns on its reuse is the presence of pathogenic bacteria. In a study carried out with restaurant wastewater, Ma et al. (2015a) observed that the typical fecal indicator bacteria provide only a rough estimation of the potentially pathogenic bacteria. The dominant potential pathogens in AS and treated wastewater were affiliated to the genera of Legionella, Clostridium and Mycobacterium. MBR treatment showed good removal of pathogenic bacteria: Arcobacter was decreased by six orders of magnitude, while Aeromonas, Enterobacter, Enterococcus, and Pseudomonas were not detected in the treated wastewater. With a similar objective, Harb and Hong (2017) examined two MBR (full and lab-scale) systems treating municipal wastewater using microfiltration polymeric membranes. High pathogens removal was performed by both MBRs, although the effluents still contained some genera associated with opportunistic pathogens, mainly Pseudomonas and Acinetobacter.
In the second half of the first decade of the 21st century, several studies dedicated to a new development in wastewater treatment technology were published: the aerobic granular sludge (AGS) predominantly used in SBR. AGS overcomes the principal limitations of the activated sludge process with flocculent biomass. The process is characterized by an excellent settling ability, allowing for a high concentration of biomass and sustaining high organic loads, while being efficient for the simultaneous removal of organic matter and nitrogen.
AGS is usually developed from flocculent activated sludge, and a number of operating parameters must be optimized for aerobic granulation. High hydraulic selection pressure, resulting in the wash-out of slow settling material is a determining factor for granule formation. The microbial community composition of the suspended and granular phases in an AGS system was analyzed by Szabo et al. (2017). Although the microbiota of the washed-out biomass was similar to the microbiota of the granules, some taxa (e.g. Flavobacterium sp. and Bdellovibrio sp.) were significantly more abundant, while other taxa (e.g. Meganema sp. and Zooglea sp.) showed lower relative abundance in the granules as compared with the washed-out biomass. Anaerobic granular sludge can be easily obtained from UASB or EGSB reactors. Since aerobic granulation requires a start-up period of up to several months, the conversion of anaerobic into aerobic granules could be a reasonable solution. Sun et al. (2017) have recently demonstrated that anaerobic granular sludge could be successfully transformed into AGS in a continuous up-flow reactor in 45 days. A shift in the bacterial community took place during the process. Bacterial sequences affiliated with the Comamonadaceae, Xanthomonadaceae, Rhodocyclaceae, Moraxellaceae, and Nitrosomonadaceae families played important roles in maintaining the structure and function of the aerobic granules. Moreover, obvious differences between the outer and the inner core of the granules were observed. The relative abundance of Proteobacteria and Bacteroidetes in the outer shell was significantly higher than that in the inner core. Other phyla such a GN04, Chloroflexi, Thermotogae, and Spirochaetes exhibited a higher abundance in the inner core relative to the outer shell. The bacterial communities in both inner and outer layers of the granules were largely different from that of the seeding anaerobic granules. Fan et al. (2018) focused their attention on the aerobic granulation in nitrifying SBR. Minor genera in the seed sludge, e.g., Arcobacter, Aeromonas, Flavobacterium and Acinetobacter, became the dominant genera in the AGS. The ability of these genera to secrete EPS, fundamental for granule formation, could be the reason for their success, showing that genera with low abundance in the seed sludge can be important for aerobic granulation. On the other hand, dominant phyla (e.g., Chloroflexi and Nitrospirae) and classes (e.g., Gammaproteobacteria, Sphingobacteriia, Acidimicrobiia and Clostridia) in the seed sludge were found in lower amounts or even disappeared in AGS.
The ability of AGS for the removal of xenobiotic compounds and the microbial communities involved has been analyzed in several studies. Wang et al. (2016d) studied the removal rates of five types of pharmaceuticals and personal care products. Proteobacteria were the most dominant bacterial phylum (more than 50% of the total bacterial sequences). Some phyla, such as Chloroflexi and Planctomycetes, were gradually enriched, whereas others, such as Bacteroidetes and Actinobacteria, decreased in number or disappeared throughout the reactor operation. The authors speculate that Zoogloea, Tolumonas, Arcobacter, Terrimonas and Singulisphaera can degrade antibacterial and anti-inflammatory compounds. The efficient removal of bisphenol was obtained by Cydzik-Kwiatkowska et al. (2017). The authors found that aerobic genera, such as Aquimonas and Pseudoxanthomonas, and anoxic and anaerobic genera, including Thauera and Azoarcus, predominated in the granules. Moreover, based on correlations between the abundances of Sphingomonas sp., Pusillimonas sp., Methylobacillus sp., and Nitrosospira sp. and the influent bisphenol concentrations, it was suggested that bisphenol was both co-metabolized and directly biodegraded. Jiang et al. (2017) studied the formation of AGS in the presence of aniline as an example of toxic aromatic pollutants. Proteobacteria were the very predominant phylum. Pseudomonas, followed by Comamonas, were responsible for aniline biodegradation.
A few trends can be pointed out: (1) the microbial composition of the membrane biofilm in MBRs is different from that in the liquid phase. The OLR and other operational parameters affect the composition of the microbial communities, although there is not enough information available to reliably assign specific genera to particular conditions; (2) similar to the MBR reactors, the microbial communities in a biofilm-like AGS and those found in the inoculum, washed-out biomass or inner/outer shell of the granules are different. Microorganisms that release EPS, like e.g. Flavobacterium, or filament-shaped bacteria, like the members of the phylum Chloroflexi, are generally abundant in the AGS. Further efforts are necessary to continue deepening the knowledge about the microbiota of both, MBRs and AGS.
We would like to finish this section with the discussion of one particular type of emerging pollutant, which affects the functioning of WWTP: the nanoparticles (NPs). NPs are particles between 1 and 100 nm in size with a surrounding interfacial layer consisting of ionic, inorganic and organic molecules. More than 1300 nanotechnology-derived products have already entered the market: e.g. Silver nanoparticles (Ag-NPs) which can be found in personal care products, laundry additives, clothes and paintings (Yang et al. 2014a); NiO NPs are extensively utilized in lithium-ion batteries, electro-chromic films, and light-emitting diodes, as catalyst or diesel–fuel additive (Wang et al. 2017c). More than 300 nanoparticle products are used in the agrifood industry. With the increasing production and application of NPs, these particles can be released into water, wastes and wastewater, entering the environment and WWTPs. The rapid increase in their use is a concern due to the possible long-term consequences of chronic exposure to nanoparticles, which are still unknown. Moreover, since NPs, like many heavy metals, can be toxic to bacteria, their effects on the activity and viability of the bacterial populations of WWTPs have been studied in recent years.
Several studies related to this subject have been conducted in AS. Ag-NPs hurt organic matter oxidation and nitrification. Because the bacterial susceptibility is different for each group of microorganisms, nitrification was more severely inhibited than organic matter removal (Jeong et al. 2014). In a similar study with oxide nanoparticles (ZnO NPs), Wang et al. (2016d) observed that polyphosphate-accumulating organisms (PAOs), i.e. Betaproteobacteria, were negatively affected in comparison with glycogen-accumulating organisms (GAOs) affiliated to the classes Alphaproteobacteria and Gammaproteobacteria. In both studies, a negative effect of NPs on nutrient removal could be detected. Silver, zinc and titanium NPs are capable of decreasing the synthesis of extracellular polymeric substances (EPS) and therefore floc formation (Eduok et al. 2015). In a set of related studies carried out with a sequencing batch reactor (SBR), Wang et al. (2017a, b) found that CuO NPs affected the richness, diversity and composition of the sludge microbiota. They reported a significantly increased abundance of Comamonas, a slight increase in Zoogloea, and a notable decrease in Flavobacterium. The shifts of the dominant groups seem to be related to the effect of NPs on the periplasmatic membrane and, again, the EPS production. Several studies have been conducted with titanium dioxide nanoparticles (TiO2 NPs) in different drinking- (Liu et al. 2016) and wastewater (Li et al. 2016a) treatment plants. The antibacterial activity of these NPs caused a lower diversity and evenness in the microbiota of the exposed activated sludge as compared with the control samples.
In general, two clear trends can be drawn from the use of NPs on WWTP: (1) the NPs antibacterial activity is well established; and (2) the sensitivity to the nanoparticles varies with the phylogenetic affiliation of the microorganisms, where selective inhibitory effects can be assigned at phylum or, even, the genus level. For example, in biological activated carbon filters, Nitrospira and Betaproteobacteria classes decreased under TiO2 NP treatment, whereas Bacilli and Gammaproteobacteria classes increased (Liu et al. 2016). At the genus level, while no sequences affiliated to the genera Nitrosomonas, Nitrobacter or Nitrospira could be retrieved, Acidovorax, Rhodoferax, Comamonas and Methanosarcina were identified as nano-tolerant species in activated sludge plants spiked with a mixture of Ag, Zn and Ti NPs (Eduok et al. 2015).
6 Nutrient removal
6.1 Nitrogen removal
The interest in revealing the microorganisms involved in the elimination of nutrients (N and P) is demonstrated by the fact that the first instances of NGS-based approaches for the analysis of microbial communities in wastewater were focused on N and P removal systems. As a result, outstanding studies using the still novel 454-pyrosequencing technique were published in the first years of the last decade, e.g.: the nitrifying communities were studied in a full-scale integrated fixed-film activated sludge reactor (Kim et al. 2011), as well as in a lab-scale nitrifying reactor and a full-scale WWTP (Ye and Zhang 2011); the ammonia-oxidizing microorganisms were analysed in six full-scale wastewater treatment reactors (Zhang et al. 2011); and the microbial community carrying out enhanced biological phosphorus removal (EBPR) was investigated in a full-scale plant (Albertsen et al. 2012).
Nitrogen removal is important to prevent a wide array of public health and environmental impacts, i.e. NH4+ can be oxidized by nitrifying microorganisms, leading to a decrease of the dissolved oxygen content in the receiving waters; inorganic N-compounds contribute to the eutrophication of rivers and lakes; ammonium is toxic to aquatic organisms; nitrate formed by nitrification can cause methemoglobinemia in infants. Kim et al. (2011) analyzed the suspended biomass and the biomass attached to the support material in a fixed-film AS reactor separately. Nitrospira-like nitrite-oxidizing bacterial sequences were retrieved from the suspended biomass, but not from the attached biomass. Sequences affiliated to Nitrobacter were not detected. The authors concluded that the suspended portion of the biomass, rather than the attached portion, played a major role in the nitrification in the WWTP. In a more recent survey, Dong et al. (2016a) investigated the nitrifying bacteria in a similar reactor to that used by Kim and co-workers. The results of Illumina MiSeq amplicon sequencing showed Proteobacteria and Bacteroidetes as the dominant phyla in both biofilm and suspended sludge. With respect to nitrifying bacteria, the results were similar: Nitrosomonas was the dominant ammonia-oxidizing bacteria (AOB), while the dominant nitrite-oxidizing bacterium (NOB) was Nitrospira. Rt-PCR (real-time polymerase chain reaction) showed that the abundance of AOB was higher in the suspended sludge than in the biofilm. It is worth mentioning that they found that, at low temperatures, nitrification was more dependent on attached growth than on suspended growth while the abundance of AOB was also higher in the biofilm than in the suspended sludge.
The analysis of six full-scale WWTP (Zhang et al. 2011) revealed that most of the AOB could be assigned to the Nitrosomonas genus, with N. ureae, N. oligotropha, N. marina, and N. aestuarii as the dominant species. The abundance of AOB in activated sludge was very low. The CGI.1b group, affiliated to the ammonia-oxidizing archaea (AOA), accounted for most of the sequences of AOA amoA genes. Using a nitrifying CSTR operating under stable conditions, Ramirez-Vargas et al. (2015) exclusively retrieved sequences affiliated to Nitrosomonas (relative abundance 11.0%) as the sole AOB and Nitrobacter (9.3%) as the sole NOB. In a work carried out with two rotating biological contactor (RBC) systems, Peng et al. (2015) found that the microbial communities of the biofilms shifted with the time and along the plug flowpath. Nitrifiers, including AOB (i.e. Nitrosomonas), NOB (i.e. Nitrospira), and AOA, increased in number along the flowpath, whereas denitrifiers (Rhodanobacter, Paracoccus, Thauera, and Azoarcus) markedly decreased. AOB prevailed over AOA in all the samples. Likewise, AOB dominated over AOA in a full-scale WWTP (Pan et al. 2018). The use of DNA-based stable isotope probing (DNA-SIP) followed by high-throughput sequencing revealed that Nitrosomonas sp. NP1, N. oligotropha and N. marina were the active AOB, and Nitrososphaera viennensis dominated the active AOA. Fan et al. (2018) found that, during aerobic granulation in an SBR, AOA were gradually washed-out, while AOB and nitrite-oxidizing bacteria were retained in the system.
Classical nitrogen removal processes include the oxidation of ammonium to nitrite as the first step of nitrification. However, the oxidation of NO2− to NO3− is an unnecessary and oxygen-consuming step. For this reason, partial nitrification to nitrite has gained increasing attention in recent years. Diverse studies have concluded that the AOB Nitrosomonas plays the primary role in the establishment and maintenance of nitritation. Members of this genus accounted for 83% of the relative abundance in a sequencing batch reactor (SBR) treating high-strength ammonia wastewater (Chen et al. 2016). Nitrosomonas, together with the AOB k-strategist Nitrosospira, accounted for 40% of the total biomass of partial nitrifyer granular sludge in an SBR (Wang et al. 2016a). High ammonium and low oxygen concentrations contributed to the inhibition of the NOB Nitrospira. In the biomass of a moving bed biofilm reactor (MBBR) using novel carriers with enhanced hydrophilicity and electrophilicity, Liu et al. (2017) reported that, together with Nitrosomonas, Comamonas might also play a vital role in ammonia oxidation.
Compared with the conventional nitrification–denitrification processes, the anaerobic ammonia oxidation (ANAMMOX) is an efficient and cost-saving process for nitrogen removal from wastewater. The relative novelty of the ANAMMOX process, compared with the knowledge that had been accumulated over many years of applying conventional nitrogen removal technology, has led many researchers to closely analyze the underlying microbial communities, taking advantage of the potential of the NGS techniques. Usually, the nitrogen in the wastewater is present in the form of ammoniacal nitrogen (N-NH4+). Because ANAMMOX needs nitrite as an electron acceptor (NO2− + NH4+→ N2), the partial nitritation from ammonia to nitrite is required. Dosta et al. (2015) studied the partial nitritation/ANAMMOX process to treat water rejected from a municipal WWTP in a granular SBR. Microbial characterization revealed, as in the papers cited above, the predominance of Nitrosomonas as the main autotrophic AOB. Planctomycetes accounted for 7% of the global community, with Brocadia spp. (sic) (1.4% of the total abundance) as the main anaerobic ammonium oxidizer detected. The authors determined an N-NH4+: N-NO2−: N-NO3− stoichiometry of 1:1.25:0.14. The deviation to the theoretical stoichiometric ratios could be attributed to the presence of heterotrophic bacteria (mainly members of Chlorobi and Chloroflexi). Similar results have been reported by Wen et al. (2017) during partial nitrification, anammox and denitrification in a sequencing batch biofilm reactor (SBBR) equipped with a fine control of oxygen supply. Here, N. europaea was detected as the predominant AOB. Its number decreased when the concentration of influent ammonia decreased from 2200 to 50 mg L−1. Candidatus Brocadia was the dominant anammox bacterium. It remained relatively stable in abundance regardless of the ammonium concentration. Thauera and Pseudomonas predominated as functional denitrifiers in the system. The biomass of the biofilm in a Moving Bed Biofilm reactor operating at low temperature (13 °C) was investigated by Persson et al. (2017). The anammox bacteria (Candidatus Brocadia) constituted a large fraction of the biomass with fewer AOB (Nitrosomonas, Nitrosospira) and even less NOB (Nitrobacter and Nitrospira) present. Still, NOB had considerable impact on the process performance. The heterotrophic bacterial community was diverse, while the majority of the sequences belonged to Planctomycetes, followed by Chloroflexi and Proteobacteria.
In wastewater with a high nitrate content, the partial denitrification from NO3− to NO2− permits energy-efficient nitrogen removal. Several studies have concluded that the genus Thauera is the denitrifying bacterium responsible for the nitrite accumulation. Thauera accounted for 50–67% of the sequence reads in a denitrifying USB reactor equipped with gas automatic circulation (Cao et al. 2016) and the 67% of total microorganism in an SBR (Du et al. 2016) as the Illumina high-throughput sequencing analysis revealed. The same research group analyzed the performance and a microbial community of a novel DEAMOX (DEnitrifying AMmonium OXidation) process, which couples partial-denitrification with ANAMMOX, for nitrogen removal in SBRs. Again, they concluded that Thauera possibly played a key role in partial denitrification. In a study with two SBR, one of which used acetate as the electron donor (R1) and the other ethanol (R2), Thauera genus was detected to be dominant in both SBRs (accounting for 61.5% in R1 and 45.2% in R2). Different ANAMMOX species were detected, for example, Candidatus Brocadia and Candidatus Kuenenia in R1, but only Candidatus Kuenenia was detected in R2 (Du et al. 2017a). In a similar work (Du et al. 2017b), Thauera genera accounted for 26.3%, while Candidatus Brocadia (1.7%) was the major ANAMMOX-related species.
The complete process of denitrification involves the reduction of NO3− to dinitrogen gas (N2) by anaerobic facultative bacteria that utilize NO3− as electron acceptor (nitrate respiration). Denitrifying bacteria are generally heterotrophic and utilize organic matter as electron donor. A limited number of bacteria are capable of chemolithotrophic denitrification, utilizing inorganic compounds, e.g. reduced sulfur compounds or hydrogen as electron donors for the reduction of nitrate. Denitrification with reduced sulfur compounds can attain the simultaneous removal of N and S contamination in a single-phase system and transform these contaminants into environmentally acceptable forms (N2 gas and sulfate or S0) (Fernandez et al. 2008).
Microbial communities in the denitrification systems may vary greatly according to the electron donor, operational conditions, sampling position, and other factors. In this regard, in a study carried out by Liu et al. (2015a) in an EGSB, changes in salinity caused a shift from the predominating heterotrophic denitrifiers at less than 10 g/L NaCl (e.g. Thauera, Thermomonas, Rhodobacteraceae family) to autotrophic denitrifiers at concentrations higher than 10 g/L NaCl (e.g. Azoarcus, Thiobacillus). Zhou et al. (2017a) compared the microbial communities in autotrophic denitrification reactors using thiosulfate, elemental sulfur, and sulfide electron donors. The well-known Thiobacillus existed abundantly in every system, even under heterotrophic conditions. Besides Thiobacillus, many other genera of denitrifiers were identified. Chlorobaculum, Dechloromonas, and Acinetobacter were the most predominant genera in thiosulfate, elemental sulfur, and sulfide systems, respectively, while Janthinobacterium was the most abundant genus in the heterotrophic reactor with ethanol as electron donor.
Hydrogenotrophic denitrification (HDN) is also regarded as an efficient alternative technology for removing nitrate from wastewater with low organic matter content. The effect of key environmental factors on HDN was investigated by Wang et al. (2015) and Li et al. (2017a). The reduction of nitrate was significantly affected by the pH, temperature, inorganic carbon content, dissolved hydrogen concentration, nitrate loading, and C/N ratio. According to Wang et al. (2015), the phylum Firmicutes, class Clostridia, was most abundant, as determined by 454-pyrosequencing read counts, with Proteiniclasticum, followed by Gemmata, Planctomyces, and Hyphomicrobium, being the predominant genera. Paracoccus (26.1%), Azoarcus (24.8%), Acetoanaerobium (11.4%), Labrenzia (7.4%), and Dysgonomonas (6.0%) were the dominant genera assessed in a comparable study using Illumina sequencing (Li et al. 2017a).
Some evident conclusions can be highlighted from the literature: (1) in the nitrification process (from ammonia to nitrite and from this one to nitrate) the genus Nitrosomonas (e.g. N. oligotropha, N. marina) is always the dominant AOB, while Nitrobacter and Nitrospira share the role of the NOB. In general, the k-strategist Nitrospira prevails over Nitrobacter in biofilm reactors; (2) Thauera is the dominant genus in the denitrification process, playing a key role in partial denitrification (from nitrate to nitrite). However, in autotrophic denitrification reactors fed with S-compounds, the most abundant sequences were affiliated to Thiobacillus, and in the reactors fed with hydrogen as the electron donor, several genera within the Clostridia class were the most abundant; (3) Candidatus Brocadia is the dominant anammox bacterium. The heterotrophic bacterial community in the ANAMMOX process includes mainly members of the Chloroflexi and the Proteobacteria; (4) the microbial community involved in nitrogen removal is independent of the reactor setup used. From all of the above it may be deduced that the most relevant bacteria related to nitrogen removal are well-known and no further sequencing effort should be necessary. However, we cannot rule out the emergence of new biochemical pathways in the nitrogen cycle with an involvement in nitrogen removal. As an example the anaerobic oxidation of methane coupled to denitrification (AOM-D) should be mentioned. Consequently, each researcher has the responsibility to decide whether or not additional sequencing efforts are necessary to expand the knowledge about the system under study.
6.2 Phosphorous removal
Phosphorous must be removed from the WWTP’s discharges to avoid eutrophication. Moreover, phosphorous is a non-renewable resource and its discharge to water bodies constitutes a loss of resources. Enhanced biological phosphorus removal (EBPR) is considered the most cost-effective and environmentally friendly technology to reduce the phosphorous content in WWTP effluents. For this reason, EBPR is widely used for the removal and recovery of phosphorus from both domestic and industrial wastewater throughout the world. Due to the interest in the EBPR process, a large body of work exists that is focused on the identification of the microbiota involved. All of these studies agree on the key role played by Candidatus Accumulibacter phosphatis, a clade formed by Candidatus Accumulibacter-like polyphosphate-accumulating organisms (PAOs) though its relative number depends on the operational and environmental conditions. Dechloromonas-related PAO and the glycogen-accumulating organism (GAO) Candidatus Competibacter sequences are also frequently retrieved from EBPR systems.
In a pioneering article, Albertsen et al. (2012) studied the metagenome of a full-scale EBPR plant by Illumina sequencing. Most of the contigs (partially reconstructed genetic sequences) in the assembled metagenome exhibited low similarity to the genomes available at that time. Only the genome of Candidatus Accumulibacter was closely related to the species present in the metagenome. Applying a quantitative method (FISH), Accumulibacter were determined to account for 4.8% of the total number of bacteria. Similarly, the metagenome of the biomass from a full-scale anaerobic-anoxic–oxic municipal sewage treatment plant was analyzed by Tian et al. (2015b). Proteobacteria, Bacteroidetes, Nitrospirae and Chloroflexi were the predominant phyla in the AS. On the genus level, Nitrospira, Thauera, Dechloromonas and Ignavibacterium were the most prevalent microorganisms. The relative proportion of Candidatus Accumulibacter (1.37%) was several times higher than that reported in other studies of anaerobic-anoxic–oxic systems.
Lv et al. (2014) operated two SBRs in anaerobic-anoxic (A–A) or anaerobic-oxic (A-O) conditions to achieve denitrifying EBPR (A–A reactor) and traditional EBPR (A-O reactor). No differences were observed in terms of phosphorous removal efficiency. However, one Dechloromonas-related OTU was detected as the main OTU in the A–A sludge while Candidatus Accumulibacter dominated in the A-O sludge. The same results were found by Dai et al. (2017) during the enrichment of denitrifying phosphorous removal sludge. Under alternative anaerobic/anoxic operational conditions, the dominant microbial population consisted of Dechloromonas-related bacteria involved in phosphorus removal. Anaerobic/oxic conditions were favorable for the enrichment of Candidatus Accumulibacter-related organisms, also known for their role in the biological removal of phosphorous.
As mentioned above, most of the studies have reported Candidatus Accumulibacter within the phylum Proteobacteria as dominant in reactors that performed EBPR. In a study published by Guo et al. (2018) in an anaerobic-anoxic–oxic (A2O) process using different electron acceptors, DNA-SIP followed by Illumina MiSeq sequencing showed that the PAO organisms were mainly affiliated to the phylum Proteobacteria, with Candidatus Accumulibacter as the major genus using oxygen and nitrate as electron acceptors. Acinetobacter was the most dominant genus using nitrate and nitrite as electron acceptors. The authors concluded that not only anaerobic P removal but also denitrifying P removal are important mechanisms of the EBPR process and deserve further study.
Mao et al. (2016) subjected an EBPR-performing SBR to a short-term pH shock. The treatment caused a bloom of GAOs (accounting for 16% of bacteria), including Candidatus Competibacter phosphatis and Defluviicoccus-related organisms, causing a drastic breakdown in EBPR efficiency. However, the EBPR performance recovered over time and the dominant Candidatus Accumulibacter shifted from Clade IIC to IIA. This distribution of different strains (or ecotypes) of Candidatus Accumulibacter phosphatis into clades is based on a classification using the ppk1 gene coding for a polyphosphate kinase. This approach is also an example of the successful application of a functional gene as a biomarker for the resolution of highly similar microorganisms.
Given the fact that the EBPR process involves aerobic-anaerobic (or anoxic) phases, Keating et al. (2016) described an alternative approach, employing a hybrid sludge bed/fixed-film reactor. The authors studied the phosphate removal during the anaerobic treatment of synthetic sewage wastewater. Efficient removal (up to 78% of influent phosphate) could be observed, mediated through the formation of intracellular polyphosphate granules within the biofilms. The dominant bacterial phyla present in the biomass were Proteobacteria and Firmicutes. The presence of PAOs, such as Rhodocyclus, Chromatiales, Actinobacter, and Acinetobacter, was recorded at low numbers.
Similar to what could be observed for N removal, during the elimination of P through the EBPR process, all studies agree on the key role played by one species: the proteobacterium Candidatus Accumulibacter phosphatis. In treatment plants which, together with the anaerobic-oxic phases of the EBPR include an anoxic stage to eliminate N-compounds, Dechloromonas and other denitrifiers are also detected.
7 Conclusions and perspectives
Due to their low cost and simplicity of use, the next-generation sequencing technologies have become very popular in microbial ecology for studying both natural and engineered ecosystems, like wastewater treatment plants. In case of 16S rDNA amplicon sequencing, all that is needed for analysis via NGS is a whole community genomic DNA extract, e.g. from a soil or sludge sample, and a pair of PCR primers that target the desired region of the 16S gene. Subjecting the resulting PCR products to NGS amplicon sequencing will produce large amounts of sequence data. Illumina sequencing, the most popular of the available NGS methods applied in microbial ecology today, easily yields thousands or millions of sequence reads from a single run. The bottleneck of a study in microbial ecology usually lies in the data processing and analysis step, as the conversion of the raw data and classification as phylogenetic information requires the use of dedicated software and expertise in bioinformatics to apply the appropriate algorithms to the data efficiently. Freely available pipeline programs like Qiime or mothur are making the handling of sequence data considerably easier but still require a thorough understanding of the individual steps of the pipeline and their interconnections to produce meaningful results in the form of taxonomic classification, compositional and structural characterization of the communities under survey and the graphical presentation of the results. As NGS techniques are used in more and more laboratories, the demand for bioinformaticians increases, both as a part of research teams and in the area of algorithm design and software development.
High-throughput amplicon sequencing applied to aerobic and anaerobic waste and wastewater treatment systems has played an important role in revealing the extraordinary microbial complexity of these ecosystems. Still, this approach does not provide all the information many microbial studies could benefit from. Often, the depth of the phylogenetic analysis that can be achieved using 16S rDNA sequences is a limiting factor for the analysis of microbial communities. Many authors present taxonomic data and the discrimination of different microorganisms in the studied systems at the level of phylum, class or order. Unfortunately, this coarse level of resolution provides information of little value, as large and diverse phyla like the Firmicutes or the Proteobacteria are present in most wastewater-related systems, not allowing for conclusions as to their possible function in these environments. Other examples are the orders Clostridiales and Bacteroidales, which typically contain hydrolytic and fermentative bacteria and are, therefore, encountered in anaerobic environments at high abundances.
Another important drawback of the NGS lies in the fact that the gained information refers to the total population present, based on the retrieved sequences of the analyzed sample. However, the taxonomic information does not explain the activity or metabolic function of the microorganisms present. De Vrieze et al. (2018) for example evaluated the microbial communities of 48 full-scale AD plants by amplicon sequencing of the 16S rRNA gene and the 16S rRNA transcripts to compare the composition of the total number of microorganisms to that of the active portion of the population. In brief, the authors observed a clear difference between the profiles of the entire microbiota (DNA-based) and the active subpopulation (RNA-based). In the case of the Archaea, a significantly higher diversity was detected based on the analysis of the DNA sequence reads, whereas on the transcript level the diversity was lower.
The presence of microorganisms in wastewater treating systems often depends on multiple factors, of which the composition and physico-chemical properties of the wastewater are essential. Accordingly, the microbiota present in these reactors can be considered case-specific, depending on the contaminant chemical compounds the water contains. On the other hand, the microorganisms involved in the elimination of nutrients via the well-described processes of nitrification, denitrification, ANAMMOX or EBPR seem to be closely related to each particular process. Aerobic activated sludge systems present an intermediate scenario: These reactors are used for the treatment of urban wastewater where it is often possible to identify a common underlying core population among the resident microbiota.
Table 2 shows the microorganisms predominantly detected in the different wastewater treatment processes, based on the information contained in the research papers discussed in this review. Excepting nitrogen and phosphorous removal, we intend to give a generalized overview, including only the most frequently detected microbes, not an exhaustive survey. Each biological reactor is a highly complex and individual ecosystem, and this review is intended to come up with an idea on the compositional aspects of microbial communities that might be expected, based on the associated metabolic process(es). Each researcher should perform the appropriate experiments to obtain precise information on the actual composition and microbial diversity in the system under study.
Finally, it should be stressed that any taxonomic information gained by amplicon sequencing is of relative importance without the interpretation of the microorganisms’ functional role in the environment studied. In this regard, NGS approaches based on single genes answer the questions ‘who is there?’, ‘how abundant are they?’, or ‘how does the community shift over time or with changing operational conditions?’ Further unanswered questions however remain, for example concerning the identity of the genes that are expressed at the time of sampling or the metabolic and ecological function of the detected microorganisms. Although the omics approaches, mainly due to their higher complexity, are currently not as widespread for the study of microbial communities in wastewater treatment systems, they will probably increase in popularity and play an important role in answering these questions in the near future. The set of omics methods taking place on the three levels of genetic information (DNA, RNA and protein) forms a relatively recent framework of techniques that surpasses amplicon-sequencing methods. Metagenomics operate on the gene level, transcriptomics on the RNA/transcript level and proteomics on the protein/expression level. Together, these methods embrace the entire genetic setup of a sampled microbial community. Here the application of the omics methods opens the much needed possibility to obtain, e.g. taxonomical as well as functional information. Transcriptomics are even more interesting for the elucidation of the metabolic processes taking place in a wastewater treatment system, as the sequence information about the set of expressed genes under certain operational conditions can be studied, eliminating the informational clutter of those genes that are not involved.
A different but powerful approach, when external parameters and compounds such as metabolites are of importance for a given study, can be found in ecological network analysis (ENA), a long established methodology for the analysis of interactions and dependencies between members or species in an environment (Hannon 1973). Lately, researchers have started to use ENA as a tool for studying the dynamics of microbiomes involved in wastewater treatment processes. In this context, data from deep amplicon, metagenome or transcriptome sequencing combined with the detailed recording of operational, environmental or geographical parameters of interest can uncover interdependencies or co-occurrences of microbial OTUs that would otherwise be difficult to detect. Very recently, microbial network analysis has been applied successfully in the investigation of the microbial response to changes in the concentration and composition of substrate in anaerobic digestion (Orellana et al. 2019) and in studying the distribution and exchange of antibiotic resistance genes across a wastewater network of clinical and domestic sources (Quintela-Baluja et al. 2019). Stable isotope Probing (SIP) presents another possibility of investigating the fate and role of chemical compounds and their interaction with the microbial population in an environment. This methodology is based on the use of substrates labeled with heavy isotopes that can be traced when incorporated into the DNA or RNA of microbial cells.
Independent of the chosen experimental and analytical techniques, the close collaboration of several scientific disciplines is necessary to further advance our understanding of bioreactors as functional ecosystems. NGS requires the results gained by the application of conventional microbiological methods, culture-based and directly assessing microbial metabolism and function, as the groundwork on which to map the already vast and ever-growing body of phylogenetic information. Probably the greatest challenge lies in answering the question posed by the engineers to the microbiologists: ‘How can the knowledge of the microbiota present in my reactor help me optimize its operational parameters and efficiency?’ Some very satisfactory examples are the identification of filamentous bacteria (BFB) to prevent bulking, the optimization and extension to industrial scale of the ANAMMOX process, or the recent development of the aerobic granular sludge. The future perspective is clear: The combined effort of microbiologists, engineers and bioinformatics will be required to describe and analyze microbial communities in industrially relevant systems like wastewater treatment plans and, in turn, allow for the optimization and further development of more efficient strategies as environmental challenges become bigger in today’s societies.
References
Albertsen M, Hansen LBS, Saunders AM et al (2012) A metagenome of a full-scale microbial community carrying out enhanced biological phosphorus removal. ISME J 6:1094–1106
Awolusi OO, Nasr M, Kumari S, Bux F (2018) Principal component analysis for interaction of nitrifiers and wastewater environments at a full-scale activated sludge plant. Int J Environ Sci Technol 15:1477–1490. https://doi.org/10.1007/s13762-017-1506-9
Bailon-Salas AM, Medrano-Roldan H, Ordaz-Diaz SV-CLA et al (2017) Review of molecular techniques for the identification of bacterial communities in biological effluent treatment facilities at pulp and paper mills. BioResources 12:4384–4409
Bakken LR (1985) Separation and purification of bacteria from soil. Appl Environ Microbiol 49:1482–1487
Berger WH, Parker FL (1970) Diversity of planktonic foraminifera in deep-sea sediments. Science 168:1345–1347
Bokulich NA, Subramanian S, Faith JJ et al (2013) Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10:57–59
Bowers RM, Clum A, Tice H et al (2015) Impact of library preparation protocols and template quantity on the metagenomic reconstruction of a mock microbial community. BMC Genom 16:856. https://doi.org/10.1186/s12864-015-2063-6
Braga JK, Motteran F, Silva EL, Varesche MBA (2015) Evaluation of bacterial community from anaerobic fluidized bed reactor for the removal of linear alkylbenzene sulfonate from laundry wastewater by 454-pyrosequence. Ecol Eng 82:231–240
Bragg L, Stone G, Imelfort M et al (2012) Fast, accurate error-correction of amplicon pyrosequences using Acacia. Nat Methods 9:425–426
Breitwieser FP, Lu J, Salzberg SL (2017) A review of methods and databases for metagenomic classification and assembly. Brief Bioinform. https://doi.org/10.1093/bib/bbx120
Buttigieg PL, Ramette A (2014) A guide to statistical analysis in microbial ecology: a community-focused, living review of multivariate data analyses. FEMS Microbiol Ecol 90:543–550. https://doi.org/10.1111/1574-6941.12437
Cabezas A, de Araujo JC, Callejas C et al (2015) How to use molecular biology tools for the study of the anaerobic digestion process? Rev Environ Sci Biotechnol 14:555–593
Cao SB, Li BK, Du R et al (2016) Nitrite production in a partial denitrifying upflow sludge bed (USB) reactor equipped with gas automatic circulation (GAC). Water Res 90:309–316
Caporaso JG, Bittinger K, Bushman FD et al (2010a) PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267
Caporaso JG, Kuczynski J, Stombaugh J et al (2010b) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Cerrillo M, Morey L, Vinas M, Bonmati A (2016) Assessment of active methanogenic archaea in a methanol-fed upflow anaerobic sludge blanket reactor. Appl Microbiol Biotechnol 100:10137–10146
Chai B, Wang Q, Kulam-Syed-Mohideen AS et al (2006) The ribosomal database project-II: the RDP pipeline. In: ASM general meeting
Chao A (1984) Nonparametric estimation of the number of classes in a population. Scand J Stat 11:265–270
Chen HB, Chang S (2017) Impact of temperatures on microbial community structures of sewage sludge biological hydrolysis. Bioresour Technol 245:502–510
Chen WJ, Dai XH, Cao DW et al (2016) Performance and microbial ecology of a nitritation sequencing batch reactor treating high-strength ammonia wastewater. Sci Rep 6:35693
Cho K, Shin SG, Kim W et al (2017) Microbial community shifts in a farm-scale anaerobic digester treating swine waste: correlations between bacteria communities associated with hydrogenotrophic methanogens and environmental conditions. Sci Total Environ 601:167–176
Clark K, Karsch-Mizrachi I, Lipman DJ et al (2016) GenBank. Nucleic Acids Res 44:D67–D72
Cole JR, Wang Q, Fish JA et al (2014) Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42:D633–D642
Cui M-H, Cui D, Liang B et al (2016) Decolorization enhancement by optimizing azo dye loading rate in an anaerobic reactor. RSC Adv 6:49995–50001
Cydzik-Kwiatkowska A, Zielinska M (2016) Bacterial communities in full-scale wastewater treatment systems. World J Microbiol Biotechnol 32:66
Cydzik-Kwiatkowska A, Bernat K, Zielinska M et al (2017) Aerobic granular sludge for bisphenol A (BPA) removal from wastewater. Int Biodeterior Biodegrad 122:1–11
Dai HL, Lu XW, Peng LH et al (2017) Enrichment culture of denitrifying phosphorus removal sludge and its microbial community analysis. Environ Technol 38:2800–2810
Dai RB, Chen XG, Xiang XY et al (2018) Understanding azo dye anaerobic bio-decolorization with artificial redox mediator supplement: considering the methane production. Bioresour Technol 249:799–808
Daims H, Taylor MW, Wagner M (2006) Wastewater treatment: a model system for microbial ecology. Trends Biotechnol 24:483–489
Delforno TP, Lacerda GV, Sierra-Garcia IN et al (2017a) Metagenomic analysis of the microbiome in three different bioreactor configurations applied to commercial laundry wastewater treatment. Sci Total Environ 587:389–398
Delforno TP, Noronha GVLMF, Sakamoto IK et al (2017b) Microbial diversity of a full-scale UASB reactor applied to poultry slaughterhouse wastewater treatment: integration of 16S rRNA gene amplicon and shotgun metagenomic sequencing. Microbiol Open 6:e443
Dennehy C, Lawlor PG, Gardiner GE et al (2017) Process stability and microbial community composition in pig manure and food waste anaerobic co-digesters operated at low HRTs. Front Environ Sci Eng 11:4
DeSantis TZ, Hugenholtz P, Larsen N et al (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072
Dhillon A, Lever M, Lloyd KG et al (2005) Methanogen diversity evidenced by molecular characterization of methyl coenzyme M reductase A (mcrA) genes in hydrothermal sediments of the Guaymas Basin. Appl Environ Microbiol 71:4592–4601
Dong B, Tan J, Yang Y et al (2016a) Linking nitrification characteristic and microbial community structures in integrated fixed film activated sludge reactor by high-throughput sequencing. Water Sci Technol 74:354–1364
Dong H, Dong H, Zhang Z et al (2016b) Microbial community dynamics in an anaerobic biofilm reactor treating heavy oil refinery wastewater. RSC Adv 6:107442–107451
dos Furtado AL, Casper P (2000) Different methods for extracting bacteria from freshwater sediment and a simple method to measure bacterial production in sediment samples. J Microbiol Methods 41:249–257
Dosta J, Vila J, Sancho I et al (2015) Two-step partial nitritation/Anammox process in granulation reactors: start-up operation and microbial characterization. J Environ Manag 164:196–205
Dray S, Dufour AB (2007) The ade4 package: implementing the duality diagram for ecologists. J Stat Softw 22:1–20. https://doi.org/10.18637/jss.v022.i04
Du R, Peng YZ, Cao SB et al (2016) Mechanisms and microbial structure of partial denitrification with high nitrite accumulation. Appl Microbiol Biotechnol 100:2011–2021
Du R, Cao SB, Li BK et al (2017a) Performance and microbial community analysis of a novel DEAMOX based on partial-denitrification and anammox treating ammonia and nitrate wastewaters. Water Res 108:46–56
Du R, Cao SB, Li BK et al (2017b) Simultaneous domestic wastewater and nitrate sewage treatment by DEnitrifying AMmonium OXidation (DEAMOX) in sequencing batch reactor. Chemosphere 174:399–407
Ducey TF, Hunt PG (2013) Microbial community analysis of swine wastewater anaerobic lagoons by next-generation DNA sequencing. Anaerobe 21:50–57
Ecologist TM (2016) NGS field guide: overview. https://www.molecularecologist.com/next-gen-fieldguide-2016/. Accessed 31 Dec 2018
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998
Edgar RC, Haas BJ, Clemente JC et al (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2195–2200
Eduok S, Hendry C, Ferguson R et al (2015) Insights into the effect of mixed engineered nanoparticles on activated sludge performance. FEMS Microbiol Ecol 91:fiv082
Efron B, Stein C (1981) The Jackknife estimate of variance. Ann Stat 9:586–596
Etchebehere C, Castello E, Wenzel J et al (2016) Microbial communities from 20 different hydrogen-producing reactors studied by 454 pyrosequencing. Appl Microbiol Biotechnol 100:3371–3384
Fan XY, Gao JF, Pan KL et al (2018) Shifts in bacterial community composition and abundance of nitrifiers during aerobic granulation in two nitrifying sequencing batch reactors. Bioresour Technol 251:99–107
Fernandez N, Sierra-Alvarez R, Field JA et al (2008) Microbial community dinamics in a chemolitotrophic denitrification reactor inoculated with methanogenic granular sludge. Chemosphere 70:462–474
FitzGerald JA, Allen E, Wall DM et al (2015) Methanosarcina play an important role in anaerobic co-digestion of the seaweed Ulva lactuca: taxonomy and predicted metabolism of functional microbial communities. PLoS ONE 10:e0142603
Gao P, Xu WL, Sontag P et al (2016) Correlating microbial community compositions with environmental factors in activated sludge from four full-scale municipal wastewater treatment plants in Shanghai, China. Appl Microbiol Biotechnol 100:4663–4673
Gomes BC, Adorno MAT, Okada DY et al (2014) Analysis of a microbial community associated with polychlorinated biphenyl degradation in anaerobic batch reactors. Biodegradation 25:797–810
Gonzalez-Martinez A, Rodriguez-Sanchez A, Lotti T et al (2016) Comparison of bacterial communities of conventional and A-stage activated sludge systems. Sci Rep 6:18786
Good IJ (1953) The population frequencies of species and the estimation of population parameters. Biometrika 40:237–264
Guo F, Zhang T (2012) Profiling bulking and foaming bacteria in activated sludge by high throughput sequencing. Water Res 46:2772–2782
Guo JH, Peng YZ, Ni BJ et al (2015) Dissecting microbial community structure and methane-producing pathways of a full-scale anaerobic reactor digesting activated sludge from wastewater treatment by metagenomic sequencing. Microb Cell Fact 14:33
Guo Y, Zeng W, Li N, Peng YZ (2018) Effect of electron acceptor on community structures of denitrifying polyphosphate accumulating organisms in anaerobic-anoxic-oxic (A(2)O) process using DNA based stable-isotope probing (DNA-SIP). Chem Eng J 334:2039–2049
Han XM, Wang ZW, Ma JX et al (2015) Membrane bioreactors fed with different COD/N ratio wastewater: impacts on microbial community, microbial products, and membrane fouling. Environ Sci Pollut Res 22:11436–11445
Han G, Shin SG, Lee J et al (2017) A comparative study on the process efficiencies and microbial community structures of six full-scale wet and semi-dry anaerobic digesters treating food wastes. Bioresour Technol 245:869–875
Hannon B (1973) The structure of ecosystems. J Theor Biol 41:535–546
Harb M, Hong PY (2017) Molecular-based detection of potentially pathogenic bacteria in membrane bioreactor (MBR) systems treating municipal wastewater: a case study. Environ Sci Pollut Res 24:5370–5380
Hartman AL, Riddle S, McPhillips T, Eisen BLJA (2010) Introducing WATERS: a workflow for the alignment, taxonomy, and ecology of ribosomal sequences. BMC Bioinform 11:317
Hill MO (1973) Diversity and evenness: a unifying notation and its consequences. Ecology 54:427–432
Hjelmsø MH, Hellmer M et al (2017) Evaluation of methods for the concentration and extraction of viruses from sewage in the context of metagenomic sequencing. PLoS ONE. https://doi.org/10.1371/journal.pone.0170199
Ibarbalz FM, Figuerola ELM, Erijman L (2013) Industrial activated sludge exhibit unique bacterial community composition at high taxonomic ranks. Water Res 47:3854–3864
Ibarbalz FM, Orellana E, Figuerola ELM, Erijman L (2016) Shotgun metagenomic profiles have a high capacity to discriminate samples of activated sludge according to wastewater type. Appl Environ Microbiol 82:5186–5196
Inaba T, Hori T, Aizawa H et al (2017) Architecture, component, and microbiome of biofilm involved in the fouling of membrane bioreactors. NPJ Biofilms Microbiomes 3:UNSP 5
Isazadeh S, Jauffur S, Frigon D (2016) Bacterial community assembly in activated sludge: mapping beta diversity across environmental variables. MicrobiologyOpen 5:1050–1060
Jabari L, Gannoun H, Khelifi E et al (2016) Bacterial ecology of abattoir wastewater treated by an anaerobic digestor. Braz J Microbiol 47:73–84
Jang HM, Ha JH, Park JM et al (2015) Comprehensive microbial analysis of combined mesophilic anaerobic-thermophilic aerobic process treating high-strength food wastewater. Water Res 73:291–303
Jang HM, Ha JH, Kim MS et al (2016) Effect of increased load of high-strength food wastewater in thermophilic and mesophilic anaerobic co-digestion of waste activated sludge on bacterial community structure. Water Res 99:140–148
Jeong E, Im WT, Kim DH et al (2014) Different susceptibilities of bacterial community to silver nanoparticles in wastewater treatment systems. J Environ Sci Health Part A Toxic Hazard Subst Environ Eng 49:685–693
Jiang XT, Guo F, Zhang T (2016) Population dynamics of bulking and foaming bacteria in a full-scale wastewater treatment plant over five years. Sci Rep 6:24180
Jiang Y, Wei L, Yang K et al (2017) Rapid formation of aniline-degrading aerobic granular sludge and investigation of its microbial community succession. J Clean Prod 166:1235–1243
Jost L (2006) Entropy and diversity. Oikos 113:363–375
Ju F, Xia Y, Guo F et al (2014) Taxonomic relatedness shapes bacterial assembly in activated sludge of globally distributed wastewater treatment plants. Environ Microbiol 16:2421–2432
Jünemann S, Kleinbölting N, Jaenicke S et al (2017) Bioinformatics for NGS-based metagenomics and the application to biogas research. J Biotechnol 261:10–23. https://doi.org/10.1016/j.jbiotec.2017.08.012
Keating C, Chin JP, Hughes D et al (2016) Biological phosphorus removal during high-rate, low-temperature, anaerobic digestion of wastewater. Front Microbiol 7:226
Kembel SW, Wu M, Eisen JA, Green JL (2012) Incorporating 16S gene copy number information improves estimates of microbial diversity and abundance. PLoS ONE 8:e1002743
Kim TS, Kim HS, Kwon S, Park HD (2011) Nitrifying bacterial community structure of a full-scale integrated fixed-film activated sludge process as investigated by pyrosequencing. J Microbiol Biotechnol 21:293–298
Kim S, Bae J, Choi O et al (2014) A pilot scale two-stage anaerobic digester treating (FWL): performance and microbial structure analysis using pyrosequencing. Process Biochem 49:301–308
Kim D-H, Lee M-K, Hwang Y et al (2016) Microbial granulation for lactic acid production. Biotechnol Bioeng 113:101–111
Kindt R, Coe R (2005) Tree diversity analysis. A manual and software for common statistical methods for ecological and biodiversity studies. World Agroforestry Centre (ICRAF), Nairobi (Kenya)
Krober M, Bekel T, Diaz NN et al (2009) Phylogenetic characterization of a biogas plant microbial community integrating clone library 16S-rDNA sequences and metagenome sequence data obtained by 454-pyrosequencing. J Biotechnol 142:38–49
Kumar S, Carlsen T, Mevik B-H et al (2011) CLOTU: an online pipeline for processing and clustering of 454 amplicon reads into OTUs followed by taxonomic annotation. BMC Bioinform 12:182. https://doi.org/10.1186/1471-2105-12-182
Lemarchand K, Berthiaume F, Maynard C et al (2005) Optimization of microbial DNA extraction and purification from raw wastewater samples for downstream pathogen detection by microarrays. J Microbiol Methods 63:115–126. https://doi.org/10.1016/j.mimet.2005.02.021
Lettinga G (2014) My anaerobic sustainability story. Lettinga Foundation, Hardinxveld-Giessendam
Li JB, Rui JP, Pei ZJ et al (2014) Straw- and slurry-associated prokaryotic communities differ during co-fermentation of straw and swine manure. Appl Microbiol Biotechnol 98:4771–4780
Li C, Zhang Z, Li Y, Cao JS (2015a) Study on dyeing wastewater treatment at high temperature by MBBR and the thermotolerant mechanism based on its microbial analysis. Process Biochem 50:1934–1941
Li JB, Rui JP, Yao MJ et al (2015b) Substrate type and free ammonia determine bacterial community structure in full-scale mesophilic anaerobic digesters treating cattle or swine manure. Front Microbiol 6:1337
Li YF, Nelson MC, Chen PH et al (2015c) Comparison of the microbial communities in solid-state anaerobic digestion (SS-AD) reactors operated at mesophilic and thermophilic temperatures. Appl Microbiol Biotechnol 99:969–980
Li DP, Li B, Wang QR et al (2016a) Toxicity of TiO2 nanoparticle to denitrifying strain CFY1 and the impact on microbial community structures in activated sludge. Chemosphere 144:1334–1341
Li L, He Q, Ma Y et al (2016b) A mesophilic anaerobic digester for treating food waste: process stability and microbial community analysis using pyrosequencing. Microb Cell Factories 15:65
Li P, Wang YJ, Zuo JE et al (2017a) Nitrogen removal and N2O accumulation during hydrogenotrophic denitrification: influence of environmental factors and microbial community characteristics. Environ Sci Technol 51:870–879
Li RR, Duan N, Zhang YH et al (2017b) Anaerobic co-digestion of chicken manure and microalgae Chlorella sp.: methane potential, microbial diversity and synergistic impact evaluation. Waste Manag 68:120–127
Liao RH, Li Y, Yu XM et al (2014) Performance and microbial diversity of an expanded granular sludge bed reactor for high sulfate and nitrate waste brine treatment. J Environ Sci 26:717–725
Lim S, Kim S, Yeon KM et al (2012) Correlation between microbial community structure and biofouling in a laboratory scale membrane bioreactor with synthetic wastewater. Desalination 287:209–215
Liu Z, Lozupone C, Hamady M et al (2007) Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res 35:e120. https://doi.org/10.1093/nar/gkm541
Liu CS, Zhao CC, Wang AJ et al (2015a) Denitrifying sulfide removal process on high-salinity wastewaters. Appl Microbiol Biotechnol 99:6463–6469
Liu Z, Dang Y, Li C, Sun DZ (2015b) Inhibitory effect of high NH4 + -N concentration on anaerobic biotreatment of fresh leachate from a municipal solid waste incineration plant. Waste Manag 43:188–195
Liu ZY, Yu SL, Heedeung P et al (2016) Impact of titanium dioxide nanoparticles on the bacterial communities of biological activated carbon filter intended for drinking water treatment. Environ Sci Pollut Res 23:15574–15583
Liu T, Mao YJ, Shi YP, Quan X (2017) Start-up and bacterial community compositions of partial nitrification in moving bed biofilm reactor. Appl Microbiol Biotechnol 101:2563–2574
Liu Y, Huang L, Dong G et al (2018) Enhanced granulation and methane recovery at low load by downflow sludge circulation in anaerobic treatment of domestic wastewater. Bioresour Technol 249:851–857
Louca S, Doebeli M, Parfrey LW (2018) Correcting for 16S rRNA gene copy numbers in microbiome surveys remains an unsolved problem. Microbiome 6:41
Lu Q, Yi J, Yang DH (2016) Comparative analysis of performance and microbial characteristics between high-solid and low-solid anaerobic digestion of sewage sludge under mesophilic conditions. J Microbiol Biotechnol 26:110–119
Lucas R, Kuchenbuch A, Fetzer I et al (2015) Long-term monitoring reveals stable and remarkably similar microbial communities in parallel full-scale biogas reactors digesting energy crops. FEMS Microbiol Ecol 91:fiv004
Lv XM, Shao MF, Li CL et al (2014) A comparative study of the bacterial community in denitrifying and traditional enhanced biological phosphorus removal processes. Microbes Environ 29:261–268
Ma JX, Wang ZW, Yang Y et al (2013) Correlating microbial community structure and composition with aeration intensity in submerged membrane bioreactors by 454 high-throughput pyrosequencing. Water Res 47:859–869
Ma JX, Wang ZW, Zang LL et al (2015a) Occurrence and fate of potential pathogenic bacteria as revealed by pyrosequencing in a full-scale membrane bioreactor treating restaurant wastewater. RSC Adv 5:24469–24478
Ma K-L, Li X-K, Wang K et al (2015b) 454-Pyrosequencing reveals microbial community structure and composition in a mesophilic UAFB system treating PTA wastewater. Curr Microbiol 71:551–558
Mao YP, Wang ZP, Li LG et al (2016) Exploring the shift in structure and function of microbial communities performing biological phosphorus removal. PLoS ONE 11:e0161506
Matheson CD, Gurney C, Esau N, Lehto R (2010) Assessing PCR inhibition from humic substances. Open Enzyme Inhib J 3:38–45
Motteran F, Braga JK, Silva EL, Varesche MBA (2017) Kinetics of methane production and biodegradation of linear alkylbenzene sulfonate from laundry wastewater anaerobic batch reactors. J Environ Sci Health Part A Toxic Hazard Subst Environ Eng 51:1288–1302
Nair HP, Vincent H, Bhat SG (2014) Evaluation of five in situ lysis protocols for PCR amenable metagenomic DNA from mangrove soils. Biotechnol Rep 4:134–138
Nilakanta H, Drews KL, Firrell S et al (2014) A review of software for analyzing molecular sequences. BMC Res Notes 7:830
Niu LH, Zhang X, Li Y et al (2017) Elevational characteristics of the archaeal community in full-scale activated sludge wastewater treatment plants at a 3660-m elevational scale. Water Sci Technol 76:531–541
Nyvad B, Crielaard W, Mira A et al (2013) Dental caries from a molecular microbiological perspective. Caries Res 47:89–102
Okada DY, Delforno TP, Etchebehere C, Varesche MBA (2014) Evaluation of the microbial community of upflow anaerobic sludge blanket reactors used for the removal and degradation of linear alkylbenzene sulfonate by pyrosequencing. Int Biodeterioration Biodegrad 96:63–70
Oksanen J, Blanchet FG, Friendly M et al (2018) vegan: community ecology package. https://github.com/vegandevs/vegan
Orellana E, Davies-Sala C, Guerrero L, Vardé I, Altina M, Lorenzo MC, Figuerola EL, Pontiggia RM, Erijman L (2019) Microbiome network analysis of co-occurrence patterns in anaerobic co-digestion of sewage sludge and food waste. Water Sci Technol 79:1956–1965
Ouyang E, Lu Y, Ouyang J, Wang L, Wang X (2017) Bacterial community analysis of anoxic/aeration (A/O) system in a combined process for gibberellin wastewater treatment. PLoS ONE. https://doi.org/10.1371/journal.pone.0186743
Pan F, Zhong XH, Xia DS et al (2017) Nanoscale zero-valent iron/persulfate enhanced upflow anaerobic sludge blanket reactor for dye removal: insight into microbial metabolism and microbial community. Sci Rep 7:44626
Pan KL, Gao JF, Li HY et al (2018) Ammonia-oxidizing bacteria dominate ammonia oxidation in a full-scale wastewater treatment plant revealed by DNA-based stable isotope probing. Bioresour Technol 256:152–159
Peng XX, Guo F, Ju F, Zhang T (2015) Shifts in the microbial community, nitrifiers and denitrifiers in the biofilm in a full-scale rotating biological contactor. Environ Sci Technol 48:8044–8052
Persson F, Suarez C, Hermansson M et al (2017) Community structure of partial nitritation-anammox biofilms at decreasing substrate concentrations and low temperature. Microb Biotechnol 10:761–772
Quast C, Pruesse E, Yilmaz P et al (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596
Quince C, Lanzén A, Curtis TP et al (2009) Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods 6:639–641
Quintela-Baluja M, Abouelnaga M, Romalde J, Su JQ, Yu Y, Gomez-Lopez M, Smets B, Zhu YG, Graham DW (2019) Spatial ecology of a wastewater network defines the antibiotic resistance genes in downstream receiving waters. Water Res 162:347–357
Rademacher A, Zakrzewski M, Schlüter A et al (2012) Characterization of microbial biofilms in a thermophilic biogas system by high-throughput metagenome sequencing. FEMS Microbiol Ecol 79:785–799
Ramette A (2007) Multivariate analyses in microbial ecology. FEMS Microbiol Ecol 62:142–160. https://doi.org/10.1111/j.1574-6941.2007.00375.x
Ramirez-Vargas R, Serrano-Silva N, Navarro-Noya YE et al (2015) 454 pyrosequencing-based characterization of the bacterial consortia in a well established nitrifying reactor. Water Sci Technol 72:990–997
Ranasinghe PD, Satoh H, Oshiki M et al (2012) Revealing microbial community structures in large- and small-scale activated sludge systems by barcoded pyrosequencing of 16S rRNA gene. Water Sci Technol 66:2155–2161
Rasool K, Mahmoud KA, Lee DS (2015) Influence of co-substrate on textile wastewater treatment and microbial community changes in the anaerobic biological sulfate reduction process. J Hazard Mater 299:453–461
Reeder J, Knight R (2010) Rapid denoising of pyrosequencing amplicon data: exploiting the rank-abundance distribution. Nat Methods 7:668–669
Rodríguez N, Sanz JL (1998) Response of an anaerobic granular sludge to chlorinated aliphatic hydrocarbons in different conditions. J Ferment Bioeng 86:226–232
Rodríguez E, García-Encina P, Stams AJM et al (2015) Meta-omics approaches to understand and improve wastewater treatment systems. Rev Environ Sci Biotechnol 14:385–406
Sanz JL, Köchling T (2007) Molecular biology techniques used in wastewater treatment: an overview. Process Biochem 42:119–133
Saunders AM, Albertsen M, Vollertsen J, Nielsen PH (2016) The activated sludge ecosystem contains a core community of abundant organisms. ISME J 10:11–20
Schloss PD, Westcott SL, Ryabin T et al (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Schlüter A, Bekel T, Diaz NN et al (2008) The metagenome of a biogas-producing microbial community of a production-scale biogas plant fermenter analysed by the 454-pyrosequencing technology. J Biotechnol 136:77–90
Shan L, Zhang Z, Yu Y et al (2017) Performance of CSTR-EGSB-SBR system for treating sulfate-rich cellulosic ethanol wastewater and microbial community analysis. Environ Sci Pollut Res 24:14387–14395
Shannon CE (1948) A mathematical theory of communication. Bell Syst Tech J 27:379–423. https://doi.org/10.1002/j.1538-7305.1948.tb01338.x
Shokralla S, Spall JL, Gibson JF, Hajibabaei M (2012) Next-generation sequencing technologies for environmental DNA research. Mol Ecol 21:1794–1805
Simpson EH (1949) Measurement of diversity. Nature 163:688
Sivagurunathan P, Anburajan P, Kumar G et al (2016) High-rate hydrogen production from galactose in an upflow anaerobic sludge blanket reactor (UASBr). RSC Adv 6:59823–59833
Smith AM, Sharma D, Lappin-Scott H et al (2014) Microbial community structure of a pilot-scale thermophilic anaerobic digester treating poultry litter. Appl Microbiol Biotechnol 98:2321–2334
Su C, Lu Y, Qin J et al (2017) Performance and microbial community structure characterization of a CIC anaerobic reactor for the treatment of cassava wastewater. Ecol Eng 108:114–122
Sun HH, Yu P, Li QL et al (2017) Transformation of anaerobic granules into aerobic granules and the succession of bacterial community. Appl Microbiol Biotechnol 101:7703–7713
Sundberg C, Al-Soud WA, Larsson M et al (2013) 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol Ecol 85:612–626
Szabo E, Liebana R, Hermansson M et al (2017) Comparison of the bacterial community composition in the granular and the suspended phase of sequencing batch reactors. AMB Express 7:168
Team RDC (2008) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Tian HL, Zhao JY, Zhang HY et al (2015a) Bacterial community shift along with the changes in operational conditions in a membrane-aerated biofilm reactor. Appl Microbiol Biotechnol 99:3279–3290
Tian M, Zhao FQ, Shen X et al (2015b) The first metagenome of activated sludge from full-scale anaerobic/anoxic/oxic (A2O) nitrogen and phosphorus removal reactor using Illumina sequencing. J Environ Sci 35:181–190
Torres K, Alvarez-Hornos FJ, San-Valero P et al (2018) Granulation and microbial community dynamics in the chitosan-supplemented anaerobic treatment of wastewater polluted with organic solvents. Water Res 130:376–387
Tuan NN, Chang YC, Yu CP, Huang SL (2014) Multiple approaches to characterize the microbial community in a thermophilic anaerobic digester running on swine manure: a case study. Microbiol Res 169:717–724
Vetrovsky T, Baldrian P (2013) The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS ONE 8:e57923
Vrieze JD, Pinto AJ, Sloan WT, Ijaz UZ (2018) The active microbial community more accurately reflects the anaerobic digestion process: 16S rRNA (gene) sequencing as a predictive tool. Microbiome 6:63
Walden C, Carbonero F, Zhang W (2017) Assessing impacts of DNA extraction methods on next generation sequencing of water and wastewater samples. J Microbiol Methods 141:10–16
Wang XH, Hu M, Xia Y et al (2012) Pyrosequencing analysis of bacterial diversity in 14 wastewater treatment systems in China. Appl Microbiol Biotechnol 78:7042–7047
Wang HY, He QL, Chen D et al (2015) Microbial community in a hydrogenotrophic denitrification reactor based on pyrosequencing. Appl Microbiol Biotechnol 99:10829–10837
Wang JF, Qian FY, Liu XP et al (2016a) Cultivation and characteristics of partial nitrification granular sludge in a sequencing batch reactor inoculated with heterotrophic granules. Appl Microbiol Biotechnol 100:9381–9391
Wang P, Yu ZS, Qi R, Zhang HX (2016b) Detailed comparison of bacterial communities during seasonal sludge bulking in a municipal wastewater treatment plant. Water Res 105:157–166
Wang ST, Wang WQ, Zhang ZR, You H (2016c) The impact of zinc oxide nanoparticles on phosphorus removal and the microbial community in activated sludge in an SBR. RSC Adv 6:96706–96713
Wang XC, Shen JM, Chen ZL et al (2016d) Removal of pharmaceuticals from synthetic wastewater in an aerobic granular sludge membrane bioreactor and determination of the bioreactor microbial diversity. Appl Microbiol Biotechnol 100:8213–8223
Wang S, Li ZW, Gao MC et al (2017a) Long-term effects of cupric oxide nanoparticles (CuO NPs) on the performance, microbial community and enzymatic activity of activated sludge in a sequencing batch reactor. J Environ Manag 187:330–339
Wang S, Li ZW, Gao MC et al (2017b) Long-term effects of nickel oxide nanoparticles on performance, microbial enzymatic activity, and microbial community of a sequencing batch reactor. Chemosphere 169:387–395
Wen X, Gong BZ, Zhou J et al (2017) Efficient simultaneous partial nitrification, anammox and denitrification (SNAD) system equipped with a real-time dissolved oxygen (DO) intelligent control system and microbial community shifts of different substrate concentrations. Water Res 119:201–211
Westerholm M, Crauwels S, Geel MV et al (2016) Microwave and ultrasound pre-treatments influence microbial community structure and digester performance in anaerobic digestion of waste activated sludge. Appl Microbiol Biotechnol 100:5339–5352
Wolters B, Ding GC, Kreuzig R, Smalla K (2016) Full-scale mesophilic biogas plants using manure as C-source: bacterial community shifts along the process cause changes in the abundance of resistance genes and mobile genetic elements. FEMS Microbiol Ecol 92:fiv163
Wu SJ, Dang Y, Qiu B et al (2015) Effective treatment of fermentation wastewater containing high concentration of sulfate by two-stage expanded granular sludge bed reactors. Int Biodeteriation Biodegrad 104:15–20
Xiao X, Li TT, Lu XR et al (2018) A simple method for assaying anaerobic biodegradation of dyes. Bioresour Technol 251:204–209
Xue Y, Zhao H, Ge L et al (2015) Comparison of the performance of waste leachate treatment in submerged and recirculated membrane bioreactors. Int Biodeteriation Biodegrad 102:73–80
Yang Y, Quensen J, Mathieu J et al (2014a) Pyrosequencing reveals higher impact of silver nanoparticles than Ag + on the microbial community structure of activated sludge. Water Res 48:317–325
Yang Y, Yu K, Xia Y et al (2014b) Metagenomic analysis of sludge from full-scale anaerobic digesters operated in municipal wastewater treatment plants. Appl Microbiol Biotechnol 98:5709–5718
Yang B, Xu H, Wang J et al (2017) Bacterial and archaeal community distribution and stabilization of anaerobic sludge in a strengthen circulation anaerobic (SCA) reactor for municipal wastewater treatment. Bioresour Technol 244:750–758
Yang B, Wang M, Wang JF et al (2018) Mechanism of high contaminant removal performance in the expanded granular sludge blanket (EGSB) reactor involved with granular activated carbon for low-strength wastewater treatment. Chem Eng J 334:1176–1185
Ye L, Zhang T (2011) Pathogenic bacteria in sewage treatment plants as revealed by 454 Pyrosequencing. Environ Sci Technol 45:7173–7179
Ye L, Zhang T (2013) Bacterial communities in different sections of a municipal wastewater treatment plant revealed by 16S rDNA 454 pyrosequencing. Appl Microbiol Biotechnol 97:2681–2690
Yeates C, Gillings MR (1998) Rapid purification of DNA from soil for molecular biodiversity analysis. Lett Appl Microbiol 27:49–53
Yeates C, Gillings MR, Davison AD et al (1997) PCR amplification of crude microbial DNA extracted from soil. Lett Appl Microbiol 25:303–307
Yi J, Dong B, Jin JW, Dai XH (2014) Effect of increasing total solids contents on anaerobic digestion of food waste under mesophilic conditions: performance and microbial characteristics analysis. PLoS ONE 9:e102548
Yu HG, Wang QY, Wang ZW et al (2014) Start-up of an anaerobic dynamic membrane digester for waste activated sludge digestion: temporal variations in microbial communities. PLoS ONE 9:e93710
Yu B, Zhang DL, Shan AD et al (2015) Methane-rich biogas production from waste-activated sludge with the addition of ferric chloride under a thermophilic anaerobic digestion system. RSC Adv 5:38538–38546
Yu D, Li C, Wang L et al (2016a) Multiple effects of trace elements on methanogenesis in a two-phase anaerobic membrane bioreactor treating starch wastewater. Appl Microbiol Biotechnol 100:6631–6642
Yu HG, Wang ZW, Zhu ZCWNCW (2016b) Enhanced waste activated sludge digestion using a submerged anaerobic dynamic membrane bioreactor: performance, sludge characteristics and microbial community. Sci Rep 6:20111
Zeng Q, Hao TW, Mackey HR et al (2017) Alkaline textile wastewater biotreatment: a sulfate-reducing granular sludge based lab-scale study. J Hazard Mater 332:104–111
Zhang T, Ye L, Tong AHY et al (2011) Ammonia-oxidizing archaea and ammonia-oxidizing bacteria in six full-scale wastewater treatment bioreactors. Appl Microbiol Biotechnol 91:1215–1225
Zhang T, Shao MF, Ye L (2012) 454 Pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME J 6:1137–1147
Zhang Y, Hu M, Li P et al (2015a) Analysis of trichloroethylene removal and bacterial community function based on pH-adjusted in an upflow anaerobic sludge blanket reactor. Appl Microbiol Biotechnol 99:9289–9297
Zhang Y, Hu M, Li P et al (2015b) Trichloroethylene removal and bacterial variations in the up-flow anaerobic sludge blanket reactor in response to temperature shifts. Appl Microbiol Biotechnol 99:6091–6102
Zhang Y, Wang X, Hu M, Li P (2015c) Effect of hydraulic retention time (HRT) on the biodegradation of trichloroethylene wastewater and anaerobic bacterial community in the UASB reactor. Appl Microbiol Biotechnol 99:1977–1987
Zhang BY, Zhao HY, Yu HR et al (2016a) Evaluation of biogas production performance and Archaeal microbial dynamics of corn straw during anaerobic co-digestion with cattle manure liquid. J Microbiol Biotechnol 26:739–747
Zhang Y, Zhen Y, Mi T et al (2016b) Molecular characterization of sulfate-reducing bacteria community in surface sediments from the adjacent area of Changjiang Estuary. J Ocean Univ China 15:107–116
Zhang JX, Loh KC, Lee J et al (2017) Three-stage anaerobic co-digestion of food waste and horse manure. Sci Rep 7:1269
Zhao JW, Gui L, Wang QL et al (2017) Aged refuse enhances anaerobic digestion of waste activated sludge. Water Res 123:724–733
Zhou J, Yang J, Yu Q et al (2017a) Different organic loading rates on the biogas production during the anaerobic digestion of rice straw: a pilot study. Bioresour Technol 244:865–871
Zhou WL, Li Y, Liu X et al (2017b) Comparison of microbial communities in different sulfur-based autotrophic denitrification reactors. Appl Microbiol Biotechnol 101:447–453
Zhu S, Wu H, Zhou L, Wei C (2017a) The resilience of microbial community involved in coking wastewater treatment system. J Next Gener Seq Appl 4:1000142
Zhu XY, Kougias PG, Treu L et al (2017b) Microbial community changes in methanogenic granules during the transition from mesophilic to thermophilic conditions. Appl Microbiol Biotechnol 101:1313–1322
Ziganshin AM, Liebetrau J, Proter J, Kleinsteuber S (2013) Microbial community structure and dynamics during anaerobic digestion of various agricultural waste materials. Appl Microbiol Biotechnol 97:5161–5174
Ziganshin AM, Ziganshina EE, Kleinsteuber S, Nikolausz M (2016) Comparative analysis of methanogenic communities in different laboratory-scale anaerobic digesters. Archaea 2016:3401272
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sanz, J.L., Köchling, T. Next-generation sequencing and waste/wastewater treatment: a comprehensive overview. Rev Environ Sci Biotechnol 18, 635–680 (2019). https://doi.org/10.1007/s11157-019-09513-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11157-019-09513-0