Abstract
Phenol and its derivatives are widespread contaminants whose sources are both natural and industrial. Phenol is massively produced and used as a starting material for synthetic polymers and fibers. Although phenolic compounds play important biochemical and physiological roles in living systems, their accumulation in the environment as a result of intensive human activity may result in drastic ecological problem. Various analytical techniques are available for the detection of phenol in environmental samples. But they need complex sample pre-treatment so as are time consuming, costly and use heavy devices. On the other hand a biosensor is a device that gives rapid detection, cost effective and easy. A review study was carried out to accumulate the possible biosensors for the detection of phenolic compounds in environmental samples. A number of biological components including microorganisms, enzymes, antibodies, antigens, nucleic acids etc. can be used for the construction of biosensors that was found to detect phenolic compounds. Of all type of biological components microorganisms and enzymes are mostly used. The microorganisms are Pseudomonas, Moraxella, Arthrobacter, Rhodococcus, and Trichosporon. The most used enzymes are tyrosinase, peroxidase, laccase, glucose dehydrogenase, cellobiose dehydrogenase etc. Antibody sensors can detect a very trace level. The biorecognition of DNA biosensors occur by hybridization of DNA. Biosensors are found to work well when the biological sensing element is immobilized. A variety of immobilization techniques were found to use as adsorption, covalent binding, entrapment, cross-linking etc. For immobilization the matrices used was polyvinyl alcohol, Osmium complex, nafion/sol–gel silicate, chitosan, silica gel etc.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
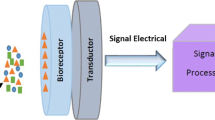
Humankind has been performing bioanalysis since the dawn of time, using the sensory nerve cells of the nose to detect scents or the enzymatic reactions in the tongue to taste food. As time has progressed, so has our level of understanding about the function of living organisms in detecting trace amounts of biochemicals in complex systems (Vo-Dinh and Cullum 2000). A biosensor is an analytical device that combines a biological sensing element with a transducer to produce a signal proportional to the analyte concentration (Blum and Coulet 1991; Turner et al. 1992; Tran 1993; Nikolelis et al. 1998). Biomolecules such as enzymes, antibodies, receptors, organelles and microorganisms as well as animal and plant cells or tissues have been used as biological sensing elements (Lei et al. 2006). Enzymes are the most widely used biological sensing element in the fabrication of biosensors (Turner et al. 1992). Microorganisms have been integrated with a variety of transducers such as amperometric, potentiometric, calorimetric, conductimetric, colorimetric, luminescence and fluorescence to construct biosensor devices (Rensing and Maier 2003; Belkin 2003).
The available classical tools of measuring phenol are chromatography (gas, liquid and capillary electrophoresis), enzyme-linked immunosorbent assay (ELISA) electro analytical techniques (polarography, cyclic voltammetry, adsorptive stripping and differential pulse voltammetry) (Zietek 1975; Hernandez et al. 1993; Barek et al. 1994; Clement et al. 1995; Rodriguez et al. 1997a, b, c) give accurate results at very low concentration, they are heavy devices and their use can induce delays which are mainly due to the transport of samples to the laboratory for analysis. Although sensitive, these techniques have limitations that limit their applicability to on-line or field monitoring (Nistor et al. 2001). The major advantages of biosensors are fastness, simplicity, and applicability to multi component solutions in situ.
2 Sources, health effect and toxicity of phenol
Phenol and phenolic compounds are natural harmful substances found in water. They are used in many industrial activities for the production of pesticides, insecticides, herbicides, and synthetic products. Phenol can enter natural water reservoirs with industrial waste. It is toxic compound and its permissible concentration limit in natural waters is 0.001 mg/L (10–2 μM); therefore, it is necessary to keep watch on phenol entering aquatic ecosystems (Makarenko et al. 2002). The annual global production of phenol compounds amounts to about 50,000 t (Riedel et al. 1993). Phenol can enter natural water reservoirs with industrial waste. It is a toxic compound, and its permissible concentration limit in natural waters is 0.001 mg/L (10–2 mM); therefore, it is necessary to keep watch on phenol entering aquatic ecosystems (Riedel et al. 1993).
Phenol is the simplest member of a family of compounds in which a hydroxyl (–OH) group is attached directly to a benzene ring. Phenols are classified as organic compounds similar to alcohols, but they form stronger hydrogen bonds. Phenols which have a structural formula (C6H5OH), that is the formula of phenol only, as a simplest member of phenols, and subsequent phenols with structure R–C6H4OH, where R represents some groups like methyl (CH3) and nitro (NO2). Pure phenol is a white crystalline solid, smelling of disinfectant. It has to be handled with great care because it causes immediate white blistering to the skin. The crystals are often rather wet and discoloured. It is toxic compound and its permissible concentration limit in natural waters is 0.001 mg/L (10–2 μM) (Makarenko et al. 2002).
Phenol poisoning can occurs in humans after skin absorption, inhalation of vapours or ingestion. Acute local effects are severe tissue irritation and necrosis. At high doses, the most prominent systemic effect is central nervous system depression can occur (IARC 1989). Excess mortality could be attributed to occupational exposure to phenol (Dosemeci et al. 1991). There is a risk of lung cancer with the exposure of phenol (Morimoto et al. 1976). However, it only slightly inhibits DNA repair synthesis and DNA replication synthesis in WI-38 human diploid fibroblasts (Poirier et al. 1975). Some phenolic compounds can be harmful when consumed in large amounts. The best known negative properties attributed to phenolic compounds are the capacity to precipitate proteins, form complexes with polysaccharides, affect lipid metabolism and interfere with the bioavailability of metal ions (Bravo 1998). Phenol and its vapors are corrosive to the eyes, the skin, and the respiratory tract. Repeated or prolonged skin contact with phenol may cause dermatitis, or even second and third-degree burns due to phenol’s caustic and defatting properties. Inhalation of phenol vapor may cause lung edema. The substance may cause harmful effects on the central nervous system and heart, resulting in dysrhythmia, seizures, and coma. The kidneys may be affected as well. Exposure may result in death and the effects may be delayed. Long-term or repeated exposure of the substance may have harmful effects on the liver and kidneys.” There is no evidence to believe that phenol causes cancer in humans. Besides its hydrophobic effects, another mechanism for the toxicity of phenol may be the formation of phenoxyl radicals. The major hazard of phenol is its ability to penetrate the skin rapidly, particularly when liquid, causing severe injury which can be fatal. Phenol also has a strong corrosive effect on body tissue causing severe chemical burns. For its local anesthetizing properties, skin burns may be painless.
3 Methods of phenol detection
Spectrophotometric, gas chromatography, liquid chromatography and capillary electrophoresis are the commonly used methods for the determination of phenolic compounds (Rogers et al. 2001). Methods to determine phenols at a very low concentration are gas chromatographic (GC) method using liquid–liquid extraction, flame ionic detection (FID) or derivatization and electron capture detection (ECD). Other method to determine phenol at slightly higher concentrations may be liquid–liquid extraction gas chromatographic/mass spectrometric method (Clesceri et al. 1998). However, these methods are time consuming including complicated sample pre-treatment (Rogers et al. 2001). In addition, the equipment is expensive and is not generally portable. Therefore, there is an interest in developing simple, sensitive, specific, accurate and portable system such as biosensor for determination of phenolic compounds.
4 Biosensor
A biosensor is an analytical device, which converts the modification of the physical or chemical properties of a biomatrix (enzyme, antibodies, receptors, organelles, microorganisms) into an electric or other kinds of signal whose amplitude depends on the concentration of defined analytes in the solution (Dzyadevych et al. 2008).
A specific “bio” element (say, enzyme) recognizes a specific analyte and the “sensor” element transduces the change in the biomolecule into an electrical signal. The bio element is very specific to the analyte to which it is sensitive. It does not recognize other analytes (Rogers et al. 2001).
4.1 Immobilization
Biosensors work best when the biological sensing element and the transducer are kept in close proximity to one another. Immobilization technology has played a major role in this direction. Immobilization not only helps in attachment of biological component to the transducer, but also helps in stabilising it for reuse. The biological material has been immobilized directly on the transducer or in membranes, which can be subsequently mounted on the transducer (Mehrvar et al. 2000; D`Souza 2001). The choice of immobilization method depends on many factors, such as the nature of the biological element, the type of transducer used, the physicochemical properties of the analyte and the operating conditions in which the biosensor is to function, and overriding all these considerations is necessary for the biological element to exhibit maximum activity in its immobilized microenvironment (Singh et al. 2008). Generally, there are five common methods of immobilization: adsorption, covalent binding, entrapment, encapsulation and cross-linking (Bickerstaff 1997).
Adsorption:
It is the simplest and fastest way to prepare immobilized enzymes. Adsorption can roughly be divided into two classes: physical adsorption and chemical adsorption. Many substances adsorb enzymes on their surfaces, e.g. Alumina, charcoal, clay, cellulose, kaolin, silica gel, glass and collagen.
Covalent bonding:
In this method, the bond occurs between a functional group in the biomaterial to the support matrix.
Entrapment:
It refers to mixture of the biomaterial with monomer solution and then polymerized to a gel, trapping the biomaterial.
Cross-linking:
For this method, usually, biomaterial is chemically bonded to solid supports or to another supporting material such as cross-linking agent to significantly increase the attachment (Zhao and Jiang 2010).
5 Microbial biosensors for phenol detection
Initiated almost 20 years ago (King et al. 1990), the engineering of microbial cells with the purpose of chemical detection has enormously expanded since. A microbial biosensor is an analytical device that couples microorganisms with a transducer to enable rapid, accurate and sensitive detection of target analytes in fields as diverse as medicine, environmental monitoring, defense, food processing and safety (Leveau and Lindow 2002). Various microorganisms can be used for the detection of phenolic compounds such as Pseudomonas, Arthrobacter, Rhodococcus, Trichosporon and Moraxella. Table 1 summarizes the list of microbial biosensors.
Most of the biosensors developed uses amperometric detection. Such as biosensors using Rhodococcus, Trichosporon, Moraxella, Pseudomonas etc. Rhodococcus using biosensors has been found to detect mainly phenol and chlorophenol (Riedel et al. 1993). Also an amperometric microbial biosensor for highly specific, sensitive and rapid quantitative determination of p-nitrophenol was developed using Moraxella sp. to specifically degrade p-nitrophenol to hydroquinone, a more electroactive compound than p-nitrophenol. The electrochemical oxidation current of hydroquinone formed in biodegradation of p-nitrophenol was measured at Moraxella sp. modified carbon paste electrode and correlated to p-phenol concentrations (Mulchandani et al. 2005). Trichosporon beigelii which is a species of fungus in the family Trichosporonaceae. An amperometric biosensor for the determination of phenol and chlorophenols using Trichosporon beigelii (cutaneum) has been developed (Riedel et al. 1995).
5.1 Microbial culture and sensing mechanism
The entrapment of Rhodococcus, Trichosporon and Pseudomonas cells can be done with polyvinyl alcohol of relative molecular weight 28,000. The culture conditions of microorganisms maintained as Rhodococcus sp. was grown on a rotating shaker at 30°C for 24 h in a mineral medium of the following composition (Straube et al. 1990), Na2HPO4·12H20, 9 g; KH2PO4, 1.5 g; NH4C1, 1.0 g; MgSO4·7H2O, 0.2 g; CaCO3, 0.02 g; FeSO4·7H2O, 0.01 g; H3BO4, 50 μg; CuSO4·5H20, 10 μg; KI, 10 μg; MnSO4·4H20, 40 μg ZnSO4·7H20 40 μg; Na2MoO4·2H20, 24 μg in 1 L distilled water. Phenol was sterilized by filtration, and added after sterilisation to the medium (0.5 g/L) (Riedel et al. 1993). The yeast Trichosporon beigelii (cutaneum) E4 was cultured under aerobic conditions (rotating shaker) at 24°C for 24 h in a medium containing 0.3% malt extract, 0.3% peptone, 0.3% yeast extract and 1.00% glucose (Riedel et al. 1995). The sensing mechanism of Rhodococcus and Trichosporon is almost same. An oxygen electrode (Pt cathode with a diameter of 0.5 mm, was covered with a polyethylene membrane (thickness 10 gm). The immobilized microorganisms were placed on the membrane and covered with a capillary membrane (diameter of pores 1 gm, Oxyphen, Dresden, FRG). The sensor was placed in a measuring cell containing 2.5 mL of phosphate buffer (0.1 mmol/L, pH 6.8, 30°C). The change in current was followed at −600 mV versus Ag/AgCl (Riedel et al. 1993, 1995). Moraxella sp. stock culture maintained on tryptic soy broth was inoculated into tryptic soy broth and incubated overnight on a gyratory incubator shaker in pH 7.2 minimal salt medium (3.73 mM K2HPO4, 1.25 mM KH2PO4, 1.4 mM NH4Cl, 0.4 mM MgSO4·7H2O and 0.02 mM FeSO4·7H2O) supplemented with 0.4 mM p-nitrophenol and 0.2% yeast extract. Amperometric measurements were done using a Bioanalytical Systems (BAS). All experiments were conducted using a three electrode electrochemical cell (10 mL volume with a working volume of 3 mL), with an Ag/AgCl reference electrode, and platinum wire auxiliary electrode. When the baseline was established then 5–10 μL of p-nitrophenol was added to the cell and the steady-state current was recorded (Mulchandani et al. 2005). The Moraxella sp. is reported to degrade p-nitrophenol using the following enzymes: p-nitrophenol monooxygenase, benzoquinine reductase, hydroquinone dioxygenase, hydroxymuconic semialdehyde dehydrogenase and maleylacetate reductase (Spain and Gibson 1991). Pseudomonas putida GFS-8 is a gram negative, polar, flagellated unicellular bacterium. The culture was maintained consistently on Luria-Broth (LB) agar plates and synthetic medium (M-9) supplemented with phenol agar plates. The growth medium used were LB medium: 1% peptone; 0.5% yeast extract; 0.5% NaCl; pH 7.0, synthetic medium supplemented with yeast extract and phenol: 0.075% (NH4)2SO4; 0.05% NH4C1; 0.05% MgSO4·7H20;0.05% KH2PO4; 0.05% K2HPO4; 0.1% yeast extract; 0.025% phenol, pH 7.0. Phenol oxidizing activity was measured amperometrically with a Clark membrane electrode. The measurements were made in a temperature-controlled 5-mL amperometric unit, containing a cell suspension or immobilized cells, and 50 mM potassium phosphate buffer, pH 7.0, at 30°C. The initiation of the reaction by phenol allowed us to take into account the endogenic respiration of free or immobilized cells. The dynamics of phenol concentration through cells incubation in the M-9 medium supplemented with phenol was measured spectrophotometrically at 270 nm (Rainina et al. 1996).
5.2 Detection
Rhodococcus and Trichosporon based sensor is more sensitive to phenol and chlorophenols, especially to mono and dichlorinated phenol. For Rhodococcus a linear relationship between the current range and the concentration of phenol, 2-, 3- and 4-chlorophenol was observed up to 20 μmol/L. The detection limit for all studied substrates was 4 μmol/L (Riedel et al. 1993). For Trichosporon a linear relationship between the current range and the concentration of 4-chlorophenol is observed up to 40 μmol/L. The detection limit for all substrates studied is 2 μmol/L (Riedel et al. 1995). Moraxella based biosensor has excellent selectivity against phenol derivatives and was able to measure as low as 20 nM (2.78 ppb) p-nitrophenol with very good accuracy and reproducibility. The biosensor was stable for approximately 3 weeks when stored at 4°C (Mulchandani et al. 2005). Pseudomonas putida is relatively sensitive to benzoate and monochlorobenzoates and is insensitive to phenol, whereas Rhodococcus shows high sensitivity to phenol and monochlorophenols (Rainina et al. 1996).
6 Enzymatic biosensors for phenol detection
Enzymes are the most frequently used biological components in biosensors, because a wide range of enzymes is suitable for acting as recognition elements and very often their catalytic properties and substrate specificity can be modified by means of genetic engineering (Rodriguez-Mozaz et al. 2006). Purified enzymes have been most commonly used in the construction of biosensors due to their high analytical specificity. There are several advantages of enzyme biosensors: ability to modify the catalytic properties or substrate specificity by means of genetic engineering and catalytic amplification of the biosensor response by modulation of the enzyme activity with respect to the target analyte (D`Souza 2001; Rogers and Mascini 2009).
Many biosensor research papers have been reported previously for the detection of phenolic compounds based on several types of enzyme such as tyrosinase, laccase, horseradish peroxidase (HRP). However, reaction mechanisms of the biosensors based on tyrosinase, laccase and HRP are different for various types of phenolic compounds. For tyrosinase and laccase, the enzyme molecules are reduced by phenolic compounds after being oxidized by oxygen (Yang et al. 2005). For HRP, it is oxidized by hydrogen peroxide after its reduction by phenolic compounds (Rosatto et al. 1999). Tyrosinase based biosensor can be used for the monitoring of phenolic compounds with ortho-position of the phenol ring free of substituent group. But for phenolic compounds with para- and meta-position free of substituent group, laccase based biosensor can be used. However its catalytic behaviour is complex (Mello et al. 2003). Alternatives to enhance enzyme stability are the use of novel immobilization methods (Scouten et al. 1995) or media (Crumbliss et al. 1994; Besombes et al. 1995; Walcarius 1998). Other attempts that have received attention are the use of additives such as positively charged soluble polymer and a neutral carbohydrate (Popescu et al. 1995; Cosnier and Popescu 1996). Table 2 summarizes the Enzyme biosensors for the detection of phenolic compounds.
Among many analytical methods for measuring phenolic compounds electrochemical biosensors based on immobilized Tyrosinase have received the major share of attention (Chen et al. 2001; Xue and Shen 2002; Yu et al. 2003). Tyrosinase is an enzyme that catalyzes phenol o-hydroxylation yielding o-diphenol, which is subsequently oxidized too-quinone (Ferreira et al. 2006). That quinone can be than reduced at the electrode surface at low potentials giving o-diphenol. This concept was already used in construction of amperometric biosensors based on Tyrosinase entrapped in different matrices (Duran et al. 2002). So, it is important to seek a suitable method or matrix for enzyme immobilization and providing essential water layer around the enzyme.
6.1 Tyrosinase based biosensor
A biosensor was fabricated by immobilizing tyrosinase in a titania sol–gel membrane which was obtained with a vapor deposition method (Yu and Ju 2004). The experimental parameters such as solvent and operating potential were optimized. At −100 mV this biosensor showed a good amperometric response to phenols in pure chloroform without any mediator and rehydration of the enzyme. For catechol determination the sensor exhibited a fast response of less than 5 s. The sensitivity of different phenols was as follows: catechol > phenol > p-cresol (Yu and Ju 2004). Also A novel tyrosinase biosensor has been developed for the subnanomolar detection of phenols, based on the immobilization of tyrosinase in a positively charged Al2O3 sol–gel membrane on a glassy carbon electrode. The Al2O3 sol–gel-containing surface displays an intrinsic electrocatalytic o-quinone response and, hence, offers a high-sensitivity (127 μA/mM−1) monitoring of phenols (Liu et al. 2000). A tyrosinase-modified solid composite biosensor has been developed. The composite transducer for amperometric biosensor was based on graphite powder modified with tyrosinase and 2-hexadecanol used as a solid binding matrix. The response of a biosensor modified with 4% of tyrosinase was linear up to 2.5 μM, the sensitivity was 0.0225 μA/μM, and the detection limit 0.2 μM (Svitel and Miertus 1998). The main advantages of biosensors based on SBM (hexadecanol, hexadecanone, cholesteryloleate, etc.) are good stability and mechanical (possibility of modification with enzymes and mediators, easy fabrication) and electrochemical (fast response, low background currents, operational and storage stability) properties (Svorc et al. 1997). Also immobilization of tyrosinase within an Os-complex functionalized electro deposition polymer can be used for the monitoring of phenolic compounds with ortho-position of the phenol ring free of substituent group (Yildiz et al. 2007). Catechol can be detected effectively using Tyrosinase entrapped in agarose-guar gum entrapped matrix (Tembe et al. 2007). A biosensor was constructed with tyrosinase immobilized in a gelatine matrix cross linked with formaldehyde. This biosensor exhibits a fast amperometric response to phenolic compounds. The linear ranges for catechol, phenol, and p-cresol determination were from 5 × 10−8 to 1.4 × 10−4 M, 5 × 10−8 to 7.1 × 10−5 M, and 1 × 10−7 to 3.6 × 10−5 M, with detection limits of 2.1 × 10−8 M, 1.5 × 10−8 M, and 7.1 × 10−8 M respectively (Li et al. 2005).
6.2 Dual enzyme based biosensors
Sometimes bi-immobilization of enzymes can be used for biosensor fabrication. Such as laccase and tyrosinase phenoloxidase enzymes has been successfully used for detection of polyphenolic compounds (Elkaoutit et al. 2007). This biosensor employs as the electrochemical transducer the Sonogel–Carbon, a novel type of electrode developed by our group. The Lac–Ty/Sonogel–Carbon electrode responds to nanomolar concentrations of flavan-3-ols, hydroxycinnamic acids, and hydroxybenzoic acids. The limit of detection, sensitivity, and linear range for caffeic acid, taken as an example, were 26 nM, 167.53 nA M−1, and 0.01–2 μM, respectively. The Sonogel–Carbon electrode (Cordero-Rando et al. 2005) was chosen because of its demonstrated high sensitivity, stability, and biocompatibility when used as an electrochemical transducer. Another biosensor developed based on tyrosinase-horseradish peroxidase, enhanced sensitivity was observed on comparing with tyrosinase or horseradish peroxidase monoenzyme modified electrodes (Dai et al. 2005). The phenol sensor exhibited a fast response of less than 10 s. The sensitivity of the biosensor for phenol was 14 mA mM−1 cm−2 with a linear range from 2 × 10−7 to 2.3 × 10−4 M and a detection limit of 4.1 × 10−9 M. The biosensor showed a good stability and reproducibility. Another tyrosinase-HRP peroxidase sensor by co immobilizing HRP and tyrosinase in poly-(carbamoylsulfonate) hydrogel (Chang et al. 2002) has also been reported.
6.3 Laccase based biosensors
Laccase is a multi-copper oxidase, which couples catalytic oxidation of phenolic substrates with four electron reductions to dioxygen to water (Solomon and Lowery 1993). An amperometric biosensor was developed for the determination of polyphenolic compounds based on the immobilization of laccasse. Reduction of the product of oxidation of several polyphenols catalysed by laccasse, was done at a potential for which the polyphenol of interest, catechol was found to respond (Gomes and Rebelo 2003). Another laccase biosensor was based on the fabrication of an optical biosensor by using stacked films where 3-methyl-2-benzothiazolinone hydrazone (MBTH) was immobilized in a hybrid nafion/sol–gel silicate film and laccase in a chitosan film for the detection of phenolic compounds was described (Abdullah et al. 2007). Quinone and/or phenoxy radical product from the enzymatic oxidation of phenolic compounds was allowed to couple with MBTH to form a colored azo-dye product for spectrophometric detection. The response of the biosensor was linear with substrate concentrations of 0.5-8.0 mM and 10 min of sensor exposure time. The use of the hybrid materials of nafion/sol–gel silicate to immobilize laccase has altered the selectivity of the enzyme to various phenolic compounds. Immobilization in this hybrid material has enabled the biosensor to be more selective to catechol compared with the non-immobilized enzyme. It was also observed to overcome the problem of film brittleness and shrinkage, which normally occurred when a pure sol–gel silicate film alone was used (Kim and Lee 2003).
6.4 Peroxidase based biosensors
Horseradish peroxidase (HRP) is a glycoprotein with 4 lysine residues for conjugation to a labeled molecule. HRP is often used in conjugates (molecules that have been joined genetically or chemically) to determine the presence of a molecular target. An electrochemical biosensor was developed using the enzyme peroxidase for the detection of phenol. It was found that if the peroxidase is immobilized in silica gel modified with titanium oxide the sensitivity improved significantly (Rosatto et al. 2001). When peroxidase is immobilized on an electrode surface, the oxidized form of enzyme, which is formed in the reaction with peroxide, can be reduced to its native form by a direct or mediated electron transfer. Phenols act as an electron mediator in the system. Based on these principles biosensors for phenol compounds were developed where HRP was immobilized on solid graphite electrodes (Ruzgas et al. 1995) or in carbon paste electrodes (Lindgren et al. 1997). The enzyme mechanism involved in a peroxidase based biosensor for phenol detection consists in the oxidation of enzyme molecules by hydrogen peroxide followed of its reduction by the phenolic compound. This last reduction converts phenol to quinone or semiquinone which is electro active and can be reduced on the electrode surface. The reduction current is proportional to the phenol concentration in the solution (Ruzgas et al. 1995; Lindgren et al. 1997). The high sensitivity of the biosensor ST-HRP and the stability obtained by DNA incorporation provided phenol determination in real samples of industrial effluent and the waters of polluted rivers.
7 Immunosensors for phenol detection
Immunosensors are detection systems comprising an antigen or antibody coupled to transducer which detects binding of complementary species (Morgan et al. 1996). Antibodies are proteins, produced by mammals in response to foreign elements (bacteria, viruses, chemicals etc.). This recognition is very specific in general. In an immunoassay, the binding of the analyte (or antigen) to an antibody reflects its concentration in the sample and, therefore, can allow its quantification. When the bioreceptor of a biosensor is an antibody or an active portion of an antibody, such a biosensor is generally called an immunosensor. One of the advantages of antibodies as recognition elements is their high affinity to target analyte which often results in very low detection limits. Disadvantage is that the antigen is not easily released from the antibody after measurement (Rogers and Mascini 2009). A range of analytes can be detected by immunosensors, e.g. hormones, drugs, bacteria and environmental pollutants such as pesticides (Morgan et al. 1996).
Antibodies, with their characteristics of high affinity and excellent selectivity, are ideal analytical tools. The availability of reliable production methods has meant a consistent source of antibody material. Improved assay standardization has resulted from the availability of monoclonal anti- bodies and recombinant antibodies (Killard et al. 1995). The advantages of immunoassay technology relative to other analytical techniques include: low detection limits, high analyte selectivity, high throughput of samples, reduced sample preparation, versatility for target analytes, cost effectiveness for large numbers of samples, adaptability to field use (Shan et al. 2002).
As is the case with every analytical method, immunoassay technology has limitations, including: interferences from sample matrices, cross reactivity to structural analogs of the target analyte, poor suitability for some multi-analyte applications, low availability of reagents, longer assay development time than some classical analytical methods and a large number of anticipated samples required to justify the development of a new assay for an analyte of interest (Shan et al. 2002).
8 Nucleic acid biosensors for phenol detection
In recent years there has been an increase in the use of nucleic acids as a tool in the recognition and monitoring of many compounds of analytical interest (Wang et al. 1997). Nucleic acid layers combined with electrochemical transducers produce a new kind of affinity biosensor. An interesting application of a DNA biosensor will be the testing of water, food, soil, and plant samples for the presence of analytes (carcinogens, drugs, mutagenic pollutants, etc.) with binding affinities for the structure of DNA (Mascini et al. 2001).
The biorecognition mechanism involves hybridization of deoxyribonucleic acid (DNA) or ribonucleic acid (RNA), which are the building blocks of genetics. In the last decade, nucleic acids have received increasing interest as bioreceptors for biosensor and biochip technologies. The complementarity of adenine:thymine (A:T) and cytosine:guanosine (C:G) pairing in DNA forms the basis for the specificity of biorecognition in DNA biosensors, often referred to as genosensors. If the sequence of bases composing a certain part of the DNA molecule is known, then the complementary sequence, often called a probe, can be synthesized and labeled with an optically detectable compound (a fluorescent label). By unwinding the double-stranded DNA into single strands, adding the probe, and then annealing the strands, the labeled probe will hybridize to its complementary sequence on the target molecule.
Moreover, DNA-based affinity biosensors can be used for the detection of selected oligonucleotide sequences. DNA biosensors can detect the presence of genes or mutant genes associated with inherited human diseases (Hashimoto et al. 1994). They can be employed to obtain early and precise diagnoses of infectious agents in various environments (Wang et al. 1997), or can be exploited for monitoring sequences for specific hybridisation events directly or by DNA intercalators (Marrazza et al. 2000), which form complexes with the nitrogenous bases of DNA.
9 Comparison of different methods for the detection of phenol
Various physical and chemical methods are available for the detection and determination of phenolic compounds in environmental samples as HPLC with fluorimetric detection GC/MS, colorimetric methods, or fluorescence excitation emission matrix (EEM). However, these methods require complex sample pre-treatment thus increasing time and cost. Consequently, there is still need for rapid, low-cost and direct methods to accurately quantifying phenol and its derivatives. Recent trend of technology approaches to the miniaturization of devices. It has been observed that biosensors provide analysis that is less costly, easy, less time consuming, highly sensitive and selective to various analyte. So, in this study the development of biosensors has been focused for a most threatening environmental pollutant, phenol, which is listed as a priority pollutant by US EPA. In this chapter working principles and conditions of various biosensors have been accumulated and compared.
9.1 Immobilization
By a careful selection of different immobilization matrices, the selectivity of a biocomponent can be modified to yield a biosensor with good selectivity towards certain targeted analytes. In this study most of the microorganisms has been seen to be immobilized by polyvinyl alcohol as it gives good sensitivity and selectivity. It has been seen that the most used enzyme tyrosinase can be immobilized by a variety of elements, each shows different selectivity. As when immobilized within an Os-complex functionalized electrodeposition polymer assures efficient catechol recycling between the enzyme and the polymer bound redox sites.
Similarly in the laccasse biosensor 3-methyl-2-benzothiazolinone hydrazone (MBTH) was immobilized in a hybrid nafion/sol–gelsilicate film and laccase in a chitosan film. When immobilized in this hybrid film, the biosensor response only to catechol and not other phenolic compounds investigated. Immobilization in this hybrid material has enabled the biosensor to be more selective to catechol compared with the non immobilized enzyme. In the peroxidase biosensor addition of DNA in the silica-titanium paste increased the sensitivity of the biosensor. Also in dual laccasse Tyrosinase based biosensor it was immobilized in sonogel-carbon because of its demonstrated high sensitivity, stability and biocompatibility.
9.2 Method detection limits
Detection limit is very important for determination techniques. The method detection level (MDL) is the minimum concentration of a substrate that can be measured and reported with 99% confidence that the value is above zero. The detection limit of phenolic compounds using various methods is shown in Figs. 1, 2 and 3.
Comparing this it has been found that the biosensor techniques have lower detection limits. So they can detect at more trace levels and are more feasible than these analytical techniques.
9.3 Selectivity
The Rhodococcus sensor was more sensitive to mono- than to di and trichlorinated phenols. Therefore the sensor is almost specific to phenol and chlorinated phenols. Selectivity of Rhodococcus sensor to phenol and their chloro derivatives are shown in Fig. 4.
The Trichosporon biosensor was more sensitive to chlorophenols, especially to mono- and dichlorinated phenols, than to phenol. Other tri-, tetra- and pentachlorophenols are toxic for this organism. Selectivity of a biosensor containing Trichosporon beigelii E4 (cutaneum) to phenol and their chloro derivatives are shown in Fig. 5.
Moraxella sp. has excellent selectivity against phenol derivatives and able to measure p-nitrophenol at a very low concentration. Pseudomonas immobilized in PVC cryogel showed a better sensitivity which is comparable to enzyme sensors. The enzyme Tyrosinase immobilized in Os-complex show a sensitivity of 6.1 nA μM−1 for catechol. The laccasse biosensor 3-methyl- 2-benzothiazolinone hydrazone (MBTH) was immobilized in a hybrid nafion/sol–gel silicate film and laccase in a chitosan film. The use of the hybrid materials of nafion/sol–gel silicate to immobilize laccase has altered the selectivity of the enzyme to various phenolic compounds such as catechol, guaicol, o-cresol and m-cresol when compared to the nonimmobilized enzyme. On the other hand laccasse has also showed a significant sensitivity of different polyphenols. Such as for catechol the value being 0.0216 mA M−1, for catechin 0.02443 mA M−1, for caffeic acid 0.02401 mA M−1. Catechol is the primary derivative of phenol and harmful, so the development and advancement of biosensor for the detection of other phenolic compounds and derivatives are equally important.
9.4 Stability and response time
The major problems that concern biosensor research are the operational and storage stability, as well as the reproducibility. Stability, a characteristic of great importance for the success of these devices as analytical instruments is mainly dependent on the lifetime or the rate of denaturation or inactivation of the employed enzyme. Stability period and responses time of different biosensors are presented in Tables 3 and 4, respectively.
For field monitoring a biosensor should be simple to operate, have short response time, and have excellent multiple use capability and have good long term storage stability. The short analysis time in conjunction with simple protocol for the biosensors are significant advantages of the reported biosensor over the more tedious and time consuming (hours) immunoassay technique. Comparing with the enzymatic biosensors the microbial biosensors have been found to have long response time. Also enzyme biosensors are more stable than microbial. The laccasse based biosensor give a half life of 15 days and its response remain 38% up to 38 days. In the peroxidase based biosensor the sensitivity decreases to 50% after 3 days. But after that remains constant for 25 days. The dual lac-tyrosinase biosensor can maintain its response to 80% until 15 days. So it shows high stability than others. While the stability of some microbial sensors was found only few hours. Comparison of current changes with analyte concentrations are shown in Table 5. In the table relationship of the current change with different concentrations of analytes have been shown. It was found that the devices showed change of current at various concentrations that was very small. It can be said that biosensors can detect at a very trace level. Sometimes the current response can be increased by increasing the volume of biorecognition element and immobilization condition. Such was observed for the tyrosinase detection of phenol and catechol. When the volume of enzyme/polymer was changed from 1:2 to 2:1 it showed a two times higher current response.
10 Conclusions
The sensitivity and selectivity of the numerous existing biosensor prototypes are very high and more improvements are expected. The trends for the development of biosensors are directed towards a miniaturization of the devices and integration of the advances in technological areas as surface chemistry and transduction systems. A variety of microbial biosensors have been developed for environmental, food, military and biomedical application, when compared to enzyme biosensors the development of highly satisfactory microbial biosensor is still hampered because they suffer from long response time, low sensitivity and poor selectivity. Still more research efforts are needed in order to find better alternatives. So, it can be recommended that:
-
1.
Bio elements and chemicals used in the biosensors need to be prevented from leaking out of the biosensor over time.
-
2.
To avoid contamination, biomolecules should attach to the transducer as strongly as possible.
-
3.
If a sterilized probe is used some sensor’s biomolecules may be destroyed whereas if non-sterile probes are used some compromises are needed.
-
4.
The biosensors should be more selective and the detection range should be large.
-
5.
Research should be focused on the development of low-cost biosensors.
At present, with the threat of bioterrorism omnipresent, the development of faster, reliable, accurate, portable and low-cost biosensors has become more important than ever. With a better understanding of the genetic information of microbes and the development of improved recombinant DNA technologies, different enzymes and proteins have been expressed on the cell surface through surface expression anchors. In this format, the microbes can serve as an enzymes support matrix. Through this way, faster response and highly sensitive biosensors can be developed. Moreover, the present great interest and the expected advancements in nanotechnology research will probably allow increasing dramatically the possibilities of use of biosensors. The results of such an evolution are still difficult to anticipate but should allow overcoming the restrictions of both the manufacturers of the area and the restriction of the users.
References
Abdullah J, Ahmad M, Karuppiah N, Heng LY, Sidek H (2005) Immobilization of tyrosinase in chitosan film for an optical detection of phenol. Sens Actuat B 114:604–609
Abdullah J, Ahmad M, Karuppiah N, Heng LY, Sidek H (2007) An optical biosensor based on immobilization of laccase and MBTH in stacked films for the detection of catechol. Sensors 7:2238–2250
Barek J, Ebertova H, Mejstrik V, Zima J (1994) Determination of 2-nitrophenol, 4-nitrophenol, 2-methoxy-5-nitrophenol, and 2,4-dinitrophenol by differential-pulse voltammetry and adsorptive stripping voltammetry. Collect Czech Chem Commun 59:1761–1771
Belkin S (2003) Microbial whole-cell sensing systems of environmental pollutants. Curr Opin Microbiol 6:206–212
Besombes JL, Cosnier S, Labbe P, Reverdy G (1995) Improvement of the analytical characteristics of an enzyme electrode for free and total cholesterol via laponite clay additives. Anal Chim Acta 317:275–280
Bickerstaff GF (1997) Immobilization of enzymes and cells, methods in biotechnology, vol 1. Humana Press Inc., Totowa
Blum LJ, Coulet PR (1991) Biosensor principles and applications. Marcel Dekker, New York
Bravo L (1998) Polyphenols: chemistry, dietary sources, metabolism, and nutritional significance. Nutr Rev 56:317–333
Chang SC, Rawson K, McNeil CJ (2002) Disposable tyrosinase-peroxidase bi-enzyme sensor for amperometric detection of phenols. Biosens Bioelectron 17:1015–1023
Chen X, Cheng G, Dong S (2001) Amperometric tyrosinase biosensor based on a sol–gel derived titanium oxide-copolymer composite matrix for detection of phenolic compounds. Analyst 126:1728–1732
Clement RE, Eiceman GA, Koester CJ (1995) Environmental analysis. Anal Chem 67:221–255
Clesceri LS, Greenberg AE, Eaton AD (1998) Standard methods for the examination of water and waste water, American Public Health Association, 20th edn, pp 6:73–6:78
Cordero-Rando MM, Naranjo-Rodríguez I, Palacios-Santander JM, Cubillana-Aguilera LM, Hidalgo–Hidalgo-de-Cisneros JL (2005) Study of the responses of a sonogel-carbon electrode towards phenolic compounds. Electroanalysis 17:806–814
Cosnier S, Popescu I (1996) Poly(amphiphilicpyrrole)-tyrosinase-peroxidase electrode for amplified flow injection-amperometric detection of phenol. Anal Chem Acta 319:145–151
Crumbliss AL, Stonehuerner J, Henkens RW, O’Daly JP, Zhao J (1994) The use of inorganic materials to control or maintain immobilized enzyme activity. New J Chem 18:327–339
D`Souza SF (2001) Microbial biosensors (review). Biosens Bioelectron 16:337–353
Dai Z, Xu X, Wu L, Ju H (2005) Detection of trace phenol based on mesoporous silica derived tyrosinase-peroxidase biosensor. Electroanalysis 17:1571–1577
Dosemeci M, Blair A, Stewart PA, Chandler J, Trush MA (1991) Mortality among industrial workers exposed to phenol. Epidemiology 2:188–193
Duran N, Rosa MA, D’Annibale A, Gianfreda L (2002) Enzyme Microb Technol 31:907–931
Dzyadevych SV, Arkhypova VN, Soldatkin AP, El’skaya AV, Martelet C, Jaffrezic-Renault N (2008) Amperometric enzyme biosensors: past, present and future. IRBM 29:171–180
Elkaoutit M, Naranjo-Rodriguez I, Temsamani KR, La Vega MD, Hidalgo–Hidalgo-de-Cisneros JL (2007) Dual laccase–tyrosinase based sonogel–carbon biosensor for monitoring polyphenols in beers. J Agric Food Chem 55:8011–8018
Ferreira M, Varela H, Toressi RM, Tremiliosi-Filho G (2006) Electrode passivation caused by polymerization of different phenolic compounds. Electrochim Acta 52:434–442
Freire RS, Durana N, Kubota LT (2002) Electrochemical biosensor-based devices for continuous phenols monitoring in environmental matrices. J Braz Chem Soc 13:119–123
Gomes SASS, Rebelo MJF (2003) A new laccasse biosensor for polyphenol determination. Sensors 3:166–175
Hashimoto K, Ito K, Ishimori Y (1994) Sequence-specific gene detection with a gold electrode modified with DNA probes and an electrochemically active dye. Anal Chem 66:3830–3833
Hernandez L, Hernandez P, Vicente J (1993) Voltammetric determination of methyl parathion, ortho, meta and para nitrophenol with a carbon paste electrode modified with C-18. Fresenius J Ana Chem 345:712–715
IARC (1989) IARC monographs on the evaluation of carcinogenic risks to humans, some organic solvents, resin monomers and related compounds, pigments and occupational exposures in paint manufacture and painting, International Agency for Research on Cancer, Lyon, 47: 263–287
Killard AJ, Deasey B, O’Kennedy R, Smyth MR (1995) Antibodies production, functions and applications in biosensors. Trends Anal Chem 14:257–266
Kim MA, Lee WY (2003) Amperometric phenol biosensor based on sol-gel silicate/nafion composite film. Anal Chim Acta 479:143–150
King JMH, DiGrazia PM, Applegate B, Burlage R, Sanseverino J, Dunbar P, Larimer F, Sayler GS (1990) Rapid sensitive bioluminescent reporter technology for naphthalene exposure and biodegradation. Science 249:778–781
Lei Y, Mulchandani P, Chen W, Wang J, Mulchandani A (2003) A microbial biosensor for p-nitrophenol using arthrobacter Sp. Electroanalysis 15:1160–1164
Lei Y, Chen W, Mulchandani A (2006) Microbial biosensors. Anal Chim Acta 568:200–210
Leveau JHJ, Lindow SE (2002) Bioreporters in microbial ecology. Curr Opin Microbiol 5:259–265
Li N, Xue MH, Yao H, Zhu JJ (2005) Reagent less biosensor for phenolic compounds based on tyrosinase entrapped within gelatine film. Anal Bioanal Chem 383:1127–1132
Lindgren A, Emneus J, Ruzgas T, Gorton L, Marko-Varga G (1997) Amperometric detection of phenols using peroxidase-modified graphite electrodes. Anal Chim Acta 347:51–62
Liu Z, Liu B, Kong J, Deng J (2000) Probing trace phenols based on mediator-free alumina sol-gel-derived tyrosinase biosensor. Anal Chem 72:4707–4712
Makarenko AA, Bezverbnaya IP, Kosheleva IA, Kuvichkina TN, Il’yasov PV, Reshetilov AN (2002) Development of biosensors for phenol determination from bacteria found in petroleum fields of West Siberia. Appl Biochem Microbiol 38:23–27
Marrazza G, Chiti G, Mascini M, Anichini M (2000) Detection of human apolipoprotein E genotypes by DNA electrochemical biosensor coupled with PCR. Clin Chem 46:31–37
Mascini M, Palchetti I, Marrazza G (2001) DNA electrochemical biosensors. Fresenius J Anal Chem 369:15–22
Mehrvar M, Bis C, Scharer J, Moo-Young M, Luong J (2000) Fiber-optic biosensors: trends and advances. Anal Sci 16:677–692
Mello LD, Sotomayor MDPT, Kubota LT (2003) HRP-based amperometric biosensor for the polyphenols determination in vegetables extract. Sens Actuat B 96:636–645
Morgan CL, Newman DJ, Price CP (1996) Immunosensors: technology and opportunities in laboratory medicine. Clin Chem 42:193–209
Morimoto K, Koizumi A, Tachibana Y, Dobashi Y (1976) Inhibition of repair of radiation induced chromosome breaks, effect of phenol in cultured human leukocytes. Jpn J Ind Health 18:478–479
Mulchandani P, Hangarter CM, Lei Y, Chen W, Mulchandani A (2005) Amperometric microbial biosensor for p-nitrophenol using Moraxella sp.-modified carbon paste electrode. Biosens Bioelectron 21:523–527
Nikolelis D, Krull U, Wang J, Mascini M (1998) Biosensors for direct monitoring of environmental pollutants in field. Kluwer Academic, London
Nistor C, Oubi˜na A, Marco MP, Barcelo D (2001) Competitive flow immunoassay with fluorescence detection for determination of 4-nitrophenol. Anal Chim Acta 426:185–195
Poirier MC, De Cicco BT, Lieverman MW (1975) Nonspecific inhibition of DNA repair synthesis by tumor promoters in human diploid fibroblasts damaged with N-acetoxy-2-acetyl aminofluorene. Cancer Res 35:1392–1397
Popescu IC, Zetterberg G, Gorton L (1995) Influence of graphite powder, additives and enzyme immobilization procedures on a mediator less HRP-modified carbon paste electrode for amperometric flow injection detection of H2O2. Biosens. Bioelectron 10:443–461
Rainina EI, Badalian IE, Ignatov OV, Fedorov AYU, Simonian AL, Varfolomeyev SD (1996) Cell biosensor for detection of phenol in aqueous solutions. Appl Biochem Biotechnol 56:117–127
Rensing C, Maier RM (2003) Issues underlying use of biosensors to measure metal bioavailability. Ecotoxicol Environ Saf 56:140–147
Riedel K, Hensel J, Rothe S, Neumann B, Scheller F (1993) Microbial sensors for determination of aromatics and their chloroderivatives. Part II: determination of chlorinated phenols using a Rhodococcus-containing biosensor. Appl Microbiol Biotechnol 38:556–559
Riedel K, Beyersdorf-Radeck B, Neumann B, Schaller F (1995) Microbial sensors for determination of aromatics and their chloro derivatives. Part II1: determination of chlorinated phenols using a biosensor containing Trichosporonbeigelii (cutaneum). Appl Microbiol Biotechnol 43:7–9
Rodriguez IN, Barea Zamora M, Barber′a Salvador JM, Mu˜nozLeyva JA (1997a) Use of a bentonite-modified carbon paste electrode for the determination of some phenols in a flow system by differential-pulse voltammetry. Analyst 122:601–604
Rodriguez IN, Mu˜noz Leyva JA, Hidalgo Hidalgo de Cisneros JL (1997b) Use of a carbon paste modified electrode for the determination of 2-nitrophenol in a flow system by differential pulse voltammetry. Anal Chim Acta 344:167–173
Rodriguez IN, Barea Zamora M, Barber′a Salvador JM, Mu˜nozLeyva JA, Hernandez-Artiga MP, Hidalgo Hidalgo de Cisneros JL (1997c) Voltammetric determination of 2-nitrophenol at a bentonite-modified carbon. Mikrochim Acta 126:87–92
Rodriguez-Mozaz S, López de Alda MJ, Marco MP, Barceló D (2006) Biosensors as useful tools for environmental analysis and monitoring. Anal Bioanal Chem 386(4):1025–1041
Rogers KR, Mascini M (2009) Biosensors for analytical monitoring—general introduction and review. In: US Environmental Protection Agency, http://www.epa.gov/heasd/edrb/biochem/intro.html Accessed 5th Jan, 2011
Rogers KR, Becker JY, Cembrano J, Chough SH (2001) Viscosity and binder composition effects on tyrosinase-based carbon paste electrode for detection of phenol and catechol. Talanta 54:1059–1065
Rosatto SS, Kubota LT, Oliveira NG (1999) Biosensor for phenol based on the direct electron transfer blocking of peroxidase immobilising on silica-titanium. Anal Chim Acta 390:65–72
Rosatto SS, Neto GO, Kubota LT (2001) Effect of DNA on peroxidase based biosensor for phenol determination in waste waters. Electroanalysis 6:445–450
Russell IM, Burton SG (1999) The development of an immobilized enzyme bioprobe for the detection of phenolic pollutants in water. Anal Chim Acta 389:161–170
Ruzgas T, Emneus J, Gorton L, Marko-Varga G (1995) The development of a peroxide biosensor for monitoring phenol and related aromatic compounds. Anal Chim Acta 311:245–253
Scouten WH, Luong JHT, Brown RS (1995) Enzyme or protein immobilization techniques for applications in biosensor design. Tibtech 13:178–185
Shan G, Lipton C, Gee SJ, Hammock BD (2002) Immunoassay, biosensors and other nonchromatographic methods. In: Lee PW (ed), Handbook of residue analytical methods for agrochemicals, pp 623–679
Singh M, Verma N, Garg AK, Redhu N (2008) Urea biosensors. Sens Actuat B-Chem 134:345–351
Solomon EI, Lowery MD (1993) Electronic structure contributions to function in bioinorganic chemistry. Science 259:1575–1581
Spain JC, Gibson DT (1991) Pathway for biodegradation of p-nitrophenol in a Moraxella sp. Appl Environ Microbiol 57:812–819
Straube G, Hensel J, Niedan AC, Straube E (1990) Kinetic studies of phenol degradation by Rhodococcus sp. P1. I. Batch cultivation. Antonie van Leeuwenhoek 57(3):29–32
Svitel J, Miertus S (1998) Development of tyrosinase-based biosensor and its application for monitoring of bioremediation of phenol and phenolic compounds. Environ Sci Technol 32:828–832
Svorc J, Miertus S, Katrlik J, Stredansky M (1997) Composite transducers for amperometric biosensors: the glucose sensor. Anal Chem 69:2086–2090
Tembe S, Inamder S, Haram S, Karvee M, D’Souza SF (2007) Electrochemical biosensor for catechol using agarose-guar gum entrapped tyrosinase. J Biotechnol 128:80–85
Timur S, Pazarlioğlu N, Pilloton R, Telefoncu A (2003) Detection of phenolic compounds by thick film sensors based on Pseudomonas putida. Talanta 61:87–93
Tran MC (1993) Biosensors. Chapman and Hall and Masson, Paris
Turner APF, Karube I, Wilson GS (1992) Biosensors: fundamentals and applications. Mir Publishers, Moscow
Vo-Dinh T, Cullum B (2000) Biosensors and biochips: advances in biological and medical diagnostics. Fresenius J Anal Chem 366:540–551
Walcarius A (1998) Analytical applications of silica-modified electrodes: a comprehensive review. Electroanalysis 10:1217–1235
Wang J, Cai X, Rivas G, Shiraishi H, Dontha N (1997) Nucleic acid immobilization, recognition and detection at chronopotentiometric DNA chips. Biosens Bioelectron 12:587–599
Xue H, Shen Z (2002) A highly stable biosensor for phenols prepared by immobilizing polyphenol oxidase into polyaniline-polyacrylonitrile composite matrix. Talanta 57:289–295
Yang S, Li Y, Jiang X, Chen Z, Lin X (2005) Horseradish peroxidase biosensor based on layer by-layer technique for the determination of phenolic compounds. Sens Actuat B 114:774–780
Yildiz HB, Castillo J, Guschin DA, Toppare L, Schuhmann W (2007) Phenol biosensor based on electrochemically controlled tyrosinase in a redox polymer. Microchimacta 159:27–34
Yu J, Ju H (2004) Pure organic phase phenol biosensor based on tyrosinase entrapped in a vapor deposited titania sol-gel membrane. Electroanalysis 16:1305–1310
Yu J, Liu S, Ju H (2003) Mediator free phenol sensor based on titania sol-gelincapsulation matrix for immobilization of Tyrosinase by a vapor deposition method. Biosens Bioelectron 19:509–514
Zhao Z, Jiang H (2010) Enzyme-based electrochemical biosensors, p 302. In: Serra PA (ed) Biosensors. ITECH, Crotia
Zietek M (1975) Polarographic-determination of parathion and its metabolite p-nitrophenol in blood extracts. Mikrochim Acta 64:463–470
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Karim, F., Fakhruddin, A.N.M. Recent advances in the development of biosensor for phenol: a review. Rev Environ Sci Biotechnol 11, 261–274 (2012). https://doi.org/10.1007/s11157-012-9268-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11157-012-9268-9