Abstract
In higher plant chloroplasts, the plastid-encoded RNA polymerase (PEP) consists of four catalytic subunits and numerous nuclear-encoded accessory proteins, including pTAC10, an S1-domain-containing protein. In this study, pTAC10 knockout lines were characterized. Two ptac10 mutants had an albino phenotype and severely impaired chloroplast development. The pTAC10 genomic sequence fused to a four-tandem MYC tag driven by its own promoter functionally complemented the ptac10-1 mutant phenotype. pTAC10 was present in both the chloroplast stroma and thylakoids. Two-dimensional blue native polyacrylamide gel electrophoresis (BN-PAGE), and immunoblotting assays showed that pTAC10:MYC co-migrates with one of the PEP core subunits, RpoB. A comprehensive investigation of the plastid gene expression profiles by quantitative RT-PCR revealed that, compared with wild-type plants, the abundance of PEP-dependent plastid transcripts is severely decreased in the ptac10-1 mutant, while the amount of plastid transcripts exclusively transcribed by NEP either barely changes or even increases. RNA blot analysis confirmed that PEP-dependent chloroplast transcripts, including psaB, psbA and rbcL, substantially decrease in the ptac10-1 mutant. Immunoblotting showed reduced accumulation of most chloroplast proteins in the ptac10 mutants. These data indicate the essential role of pTAC10 in plastid gene expression and plastid development. pTAC10 interacts with chloroplast-targeted casein kinase 2 (cpCK2) in vitro and in vivo and can be phosphorylated by Arabidopsis cpCK2 in vitro at sites Ser95, Ser396 and Ser434. RNA-EMSA assays showed that pTAC10 is able to bind to the psbA, atpE and accD transcripts, suggesting a non-specific RNA-binding activity of pTAC10. The RNA affinity of pTAC10 was enhanced by phosphorylation and decreased by the amino acid substitution Ser434-Ala of pTAC10. These data show that pTAC10 is essential for plastid gene expression in Arabidopsis and that it can be phosphorylated by cpCK2.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chloroplasts originated from a cyanobacterium-like ancestor that retained its own genome (Sato et al. 1999; Wakasugi et al. 2001; Berry et al. 2013). The transcription of plastid genes in higher plants involves two types of RNA polymerases: a plastid-encoded eubacterial-type RNA polymerase (PEP) and a nuclear-encoded phage-type RNA polymerase (NEP) (Hu and Bogorad 1990; Link 1996). Previously, a general concept of plant development was that higher levels of NEP are present in young, non-green tissues in early leaf development. NEP transcribes housekeeping genes, including rpoA, rpoB, rpoC1 and rpoC2, which encode the PEP core subunits. Once the PEP complex is formed, it transcribes photosynthesis-related genes (Hajdukiewicz et al. 1997; Lopez-Juez and Pyke 2005) and plastid tRNAs (Williams-Carrier et al. 2014) during later developmental stages. In mature chloroplasts, PEP activity is maintained at high levels for chloroplast development and plant growth, while NEP activity is hardly detected. Nonetheless, recent work shows that both NEP and PEP are present in seeds, suggesting that PEP is also important for efficient germination and plant development at an early stage of development (Demarsy et al. 2006).
The PEP transcriptional machinery in higher plants has two distinct forms: PEP-A and PEP-B (Pfalz and Pfannschmidt 2013). PEP-B is composed of four plastid-encoded core subunits (α2ββ′β″) and the sigma factor, a promoter recognition subunit. This form is present in both etioplasts and greening chloroplasts. PEP-A is the major RNA polymerase in mature chloroplasts of higher plants. The core subunits of PEP-A are associated with numerous accessory proteins (Pfannschmidt and Link 1994; Pfalz et al. 2006; Pfalz and Pfannschmidt 2013; Yang et al. 2015). The characterization of these accessory proteins in the PEP complex is essential for understanding the plastid transcription mechanisms. Over the years, many research groups have attempted to isolate and identify these proteins. Among those studies, a breakthrough was achieved through the isolation of insoluble PEP polymerase, named transcriptionally active chromosome (TAC), by mass spectrometry in both Arabidopsis thaliana and mustard (Sinapis alba) chloroplasts (Suzuki et al. 2004; Pfannschmidt et al. 2000; Pfalz et al. 2006; Steiner et al. 2011; Melonek et al. 2012). Pfalz et al. (2006) identified 35 proteins from purified TAC, and 18 of them were named pTACs (pTAC1 to pTAC18). Most of these pTACs are encoded by the nuclear genome, and they are confined to DNA transcription and RNA stabilization during plastid division and inheritance (Pfalz and Pfannschmidt 2013). In 2011, Steiner et al. identified 12 proteins that were tightly associated with PEP core subunits. These were named polymerase-associated proteins (PAPs) (Steiner et al. 2011; Pfalz and Pfannschmidt 2013). The inactivation of PAP genes leads to an albino/ivory or pale-green phenotype and impaired chloroplast development in Arabidopsis. The abundance of PEP-dependent plastid transcripts is reduced in PAP-deficient lines (Pfalz et al. 2006; Garcia et al. 2008; Myouga et al. 2008; Arsova et al. 2010; Schröter et al. 2010; Gao et al. 2011; Steiner et al. 2011; Yagi et al. 2012; Gilkerson et al. 2012; Yu et al. 2013, 2014). Besides the PAPs in the plastid transcriptional machinery, the ATase2 enzyme which catalyses the first step of de novo purine biosynthesis in plastids is also involved in regulating PEP activity. Taken together, all these proteins are essential for maintaining PEP activity (Yang et al. 2015). These proteins might have conserved functions in plastid transcription in higher planta. Silencing of the STF (S1-domain-containing Transcription-stimulating Factor), the orthologue of pTAC10, in Nicotiana benthamiana plants by virus-induced gene silencing (VIGS), leads to abnormal chloroplast development and reductions of PEP-dependent chloroplast transcripts. Tobacco STF has been reported to bind RNA through the S1 domain (Jeon et al. 2012). Recently, Chang et al. (2017) showed that pTAC10 interacts with other PAPs through its C-terminal domain. Deletion of this domain prevented the truncated pTAC10 to complement the mutant phenotype, suggesting that pTAC10-PAP interactions are essential for the formation of the PEP complex in Arabidopsis thaliana. However, the analysis of ptac10 mutant features is still very limited. Therefore, the comprehensive characterization of Arabidopsis ptac10 mutants in this study is essential for an improved understanding of the function of pTAC10.
Protein phosphorylation is one of the most important post-translational modifications, leading to the activation or deactivation of many basic cellular signals in all sub-cellular compartments (Baginsky and Gruissem 2009). Casein kinase 2 (CK2) is a ubiquitous serine–threonine protein kinase that is highly conserved in all eukaryotes (Ghavidel and Schultz 2001). CK2 exists as a tetrameric complex formed by two catalytic subunits (α) and two regulatory subunits (β) (Niefind et al. 2001; Litchfield 2003). In Arabidopsis, four catalytic subunit genes and four regulatory subunit genes of CK2 have been identified (Salinas et al. 2006). Among these subunits, only one catalytic subunit, named cpCK2, is localized in chloroplasts (Salinas et al. 2006). cpCK2 was initially named plastid transcription kinase (PTK), which was originally co-purified with the plastid RNA polymerase complex by heparin-sepharose chromatography and glycerol density-gradient centrifugation (Pfannschmidt and Link 1994; Baginsky et al. 1997; Schröter et al. 2010; Lu et al. 2015). In vitro transcription analysis showed that cpCK2 regulates transcription via the phosphorylation of RNA polymerase/TAC subunits (Baginsky et al. 1999). For instance, the phosphorylation of sigma factor 6 (AtSIG6) by cpCK2 was reported to regulate PEP promoter recognition during chloroplast gene transcription (Schweer et al. 2010). Schönberg et al. (2014) developed a microarray with 905 peptides based on previously determined phosphorylation sites in chloroplast proteins, and most of the targets identified in this study are involved in plastid gene expression. However, the mechanisms through which cpCK2 regulates PEP polymerase and plastid transcription remain largely unknown. Based on phosphoproteomic studies and the CK2 substrate motif, it was also proposed that pTAC10 could be a cpCK2 substrate (Reiland et al. 2009). In this study, the potential mechanism by which pTAC10 phosphorylation (by cpCK2) regulates plastid gene transcription has also been evaluated.
Here, we have characterized two allelic null ptac10 mutants of Arabidopsis and demonstrate that pTAC10 is essential for plastid gene expression and plastid development in Arabidopsis thaliana. pTAC10 interacts with cpCK2 in vitro and in vivo, and it can be phosphorylated by this kinase in vitro. RNA-EMSA assays show that pTAC10 can bind to psbA, atpE and accD transcripts, which suggests that pTAC10 binds RNA non-specifically. The RNA affinity of pTAC10 is enhanced by its phosphorylation and decreased by amino acid substitution Ser434-Ala.
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana wild-type (ecotype Columbia) and two null ptac10 mutants (CS16115 and CS16140) were obtained from the Arabidopsis Biological Resource Center (ABRC; Ohio State University) and grown in a growth chamber with a 16-h light/8-h dark photoperiod at a constant temperature of 22 °C. The light intensity was 120 μmol m−2 s−1. For growth on agar plates, the seeds were surface-sterilized with 75% alcohol and sown on Murashige–Skoog (MS) medium containing 2% sucrose and 0.7% (w/v) phytoagar.
Genomic complementation
Both the 4299 and the 3836 kb wild-type genomic fragments of pTAC10 were amplified using KOD plus polymerase (TOYOBO, http://www.toyobo-global.com/) with gene-specific primers, and then sub-cloned into the pCAMBIA1300 binary vector (CAMBIA, http://www.cambia.org.au) and the modified pCAMBIA1300 vector with four tandem MYC tags, respectively. Both constructs were introduced into pTAC10/ptac10-1, and the genomic backgrounds of the hygromycin-resistant transgenic lines were further analysed. All binary vectors were introduced into Arabidopsis through Agrobacterium tumefaciens (Clough and Bent 1998). Transformants were screened on MS medium with 80 mg L−1 hygromycin B (Roche, http://www.roche.com). The primer sequences for the constructs are listed in supporting information Table S1.
Transmission electron microscopy
Samples were prepared as described in our previous report (Yu et al. 2013). Thin sections were prepared with an ultramicrotome and stained with uranyl acetate as well as lead citrate. The ultrastructure was examined using a Hitachi H7500 transmission electron microscope (Hitachi, http://www.hitachi.com).
RNA isolation, cDNA synthesis, RT-PCR, real-time PCR and RNA gel-blot hybridization
Procedures for the purification of total RNA for cDNA synthesis, RT-PCR and real-time PCR were performed as described (Yu et al. 2009). Total RNA was extracted from approximately 14-day-old seedlings. Next, 10 μg total RNA sample in each lane was used to perform RNA gel-blot hybridizations. Hybridization with digoxygenin (DIG)-labelled probes was carried out following the Roche manual (Roche, http://www.roche.com). The primer sequences for the DNA probes are provided in our previous reports (Gao et al. 2011; Yu et al. 2013; Zhang et al. 2015). The primer sequences for qRT-PCR analysis are listed in supporting information Table S2.
Sequence alignment and phylogenetic tree construction
Protein sequences were aligned with ClustalW (Thompson et al. 1994). The phylogenetic tree was constructed and tested using MEGA3.1 (http://www.megasoftware.net) based on the neighbour-joining method.
Purification of recombinant proteins and phosphorylation assay
Both full-length pTAC10 and cpCK2 lacking the N-terminal transpeptides were cloned into the protein expression vectors, pMAL-c5x (NEB, http://www.neb.com) and pMAL-c2x (NEB, http://www.neb.com), respectively. They were expressed in Escherichia coli BL21 (DE3) pLysS strain (Promega, http://www.promega.com), respectively. Recombinant proteins were purified on amylose resin according to the manual instruction. The purified pTAC10 protein (15–20 μg) was phosphorylated by either commercial CK2 kinase (NEB, http://www.neb.com) or the purified cpCK2 protein in 50-μL reactions containing 20 mM Tris–HCl, pH 7.5, 50 mM KCl, 10 mM MgCl2, 0.1 mM ATP and 10 μg (cp) CK2 at 30 °C for 30 min, as described by Ogrzewalla et al. (2002). Phosphorylated pTAC10 protein was analysed using anti-phospho antibodies from Invitrogen (http://www.invitrogen.com/).
Mass spectrometry
Purified pTAC10 protein was phosphorylated using commercial CK2 kinase (NEB, http://www.neb.com) or purified cpCK2 protein. Dried proteins were digested with trypsin at 37 °C for 20 h. Peptides were concentrated using the nano-HPLC system (Agilent Technologies, http://www.agilent.com/) equipped with a RP-C18 column (Column Technology Inc., http://www.columntechnology.com/). MS were recorded with a LTQ ORBITRAP mass spectrometer (Thermo Finnigan, http://www.thermoscientific.com/). The original data were searched using the Mascot 2.2 data base (Matrix Science, http://www.matrixscience.com/). The mass spectrometry and the peptide analysis were conducted by Shanghai Applied Protein Technology Co. Ltd (http://www.aptbiotech.com/) and Hangzhou Jingjie Protein Technology Co. Ltd (http://www.ptm-biolab.com.cn/), respectively.
RNA-protein EMSA
The probes were prepared using a cloned fragment into pGEM T-Easy (Promega, http://www.promega.com). After cutting out the fragment with SalI (Takara, http://www.takara-bio.com) and purifying it electrophoretically, it was transcribed using T7 RNA polymerase and then labelled using the RNA 3′ End Biotinylation Kit (Thermo, http://www.thermoscientific.com/pierce). EMSA analysis was carried out using the LightShift Chemiluminescent RNA EMSA Kit (Thermo, http://www.thermoscientific.com/pierce). The binding reactions for the interactions between recombinant pTAC10 and RNA included the following reagents 1× binding buffer, 5% glycerol, 25 mM DTT, 0.1 M KCl, 0.05 M MgCl2, 0.2 mg mL−1 tRNA, unphosphorylated or phosphorylated pTAC10 protein that was treated by the CK2 kinase, and the RNA probe in a 20-μL volume. To ensure the equal concentration of the phosphorylated and unphosphorylated pTAC10 in the RNA-EMSA reaction, equal amounts of protein from the phosphorylation reaction and the control reaction were incubated with the probes. Compared with the phosphorylation reaction, there was no ATP in the control reaction.
Chloroplast isolation, immunoblot, SDS–PAGE and blue native polyacrylamide gel electrophoresis (BN-PAGE) analyses
2- to 3-week-old seedlings were homogenized in ice-cold STN2 buffer (0.8 M sucrose, 0.5 M Tris–HCl pH 7.6, 1 M MgCl-6H2O, 5 M NaCl, 10% BSA). The homogenate was filtered successively through a double layer of 100- and 40-µm Miracloth sieves. The filtrate was centrifuged in a swing-out rotor at 2000×g for 5 min at 4 °C, and the intact chloroplasts were resuspended in STN2 and centrifuged. Finally, the intact chloroplasts were resuspended in STN2 buffer. Thylakoid membranes and stromal proteins were prepared from isolated intact chloroplasts according to Stöckel et al. (2006).
Total protein preparation and immunoblot analyses were performed according to our previous studies (Yu et al. 2013). The polyclonal antibodies against photosynthesis-related protein used in this study were obtained from Agrisera (http://www.agrisera.com). For BN-PAGE analysis, the isolated thylakoid pellets were suspended in resuspension buffer (25 mM Bis–Tris–HCl, pH 7.0, 1% n-dodecyl b-d-maltoside and 20% glycerol [w/v]) at 1.0 mg chlorophyll mL−1. After incubation at 4 °C for 5 min and centrifugation at 12,000×g for 10 min, the supernatant was added to one-tenth volume of loading buffer (100 mM Bis–Tris–HCl, pH 7.0, 0.5 M 6-amino-n-caproic acid, 5% Serva blue G and 30% [w/v] glycerol) and applied to 0.75-mm-thick 4–12% acrylamide gradient gels in a Tannon vertical electrophoresis apparatus at 4 °C. For two-dimensional analysis, excised BN-PAGE lanes were soaked in SDS sample buffer for 30 min and layered onto 1-mm-thick 10% SDS polyacrylamide gels. After electrophoresis, the proteins were transferred to nitrocellulose membranes, probed with the MYC antibodies and RpoB, and visualized using the enhanced chemiluminescence method.
Results
pTAC10 knock-out lines display an albino phenotype and impaired chloroplast development
The full-length pTAC10 (At3g48500) coding sequence consists of 2007 base pairs (bp) encoding a 669-amino-acid polypeptide with a molecular mass of 78.82 kDa. The TargetP (http://www.cbs.dtu.dk/services/TargetP/) algorithm predicts that pTAC10 is localized in the chloroplast with a 40-amino-acid chloroplast transit peptide (cTP) at its N-terminus (Fig. 1a). pTAC10 contains a single S1 RNA-binding domain from residue 268 to 340 which is highly conserved among its orthologs. Database searching revealed that there is only one copy of TAC10 in Arabidopsis, and orthologs are present in various terrestrial plants from mosses to angiosperms but not in photosynthetic bacteria (Supplementary Fig. 1). The expression data from the Genevestigator database (Zimmermann et al. 2004; http://www.genevestigator.com) show that pTAC10 is widely expressed in Arabidopsis tissues. Consistent with these data, semi-quantitative reverse transcription (RT)-PCR showed that pTAC10 is expressed in roots, stems, leaves and flowers of 14-day-old wild-type seedlings of Arabidopsis (Fig. 1b). Quantitative real-time RT-PCR further showed that pTAC10 mRNA is highly abundant in leaves, but at lower levels in both stems and roots (Fig. 1c).
Sequence and expression analysis of pTAC10 protein. a Schematic representation of pTAC10 protein. cTP (chloroplast transit peptide) and the S1 domain are indicated. b and c expression of the pTAC10 mRNA determined by RT-PCR and quantitative real-time RT-PCR in root (Rt), stem (St), leaf (Le), flower (Fl) and 14-day-old Arabidopsis seedling (Se). The tubulin gene of Arabidopsis was used as a control
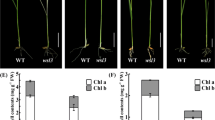
To investigate the function of pTAC10, we obtained two independent homozygous T-DNA insertion lines for pTAC10. The T-DNAs were inserted in the seventh exon (position 17,964,170 in chromosome 3) of CS16115 (ptac10-1) and in the second intron (position 17,962,469 in chromosome 3) of the CS16140 line (ptac10-2) (Fig. 2a). Both homozygous mutants display albino cotyledons and primary leaves, and thus they are seedling lethal when grown on either soil or MS medium supplemented with 2% sucrose (Fig. 2b). The plastid ultrastructure of ptac10 mutants is highly vacuolated with fewer thylakoids compared with the organized internal membranes in wild-type plastids (Fig. 2c). These results indicate that the two allelic null ptac10 mutants are seriously impaired in chloroplast development. Genetic analysis showed that the albino phenotype of ptac10 mutants is controlled by a single recessive nuclear gene that co-segregates with the T-DNA insertion. The pTAC10 transcripts were present in wild-type leaves but not in either ptac10 mutant lines (Fig. 2d). To further confirm that the knockout of the pTAC10 gene leads to an albino phenotype, the AT3G48500 genomic sequence, including its 907-bp upstream and 232-bp downstream sequences, was introduced into heterozygous plants (pTAC10-1/ptac10-1) by Agrobacterium tumefaciens-mediated transformation (Clough and Bent 1998). 5 out of 41 independent T1 transgenic plants were identified as ptac10-1/ptac10-1 genotypes. These lines had the same appearance as the wild-type plants (Fig. 2b). The transcripts of pTAC10 were detected in those complemented plants by RT-PCR analysis (Fig. 2d). The plastid ultrastructure of two independent complemented lines revealed organized thylakoids as in wild-type (Fig. 2c). These results indicate that the deficiency of pTAC10 (AT3G48500) is responsible for the albino phenotype, which could be rescued by genomic complementation.
Characterization of ptac10 knockout lines and chloroplast gene expression in the ptac10-1 line. a Gene structure of pTAC10 with T-DNA insertions indicated. Black boxes, exons; grey boxes, untranslated region. Triangles, the T-DNA insertion sites in ptac10-1 and ptac10-2. b Phenotype of wild type, ptac10-1, ptac10-2 and the complemented lines (TC) grown on soil or MS medium containing 2% sucrose for 14 days. Scale bar 2 mm. c Chloroplast ultrastructure of 14-day-old wild type and ptac10-1. Scale bars 0.5 µm. d RT-PCR analysis of pTAC10 expression in WT, ptac10-1, ptac10-2 and the complemented lines (TC)
pTAC10 is localized in both chloroplast stroma and thylakoids, and co-migrates with the PEP complex
Previous work has shown that the pTAC10 ortholog in tobacco, STF (Jeon et al. 2012), is localized in the chloroplast, which is consistent with previous chloroplast proteomics data (Pfalz et al. 2006). Here, we investigated the sub-cellular location of pTAC10 in Arabidopsis. The full-length pTAC10 genomic sequence was fused to four tandem MYC-tags at the C-terminus (Fig. 3a). The recombinant plasmid was transferred into pTAC10/ptac10-1 heterozygous Arabidopsis. Genetic screening identified seven transgenic lines with the ptac10-1/ptac10-1 background, and those transgenic plants displayed a wild-type-like phenotype (Fig. 3d). Immunoblotting was performed with anti-MYC antibody using total protein from the transgenic seedlings and wild-type controls. A specific band at approximately 100 kDa was observed in the transgenic lines that was absent in the wild-type controls (Fig. 3b). These results show that the pTAC10:MYC construct can functionally complement the ptac10-1 mutant.
pTAC10:MYC transgenic line and pTAC10 localization in Arabidopsis chloroplast. a The schematic construct of the pTAC10 genomic sequence fused to MYC. b Phenotypes of wild-type and pTAC10:MYC/ptac10 transgenic seedling grown on MS medium containing 2% sucrose for 14 days. Scale bar 2 cm. Genotype identification of wild-type and transgenic seedling is shown with the specific primers. c Immuno-blotting analysis of total protein from both wild-type and pTAC10:MYC/ptac10 transgenic seedling with MYC antibody. d pTAC10 is present in both stroma and thylakoids. Fractionation of chloroplasts from transgenic Arabidopsis seedling was performed and cellular fractions were examined by immunoblotting with anti-MYC, anti-PsaD, anti-Lhcb3 and anti-RbcS antibodies. Tc Total chloroplast protein, Str stromal, Thy thylakoid fraction. e Immunoblot detection of pTAC10 and RpoB by two-dimensional gel electrophoresis. Thylakoid membrane proteins from 3-week-old pTAC10:MYC seedlings were fractionated by BN-PAGE in the first dimension and by SDS–PAGE in the second dimension. The approximate molecular masses of the labelled protein complexes are indicated above
To further determine the localization of pTAC10, chloroplast stroma and thylakoids were isolated from the pTAC10:MYC complemented plants. Featured proteins of each compartment, such as the light-harvesting complex of PSII (Lhcb3) and PsaD in the thylakoids and Rubisco (RbcS) in the stroma, were used as controls to ensure the reliability of our isolation protocol (Fig. 3c). Immunoblotting using anti-MYC antibody showed that pTAC10:MYC localizes in both the chloroplast stroma and thylakoids (Fig. 3c). Moreover, the distribution of pTAC10 was assessed by two-dimensional (2D) gel electrophoresis. Thylakoid membrane proteins from 3-week-old pTAC10:MYC transgenic seedlings were fractionated on blue native (BN) gels and subsequently analysed by SDS–PAGE and immunoblotting (Fig. 3e). One of the PEP core subunits, RpoB, has been reported to migrate close to 669 kDa, considered to be the PEP complex (Zhong et al. 2013). Our results show that pTAC10:MYC migrates at the same position as RpoB on the 2D gel, suggesting its co-migration with the PEP complex (Fig. 3e). Taken together, pTAC10 localizes in both stroma and thylakoids, and it co-migrates with one of the core subunits of the PEP complex, RpoB, suggesting that pTAC10 is indeed associated with the PEP complex.
Chloroplast gene expression patterns in the ptac10-1 mutant
pTAC10 is one essential component of the PEP complex (Pfalz et al. 2006; Steiner et al. 2011). We systematically examined the plastid gene expression patterns in the ptac10-1 mutant by qRT-PCR. As shown clearly in Fig. 4, the general plastid gene expression pattern of the ptac10-1 mutant was severely perturbed compared to that of wild type. In detail, plastid genes encoding subunits of photosystem I (PS I), photosystem II (PS II) and the cytochrome b6/f complex had greatly decreased transcript levels in the ptac10-1 mutant compared with wild type. For example, two typical PS I genes, psaA and psaB, which are exclusively transcribed by the PEP complex, had RNA levels that were substantially reduced to 10 and 20% of wild-type in the ptac10-1 mutant. Another typical PEP-transcribed gene is rbcL (large subunit of ribulose-1,5-bis-phosphate carboxylase/oxygenase). The accumulation of rbcL transcripts in the ptac10-1 mutants was less than 25% of that in wild type. Other PEP-transcribed genes displayed similar expression trends in the ptac10-1 mutants. By contrast, typical NEP-transcribed genes exhibited a completely different expression pattern. For instance, the abundance of accD was only slightly decreased, while the abundances of rpo transcripts (for subunits of RNA polymerase) were increased. Additionally, three nuclear-encoded photosynthesis genes, Lhcb1.1, Lhcb1.2 and RbcS were investigated. The qRT-PCR showed that the abundance of the Lhcb1.1 and Lhcb1.2 transcripts was approximately 1.5-fold greater than those in wild type, while the RbcS transcript level was slightly decreased (Fig. 4). These data indicate that the loss of functional pTAC10 drastically alters the transcription of plastid genes.
To further confirm the qRT-PCR data, three different types of chloroplast genes were chosen to perform RNA blotting in wild type and the ptac10-1 mutant. In agreement with the qRT-PCR data, the expression of the PEP-dependent photosynthesis genes, psbA, psaB and rbcL, was significantly reduced in the ptac10-1 mutant compared with the wild-type (Fig. 5a). In contrast, the levels of NEP-dependent plastid transcripts, such as rpoB, rpl16 and accD, were unchanged or slightly increased in ptac10-1 (Fig. 5c). Among the tested chloroplast genes, rrn16 and clpP, are transcribed by both PEP and NEP. Their transcripts were slightly decreased in the ptac10-1 mutant (Fig. 5b). It is noteworthy that in wild type, atpB transcripts of 2.0 and 2.6 kb were transcribed by PEP and NEP, respectively (Ishizaki et al. 2005) whereas in the ptac10-1 mutant, a 4.8-kb instead of a 2.6-kb transcript was detected (Fig. 5b, left panel). The expression pattern of the plastid genes in ptac10-1 is quite similar with those in Δrpo mutants (Hess et al. 1993; Allison et al. 1996; Hajdukiewicz et al. 1997; Silhavy and Maliga 1998; De Santis-Maciossek et al. 1999; Krause et al. 2000; Legen et al. 2002) and other reported PEP-deficient mutants (Pfalz et al. 2006; Garcia et al. 2008; Myouga et al. 2008; Arsova et al. 2010; Gao et al. 2011; Steiner et al. 2011; Yagi et al. 2012; Gilkerson et al. 2012; Yu et al. 2013, 2014). Taken together, the plastid gene transcription patterns in the ptac10-1 mutant further confirm that pTAC10 is essential for PEP-dependent transcription.
Chloroplast protein accumulation in ptac10
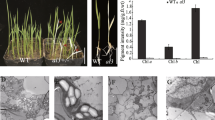
In addition to plastid gene transcription, it is also necessary to examine the accumulation of photosynthetic-related proteins in ptac10 mutants since the mutation affects the chloroplast ultrastructure. A total of 34 photosynthetic proteins were detected by immuno-blotting in wild type, ptac10 mutants and the pTAC10:MYC complemented line. Among those proteins, most chloroplast-encoded proteins, including PsaA/B, PsaC, PsaD, PsaK, PsaE, D1, D2, CP43, CP47, PsbI, AtpA and AtpE, were barely detectable in either the ptac10-1 or the ptac10-2 mutant. Other chloroplast genome-encoded proteins, such as AtpB, were clearly reduced in both ptac10 mutants (Fig. 5). These data are in agreement with their transcript abundance shown in Fig. 4. For the nuclear-encoded photosynthetic proteins, the levels of six LHC components were undetectable in the ptac10 mutants. Other proteins, such as PsbO, PsbQ, PC, FNR and FtsH2, also accumulated in reduced amounts. The level of ClpP6, a subunit of the CLP protease, was similar in the ptac10 mutants and wild type (Fig. 6). The amount of all detected proteins in the complemented lines was similar to that in wild-type plants (Fig. 6). Taken together, our data show that the accumulation of most chloroplast proteins, either encoded by the chloroplast or the nuclear genome, is decreased in ptac10 mutants.
Accumulation of chloroplast proteins in wild type and the ptac10-1 mutant. Immunoblot analysis of chloroplast- and nuclear-encoded chloroplast proteins between WT, the ptac10-1 mutant and the complemented lines. Total protein was used to perform immunoblotting analysis. Actin protein was used as a control
pTAC10 interacts with cpCK2 in vitro and in vivo
pTAC10 was predicted to be a potential substrate of cpCK2 (Reiland et al. 2009). To investigate their interaction, we expressed both proteins in E. coli with different tags. pTAC10 lacking its 36-amino-acid N-terminal transit peptide at the N-terminus was fused to maltose-binding protein (MBP), while cpCK2 protein lacking its 55-amino-acid transit peptide was tagged with glutathione S-transferase (GST). These fusion proteins were used in a pull-down assay. As shown in Fig. 7a, GST-cpCK2 could pull down pTAC10-MBP using a GST affinity column. No pTAC10-MBP was pulled down with GST alone. To confirm the interaction of pTAC10 and cpCK2 in vivo, we performed coimmunoprecipitation assays using leaf cell extracts. pTAC10 carrying a C-terminal FLAG epitope tag (pTAC10-FLAG) and cpCK2 with a C-terminal MYC epitope tag (cpCK2-MYC) were synthesized together in Nicotiana benthamiana leaves following agro-infiltration. Leaf extracts were subsequently immunoprecipitated with the anti-MYC antibody. Immunoblotting of the immunoprecipitate with anti-FLAG antibody revealed the presence of pTAC10-FLAG (Fig. 7b). Our results indicate that pTAC10 interacts with cpCK2 both in vitro and in vivo.
pTAC10 interacts with cpCK2 in vitro and in vivo. a Pull-down assay revealing in vivo interactions between pTAC10 and cpCK2. A GST affinity column was used to detect the interaction. b Coimmunoprecipitation assay revealing in vivo interactions between pTAC10 and cpCK2. The constructs indicated above each blot were transiently expressed together in N. benthamiana. Proteins detected by immunoblotting are indicated on the right. pTAC10-Flag coimmunoprecipitated with cpCK2-MYC using the anti-Flag antibody (FLAG IP), and immunoblotting was performed with anti-MYC antibody
pTAC10 can be phosphorylated by cpCK2 in vitro
We next checked whether pTAC10 could be phosphorylated by cpCK2. First, we investigated whether a commercial CK2 could phosphorylate recombinant pTAC10 protein in vitro. For this assay, the MBP-tagged recombinant pTAC10 protein without transit peptide was expressed and purified (Fig. 8a). As shown in Fig. 8b, commercial CK2 could clearly phosphorylate the recombinant pTAC10 protein in a complete phosphorylation reaction system. In contrast, pTAC10 phosphorylation could not be detected when the reaction mixture lacked either CK2 or ATP (Fig. 8b). To further determine the pTAC10 sites phosphorylated by CK2 in vitro, phosphorylated pTAC10 was analysed by mass spectrometry. Six phosphorylation sites (Thr-64, Ser-95, Thr-131, Ser-434, Ser-600 and Tyr-136) were identified (Supplementary Fig. 2). Among the six phosphorylation sites, five sites (Thr-64, Ser-95, Thr-131, Ser-434 and Ser-600) conform to the typical CK2 phosphorylation site motif S*/T*xxEx or S*/T*xxDx (Schweer et al. 2010; Supplementary Fig. 2). Thus, pTAC10 can be phosphorylated by commercial CK2 in vitro.
Phosphorylation of pTAC10 by (cp)CK2 in vitro. a SDS–PAGE analysis of total protein from non-induced (1) and induced (2) E. coli containing pMAL-C5X-pTAC10, and purified pTAC10 protein (3) with molecular markers(Prestained Protein Marker I, Genview) indicated (4). b Immunoblot analysis of pTAC10 phosphorylation by CK2 in vitro using anti-phospho-Ser, Thr and Tyr antibodies. c Immunoblot analysis of pTAC10 phosphorylation by cpCK2 in vitro using anti-phospho-Ser, Thr and Tyr antibodies. CK2 was used as positive control. ‘+’ indicates the presence of reagent in the reaction system, and ‘−’ indicates the absence of reagent in the reaction system. ‘*’ Indicates the band corresponding to pTAC10. M molecular markers (PageRuler Prestained Protein Ladder, Fermentas). d A scheme representing the protein architecture is shown. The red box in this scheme corresponds to the S1 domain. The green box which corresponds to the first 41 amino acids at the N-terminal, represents the putative transpeptide of pTAC10. Three phosphorylation sites (S95, S396 and S434) are shown in the scheme. e Phosphorylation sites determined by mass spectrometry and the amino acid sequence around the phosphorylation sites in pTAC10 are shown. The phosphoacceptors are denoted by ‘*’, and the acidic residues at n + 3 are highlighted in grey
To characterize pTAC10 phosphorylation by cpCK2 from Arabidopsis, we expressed the chloroplast-targeted CK2 protein lacking its transit peptide in vitro (Supplementary Fig. 2). The same phosphorylation assay was performed with purified cpCK2 protein and pTAC10. Like the commercial CK2, purified cpCK2 protein could clearly phosphorylate the recombinant pTAC10 protein, and the phosphorylated pTAC10 was undetectable when the reaction system lacked either cpCK2 or ATP (Fig. 8c). To identify the phosphorylated sites, mass spectrometry was used for pTAC10 phosphorylated by cpCK2. Three sites, S95, S396 and S434, were identified (Fig. 8e and Supplemental Fig. 3), which are out of the S1-domain of the pTAC10 protein (Fig. 8d). In agreement with our data, the peptide microarray “ChloroPhos1.0” software developed by Schönberg et al. (2015) also showed that S434 of pTAC10 is a target phosphorylation site of cpCK2 in Arabidopsis thaliana. We also attempted to identify the phosphorylation sites of pTAC10 in planta, but because of the low abundance of pTAC10 we were not able to obtain conclusive results. However, previous studies from two independent groups have identified one pTAC10 phosphorylation site, S434, in vivo (Reiland et al. 2009; Ingelsson and Vener 2012), suggesting that pTAC10 is indeed a substrate of cpCK2 in planta.
Phosphorylation of pTAC10 enhances its RNA-binding activity
pTAC10 contains a single S1 RNA-binding domain, suggesting its potential to bind to RNA. Consequently, a RNA electrophoretic mobility shift assay (RNA-EMSA) was carried out to analyse the RNA-binding ability of pTAC10. Purified MBP:pTAC10 protein was incubated with biotinylated RNA probes of psbA, atpE and accD, together with MBP:pTAC10. A MBP:pTAC10-RNA complex was detected as a shifted band that migrated much slower than free RNA in the gel. Notably, MBP alone did not bind to the RNA probes (Fig. 9a–c), indicating that the pTAC10 protein binds to RNA. Phosphorylated pTAC10 was also used for RNA-EMSA analysis. Interestingly, the RNA-binding activity of phosphorylated pTAC10 protein was higher than that of the unphosphorylated protein (Fig. 9a–c). This result indicates that phosphorylation of pTAC10 can enhance its RNA-binding activity.
Phosphorylation of pTAC10 affects its RNA-binding ability. a–c RNA-EMSA assays using RNA probes for three chloroplast genes (psbA, atpE and accD). Labelled RNA probes were incubated with increasing concentrations of recombinant MBP:pTAC10 (MBP:pTAC10) or phosphorylated MBP:pTAC10 (MBP:pTAC10-P) with free biotinylated RNA probes only (Mock), or incubated with empty MBP protein (MBP) as negative controls. d The RNA-EMSA analysis was conducted with biotinylated psbA probe with MBP:pTAC10 (pTAC10) and MBP:pTAC10S434A (S434A) as well as their phosphorylated proteins (pTAC10-P and S434A-P), using psbA probe only as a control (Mock)
Since Ser434 is a pTAC10 phosphorylation site both in vitro (Fig. 8) and in vivo (Reiland et al. 2009; Ingelsson and Vener 2012), we next investigated whether a change of Ser434 affects the RNA binding activity of pTAC10. pTAC10-S434A, a version of pTAC10 containing the amino acid substitution Ser434-Ala, was expressed and purified. This recombinant protein and its phosphorylated version were co-incubated with the psbA probe, respectively. The results show that the amino acid substitutions Ser434-Ala decreases RNA binding of pTAC10 regardless of whether it is phosphorylated or not. Interestingly, the RNA binding level of phosphorylated pTAC10-S434A was still slightly stronger than that of unphosphorylated pTAC10-S434A (Fig. 9d), which might due to the phosphorylation at other sites. Overall, our results suggest that the phosphorylation of pTAC10 at Ser434 might play a role in modulating its RNA binding activity.
Discussion
In this study, we characterized an S1-domain-containing protein, pTAC10, and its knockout lines in Arabidopsis. Loss of pTAC10 impairs chloroplast development and dramatically reduces the levels of PEP-dependent transcripts and of chloroplast proteins indicating an essential role of pTAC10 in plastid gene expression and plastid development. pTAC10 interacts with cpCK2 and is phosphorylated by cpCK2 at multiple sites. Moreover, phosphorylation of pTAC10 enhances its RNA-binding activity. It is therefore possible that the status of pTAC10 phosphorylation could play some role in the regulation of plastid gene expression by modulating the RNA-binding activity of this protein.
pTAC10 was originally identified in Arabidopsis as one component of the TAC by affinity chromatography purification and mass spectrometry analysis (Pfalz et al. 2006). It was recently considered as a PEP-associated protein, PAP3, because it was tightly associated with the core subunits of the PEP complex (Steiner et al. 2011). PAPs play essential roles in the transcription of the PEP-dependent plastid genes (Pfalz et al. 2006; Steiner et al. 2011). Most of the paps mutants were recently characterized (Pfalz et al. 2006, 2015; Garcia et al. 2008; Myouga et al. 2008; Arsova et al. 2010; Gao et al. 2011; Yagi et al. 2012; Yu et al. 2013). The reported pap mutants include ptac2 (Pfalz et al. 2006), ptac6 (Pfalz et al. 2006), ptac12 (Pfalz et al. 2006, 2015), atmurE (Garcia et al. 2008), fsd2 (Myouga et al. 2008), fsd3 (Myouga et al. 2008), trxz (Arsova et al. 2010), ptac14 (Gao et al. 2011), ptac3 (Yagi et al. 2012) and ptac7 (Yu et al. 2013). They display albino or pale green phenotypes and arrested chloroplast development. They also show reduced accumulation of PEP-dependent plastid transcripts (Pfalz et al. 2006, 2015; Garcia et al. 2008; Myouga et al. 2008; Arsova et al. 2010; Gao et al. 2011; Yagi et al. 2012; Yu et al. 2013). In this work, we characterized the ptac10 mutant in detail. The pTAC10 knockout lines displayed an albino phenotype similar to those of the pap mutants (Fig. 2). A comprehensive plastid transcription profile of the ptac10-1 mutant revealed that the plastid-encoded photosynthetic-related genes, especially the photosystem genes psaA and psaB, which are exclusively transcribed by the PEP-dependent RNA polymerase, are severely reduced in the ptac10-1 mutant (Figs. 4, 5). In contrast, the amounts of NEP-dependent plastid transcripts either increase or slightly decrease (Figs. 4, 5). The expression patterns of plastid genes in the ptac10-1 mutant are expected and are similar to those of the Δrpo mutants (Hess et al. 1993; Allison et al. 1996; De Santis-Maciossek et al. 1999; Krause et al. 2000; Legen et al. 2002) and reported PAP mutants (Pfalz et al. 2006, 2015; Garcia et al. 2008; Myouga et al. 2008; Arsova et al. 2010; Gao et al. 2011; Yagi et al. 2012; Yu et al. 2013). Our results demonstrate that ptac10 is also a PEP-deficient mutant, similar to other paps mutants. In agreement with our data, the virus-induced silenced STF, an ortholog of pTAC10 in Nicotiana benthamiana, also leads to severe leaf chlorosis and to a reduced accumulation of PEP-dependent transcripts (Jeon et al. 2012). Therefore, pTAC10 is essential for PEP-dependent plastid transcription and chloroplast development, and is conserved among angiosperms. Chang et al. (2017) recently demonstrated interactions of pTAC10 protein with multiple PAPs that are required for PEP complex formation. Thus, it is possible that the knockout of pTAC10 perturbs the configuration of this PEP complex, which ultimately results in reduced levels of PEP-dependent transcripts (Steiner et al. 2011). In turn, this process subsequently interferes with chloroplast development. In addition, the chloroplast-encoded photosynthetic proteins were also reduced in the ptac10 mutants (Fig. 5) probably as a result of the corresponding decrease in plastid transcripts (Fig. 4).
pTAC10 is tightly associated with the core subunits of the PEP complex bound to the thylakoid membrane (Steiner et al. 2011). Our result reveal that this protein is localized both in the chloroplast stroma and associated with the thylakoid membrane (Fig. 2d). Its association with chloroplast thylakoid membranes is consistent with the known localization of the PEP complex (Pfalz et al. 2006). Moreover, BN gel and subsequent 2D SDS–PAGE analysis revealed the co-migration of pTAC10 and RpoB (Fig. 2e). Therefore, our results indicate that pTAC10 is indeed associated with the PEP complex that includes the core subunits of PEP. In agreement with these results, our previous publications showed that pTAC10 interacts with pTAC7 (Yu et al. 2013) through the downstream S1 domain (Chang et al. 2017). A recent study revealed that pTAC10 also interacts with Trx Z, and FSD2, FSD3 and pTAC14, in addition to pTAC7. These interactions are probably essential for the formation of the PEP complex, as previously proposed (Pfalz and Pfannschmidt 2013; Chang et al. 2017). Alternatively, these interactions suggest that these proteins compose the functional modules of the PEP transcriptional machinery. Additionally, pTAC10 is localized in the chloroplast stroma, suggesting that it may have functions other than plastid transcription.
Phosphorylation is one of the most common post-translational modifications. Several chloroplast kinases have been reported (Reiland et al. 2009). cpCK2, as a highly conserved multifunctional Ser/Thr kinase, was originally identified within the chloroplast transcriptional apparatus (Baginsky et al. 1999). Several phosphorylation targets of cpCK2 in Arabidopsis thaliana were identified by the peptide microarray “ChloroPhos1.0”, and most of them are involved in plastid gene expression (Schönberg et al. 2014). These findings are consistent with the earlier denomination of cpCK2 as a plastid transcription kinase (PTK) (Baginsky et al. 1999). Recent biochemical and genetic analyses have suggested that AtSIG6 is one substrate of this kinase (Schweer et al. 2010). pTAC10 has also been predicted to be a substrate of cpCK2 (Reiland et al. 2009). The present in vitro and in vivo analysis shows that cpCK2 co-immunoprecipitates with pTAC10. Moreover, pTAC10 is not only phosphorylated by commercial CK2 but also by recombinant cpCK2 from Arabidopsis in vitro (Fig. 8, Supplemental Figs. 2 and 3). These data suggest that, as expected, pTAC10 is indeed a substrate of the chloroplast-targeted CK2 kinase (Reiland et al. 2009). Among the three identified phosphorylation sites, two sites (S95 and S434) conform to the reported CK2 target sequence motifs S*/T*xxEx or S*/T*xxDx, respectively, strengthening the view that they are authentic phosphorylation sites (Reiland et al. 2009). The motif of the remaining phosphorylation site, S396, is distinct from that of the two others, raising the possibility that the cpCK2 substrates contain additional target motifs. CK2-mediated phosphorylation is involved in the modulation of protein function, including changes in DNA-binding affinity, dimerization, stability, protein interactions and sub-cellular localization (Mulekar and Huq 2013). Nicotiana benthamiana STF is able to bind RNA (Jeon et al. 2012). Our present results also show that pTAC10 can bind to RNAs in a non-specific manner (Fig. 9). Nevertheless, the affinity of pTAC10 for RNA was improved by phosphorylation (Fig. 9). Two other chloroplast RNA-binding proteins have been reported to be phosphorylated, including RNP28 (Lisitsky et al. 1995) and 24RNP (Loza-Tavera et al. 2006), although their corresponding kinase remains unknown. Their phosphorylation mediates the in vitro interplay between petD precursor stability and 3′-UTR processing and degradation, suggesting their involvement in chloroplast mRNA metabolism (Vargas-Suárez et al. 2013). Interestingly, three identified phosphorylation sites are out of the S1 RNA binding domain (Fig. 8d). It is possible that the phosphorylation might induce the change of its configuration, which leads to the improvement of its RNA-binding ability. Phosphorylation of pTAC10 could enhance its RNA-binding activity (Fig. 9), which suggests that the phosphorylation may regulate gene transcription in the chloroplast. Further investigations of the modification of pTAC10 phosphorylation in plants will help to improve our understanding of phosphorylation-mediated regulatory mechanisms in chloroplasts.
References
Allison LA, Simon LD, Maliga P (1996) Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J 15:2802–2809
Arsova B, Hoja U, Wimmelbacher M, Greiner E, Ustun S, Melzer M, Petersen K, Lein W, Bornke F (2010) Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. Plant Cell 22:1498–1515
Baginsky S, Gruissem W (2009) The chloroplast kinase network: new insights from large-scale phosphoproteome profiling. Mol Plant 2:1141–1153
Baginsky S, Tiller K, Link G (1997) Transcription factor phosphorylation by a protein kinase associated with chloroplast RNA polymerase from mustard (Sinapis alba). Plant Mol Biol 34:181–189
Baginsky S, Tiller K, Pfannschmidt T, Link G (1999) PTK, the chloroplast RNA polymerase-associated protein kinase from mustard (Sinapis alba), mediates redox control of plastid in vitro transcription. Plant Mol Biol 39:1013–1023
Berry JO, Yerramsetty P, Zielinski AM, Mure CM (2013) Photosynthetic gene expression in higher plants. Photosynth Res 117(1–3):91–120
Chang SH, Lee S, Um TY, Kim JK, Choi YD, Jang G (2017) pTAC10, a key subunit of plastid-encoded RNA polymerase, promotes chloroplast development. Plant Physiol. https://doi.org/10.1104/pp.17.00248
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
De Santis-MacIossek G, Kofer W, Bock A, Schoch S, Maier RM, Wanner G, Rüdiger W, Koop HU, Herrmann RG (1999) Targeted disruption of the plastid RNA polymerase genes rpoA, B and C1: molecular biology, biochemistry and ultrastructure. Plant J 18:477–489
Demarsy E, Courtois F, Azevedo J, Buhot L, Lerbs-Mache S (2006) Building up of the plastid transcriptional machinery during germination and early plant development. Plant Physiol 142:993–1003
Gao ZP, Yu QB, Zhao TT, Ma Q, Chen GX, Yang ZN (2011) A functional component of the transcriptionally active chromosome complex, Arabidopsis pTAC14, interacts with pTAC12/HEMERA and regulates plastid gene expression. Plant Physiolgy 157:1733–1745
Garcia M, Myouga F, Takechi K, Sato H, Nabeshima K, Nagata N, Takio S, Shinozaki K, Takano H (2008) An Arabidopsis homolog of the bacterial peptidoglycan synthesis enzyme MurE has an essential role in chloroplast development. Plant J 53:924–934
Ghavidel A, Schultz MC (2001) TATA binding protein-associated CK2 transduces DNA damage signals to the RNA polymerase III transcriptional machinery. Cell 106:575–584
Gilkerson J, Perez-Ruiz JM, Chory J, Callis J (2012) The plastid-localized pfkB-type carbohydrate kinases FRUCTOKINASE-LIKE 1 and 2 are essential for growth and development of Arabidopsis thaliana. BMC Plant Biol 12:102
Hajdukiewicz PT, Allison LA, Maliga P (1997) The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J 16:4041–4048
Hess WR, Prombona A, Fieder B, Subramanian AR, Bo¨rner T (1993) Chloroplast rps15 and the rpoB/C1/C2 gene cluster are strongly transcribed in ribosome-deficient plastids: evidence for a functioning nonchloroplast- encoded RNA polymerase. EMBO J 12:563–571
Hu J, Bogorad L (1990) Maize chloroplast RNA polymerase: the 180-, 120-, and 38-kilodalton polypeptides are encoded in chloroplast genes. Proc Natl Acad Sci USA 87:1531–1535
Ingelsson B, Vener AV (2012) Phosphoproteomics of Arabidopsis chloroplasts reveals involvement of the STN7 kinase in phosphorylation of nucleoid protein pTAC16. FEBS Lett 586(9):1265–1271
Ishizaki Y, Tsunoyama Y, Hatano K, Ando K, Kato K, Shinmyo A, Kobori M, Takeba G, Nakahira Y, Shiina T (2005) A nuclear-encoded sigma factor, Arabidopsis SIG6, recognizes sigma-70 type chloroplast promoters and regulates early chloroplast development in cotyledons. Plant J 42:133–144
Jeon Y, Jung HJ, Kang H, Park YI, Lee SH, Pai HS (2012) S1 domain-containing STF modulates plastid transcription and chloroplast biogenesis in Nicotiana benthamiana. New Phytol 193:349–363
Krause K, Maier RM, Kofer W, Krupinska K, Herrmann RG (2000) Disruption of plastid-encoded RNA polymerase genes in tobacco: expression of only a distinct set of genes is not based on selective transcription of the plastid chromosome. Mol Genet Genomics 263:1022–1030
Legen J, Kemp S, Krause K, Profanter B, Herrmann RG, Maier RM (2002) Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. Plant J 31:171–188
Link G (1996) Green life: control of chloroplast gene transcription. Bioessays 18:465–471
Lisitsky I, Liveanu V, Schuster G (1995) RNA-binding characteristics of a ribonucleoprotein from spinach chloroplast. Plant Physiol 107:933–941
Litchfield DW (2003) Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem J 369:1–15
Lopez-Juez E, Pyke KA (2005) Plastids unleashed: their development and their integration in plant development. Int J Dev Biol 49:557–577
Loza-Tavera H, Vargas-Suárez M, Díaz-Mireles E, Torres-Márquez ME, González de la Vara LE, Moreno-Sánchez R, Gruissem W (2006) Phosphorylation of the spinach chloroplast 24 kDa RNA-binding protein (24RNP) increases its binding to petD and psbA 3′ untranslated regions. Biochimie 88:1217–1228
Lu Q, Ding S, Reiland S, Rödiger A, Roschitzki B, Xue P, Gruissem W, Lu C, Baginsky S (2015) Identification and characterization of chloroplast casein kinase II from Oryza sativa (rice). J Exp Bot 66(1):175–187
Melonek J, Matros A, Trösch M, Mock HP, Krupinska K (2012) The core of chloroplast nucleoids contains architectural SWIB domain proteins. Plant Cell 24:3060–3073
Mulekar JJ, Huq E (2013) Expanding roles of protein kinase CK2 in regulating plant growth and development. J Exp Bot 65:2883–2893
Myouga F, Hosoda C, Umezawa T, Iizumi H, Kuromori T, Motohashi R, Shono Y, Nagata N, Ikeuchi M, Shinozaki K (2008) A heterocomplex of iron superoxide dismutases defends chloroplast nucleoids against oxidative stress and is essential for chloroplast development in Arabidopsis. Plant Cell 20:3148–3162
Niefind K, Guerra B, Ermakowa I, Issinger OG (2001) Crystal structure of human protein kinase CK2: insights into basic properties of the CK2 holoenzyme. EMBO J 20:5320–5331
Ogrzewalla K, Piotrowski M, Reinbothe S, Link G (2002) The plastid transcription kinase from mustard (Sinapis alba L.). A nuclear-encoded CK2-type chloroplast enzyme with redox-sensitive function. Eur J Biochem 269:3329–3337
Pfalz J, Pfannschmidt T (2013) Essential nucleoid proteins in early chloroplast development. Trends Plant Sci 18:186–194
Pfalz J, Liere K, Kandlbinder A, DietzK J, Oelmuller R (2006) pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18:176–197
Pfalz J, Holtzegel U, Barkan A, Weisheit W, Mittag M, Pfannschmidt T (2015) ZmpTAC12 binds single-stranded nucleic acids and is essential for accumulation of the plastid-encoded polymerase complex in maize. New Phytol 206:1024–1037
Pfannschmidt T, Link G (1994) Separation of two classes of plastid DNA-dependent RNA polymerases that are differentially expressed in mustard (Sinapis alba L.) seedlings. Plant Mol Biol 25:69–81
Pfannschmidt T, Ogrzewalla K, Baginsky S, Sickmann A, Meyer HE, Link G (2000) The multisubunit chloroplast RNA polymerase A from mustard (Sinapis alba L.): integration of a prokaryotic core into a larger complex with organelle-specific functions. Eur J Biochem 267:253–261
Reiland S, Messerli G, Baerenfaller K, Gerrits B, Endler A, Grossmann J, Gruissem W, Baginsky S (2009) Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol 150:889–903
Salinas P, Fuentes D, Vidal E, Jordana X, Echeverria M, Holuigue L. 2006. An extensive survey of CK2 alpha and beta subunits in Arabidopsis: multiple isoforms exhibit differential subcellular localization. Plant Cell Physiol 47:1295–1308
Sato S, Nakamura Y, Kaneko T, Asamizu E, Tabata S (1999) Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res 6:283–290
Schönberg A, Bergner E, Helm S, Agne B, Dünschede B, Schünemann D, Schutkowski M, Baginsky S (2014) The peptide microarray “ChloroPhos1.0” identifies new phosphorylation targets of plastid casein kinase II (pCKII) in Arabidopsis thaliana. PLoS ONE 9:e108344
Schröter Y, Steiner S, Matthäi K, Pfannschmidt T (2010) Analysis of oligomeric protein complexes in the chloroplast sub-proteome of nucleic acid-binding proteins from mustard reveals potential redox regulators of plastid gene expression. Proteomics 10:2191–2204
Schweer J, Türkeri H, Link B, Link G (2010) AtSIG6, a plastid sigma factor from Arabidopsis, reveals functional impact of cpCK2 phosphorylation. Plant J 62:192–202
Silhavy D, Maliga P (1998) Mapping of promoters for the nucleus-encoded plastid RNA polymerase (NEP) in the iojap maize mutant. Curr Genet 33:340–344
Steiner S, Schroter Y, Pfalz J, Pfannschmidt T (2011) Identification of essential subunits in the plastid-encoded RNA polymerase complex reveals building blocks for proper plastid development. Plant Physiol 157:1043–1055
Stöckel J, Bennewitz S, Hein P, Oelmüller R (2006) The evolutionarily conserved tetratrico peptide repeat protein pale yellow green7 is required for photosystem I accumulation in Arabidopsis and copurifies with the complex. Plant Physiol 141:870–878
Suzuki JY, Ytterberg AJ, Beardslee TA, Allison LA, Wijk KJ, Maliga P (2004) Affinity purification of the tobacco plastid RNA polymerase and in vitro reconstitution of the holoenzyme. Plant J 40:164–172
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Vargas-Suárez M, Castro-Sánchez A, Toledo-Ortiz G, González de la Vara LE, García E, Loza-Tavera H (2013) Protein phosphorylation regulates in vitro spinach chloroplast petD mRNA 3′-untranslated region stability, processing, and degradation. Biochimie 95:400–409
Wakasugi T, Tsudzuki T, Sugiura M (2001) The genomics of land plant chloroplasts: gene content and alteration of genomic information by RNA editing. Photosynth Res 70(1):107–118
Williams-Carrier R, Zoschke R, Belcher S, Pfalz J, Barkan A (2014) A major role for the plastid-encoded RNA polymerase complex in the expression of plastid transfer RNAs. Plant Physiol 164:239–248
Yagi Y, Ishizaki Y, Nakahira Y, Tozawa Y, Shiina T (2012) Eukaryotic-type plastid nucleoid protein pTAC3 is essential for transcription by the bacterial-type plastid RNA polymerase. Proc Natl Acad Sci USA 109:7541–7546
Yang Z, Shang Z, Wang L, Lu Q, Wen X, Chi W, Zhang L, Lu C (2015) Purine biosynthetic enzyme ATase2 is involved in the regulation of early chloroplast development and chloroplast gene expression in Arabidopsis. Photosynth Res 126(2–3):285–300
Yu QB, Jiang Y, Chong K, Yang ZN (2009) AtECB2, a pentatricopeptide repeat protein, is required for chloroplast transcript accD RNA editing and early chloroplast biogenesis in Arabidopsis thaliana. Plant J 59:1011–1023
Yu QB, Lu Y, Ma Q, Zhao TT, Huang C, Zhao HF, Zhang XL, Lv RH, Yang ZN (2013) TAC7, an essential component of the plastid transcriptionally active chromosome complex, interacts with FLN1, TAC10, TAC12 and TAC14 to regulate chloroplast gene expression in Arabidopsis thaliana. Physiol Plant 148:408–421
Yu QB, Huang C, Yang ZN (2014) Nuclear-encoded factors associated with the chloroplast transcription machinery of higher plants. Front Plant Sci 5:316
Zhang HD, Cui YL, Huang C, Yin QQ, Qin XM, Xu T, He XF, Zhang Y, Li ZR, Yang ZN (2015) PPR protein PDM1/SEL1 is involved in RNA editing and splicing of plastid genes in Arabidopsis thaliana. Photosynth Res 126(2–3):311–321
Zhong L, Zhou W, Wang H, Ding S, Lu Q, Wen X, Peng L, Zhang L, Lu C (2013) Chloroplast small heat shock protein HSP21 interacts with plastid nucleoid protein pTAC5 and is essential for chloroplast development in Arabidopsis under heat stress. Plant Cell 25:2925–2943
Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136:2621–2632
Acknowledgements
We thank ABRC Bioresources, which kindly provided the transgenic Arabidopsis lines (CS16115 and CS16140). We would like to thank Prof. CM Lu from IBCAS for the kind donation of RpoB antibody. We appreciate that Prof. XW Sun from SHNU for his help in western blotting. We also thank Prof. Jean-David Rochaix for his help in the language editing of this manuscript. This work was supported by grants from the National Natural Science Foundation of China (31570232 and 31370271) and by the Innovation Program of Shanghai Municipal Education Commission (14YZ066).
Author information
Authors and Affiliations
Corresponding author
Additional information
Qing-Bo Yu and Tuan-Tuan Zhao have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1. Alignment of the amino acid sequences of the pTAC10 orthologs
. The alignment was generated using ClustalW (Thompson et al., 1994) with default parameters and BoxShade 3.21 (http://www.ch.embnet.org/software/BOX_form.html). The positions of identical and similar sequences are indicated in black and grey boxes, respectively. All the sequences used to build the alignment were obtained from NCBI (http://www.ncbi.nlm.nih.gov/). (PDF 581 KB)

Supplementary Figure 2. Identification of phosphorylation sites of pTAC10 by CK2 in vitro
. (A) Phosphorylation sites determined by mass spectrometry are shown. (B) The amino acid sequence around the phosphorylation sites in pTAC10. The phosphoacceptors are indicated by “*”, and the acidic residues at n + 3 are highlighted in grey; (C) An independent phosphorylation analysis of pTAC10. Immunoblot analysis of pTAC10 phosphorylation by cpCK2 in vitro using anti-phospho-Ser, Thr and Tyr antibodies. CK2 was used as positive control. “+” indicates the presence of reagent in the reaction system, and “-” indicates the absence of reagent in the reaction system. “*” indicates the band corresponding to pTAC10. (D) SDS–PAGE analysis of total proteins from unpurified (1) and purified (2) E. coli containing MBP:cpCK2 protein (M) with molecular markers (PageRuler Prestained Protein Ladder, Fermentas) indicated. (JPG 2750 KB)Supplementary material 2 (JPG 843 KB)

Supplementary Figure 3. Mass spectrometric analyses of phosphorylated peptides from of pTAC10 bycpCK2 in vitro
. (A) to (C) LC–MS extracted ion chromatograms showing intensities of the phosphorylated peptide signals from the phosphorylated peptide corresponding to the pTAC10 protein. The phosphorylated Ser residue in the peptide sequence is marked with "ph" on the top of the corresponding amino acid. (JPG 1135 KB)Supplementary material 2 (JPG 843 KB)
Rights and permissions
About this article
Cite this article
Yu, QB., Zhao, TT., Ye, LS. et al. pTAC10, an S1-domain-containing component of the transcriptionally active chromosome complex, is essential for plastid gene expression in Arabidopsis thaliana and is phosphorylated by chloroplast-targeted casein kinase II. Photosynth Res 137, 69–83 (2018). https://doi.org/10.1007/s11120-018-0479-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11120-018-0479-y