Abstract
Ferredoxins are soluble iron sulphur proteins which function as electron donors in a number of metabolic pathways in a broad range of organisms. In photosynthetic organisms, PETF, or ferredoxin 1 (FDX1), is the most studied ferredoxin due to its essential role in photosynthesis, where it transfers electrons from photosystem I to ferredoxin-NADP+ oxidoreductase. However, PETF can also transfer electrons to a large number of other proteins. One important PETF electron acceptor found in green microalgae is the biologically and biotechnologically important [FeFe]-hydrogenase HYDA, which catalyses the production of molecular hydrogen (H2) from protons and electrons. The interaction between PETF and HYDA is of considerable interest, as PETF is the primary electron donor to HYDA and electron supply is one of the main limiting factors for H2 production on a commercial scale. Although there is no three dimensional structure of the PETF–HYDA complex available, protein variants, nuclear magnetic resonance titration studies, molecular dynamics and modelling have provided considerable insight into the residues essential for forming and maintaining the interaction. In this review, we discuss the most recent findings with regard to ferredoxin-HYDA interactions and the evolution of the various Chlamydomonas reinhardtii ferredoxin isoforms. Finally, we provide an outlook on new PETF-based biotechnological approaches for improved H2 production efficiencies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The ferredoxin family, present in bacteria, algae, higher plants and animals, consists of a diverse range of small, soluble iron sulphur (FeS) proteins, which are characterised by their acidity and low redox potentials. Ferredoxins act as electron acceptors (e.g. for photosynthesis) and donors (e.g. for sulphur and nitrogen assimilation, biosynthesis of chlorophyll, phytochromes and fatty acids) and participate in redox signalling (Fukuyama 2004; Hanke and Mulo 2013; Hemschemeier and Happe 2011). Green microalgae contain multiple plant-type (2Fe2S) ferredoxin isoforms (Yang et al. 2015). However, ferredoxin 1 (FDX1), more commonly known as PETF, is the most studied ferredoxin due to its essential role as the first stromal electron acceptor in the photosynthetic electron transport chain, where it transfers electrons from photosystem I (PSI) to a number of proteins. The principal redox partner of PETF is the ferredoxin-NADP+ oxidoreductase (FNR), which catalyses the production of NADPH from NADP+ and protons (Shin 2004; Winkler et al. 2009).
In green microalgae, an electron acceptor of PETF of both biological and biotechnological interest is the [FeFe]-hydrogenase, HYDA, which catalyses the production of molecular hydrogen (H2) from protons and electrons. The Chlamydomonas reinhardtii [FeFe]-hydrogenase, HYDA1, was the first eukaryotic hydrogenase isolated (Happe et al. 1994; Happe and Naber 1993). This, in addition to the large knowledgebase and molecular toolset available for C. reinhardtii (Jinkerson and Jonikas 2015; Mussgnug 2015), has led to this hydrogenase being the best characterised eukaryotic hydrogenase. As H2 production efficiencies are dependent on electron supply, the PETF–HYDA1 interaction has been the focus of many studies. Although there is no three dimensional (3D) structure of this complex to date, site-directed mutagenesis and nuclear magnetic resonance (NMR) titration studies have provided considerable insight into the residues of both proteins crucial for complex formation and electron transfer. This knowledge has in turn informed genetic engineering approaches.
Chlamydomonas ferredoxins
Thirteen ferredoxin genes have been identified in the C. reinhardtii genome (Yang et al. 2015); however, only six (PETF and FDX2-6) have been characterised in any detail. FDX2 is the most similar to PETF, with 68% sequence identity according to BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi), while FDX8, FDX10, FDX11, FDX12 and MFDX are the most divergent, displaying low amino acid sequence identity. According to PROSITE (http://prosite.expasy.org/), all but FDX10 are predicted to contain the conserved 2Fe2S ferredoxin-type FeS binding domain and the characteristic 2Fe2S cluster binding motif of plant-type ferredoxins (Cx4Cx2CxnC). However, this plant-type ferredoxin motif is slightly different in FDX8, FDX11 and MDX, in that it contains an additional amino acid between the first two cysteines, resulting in Cx5Cx2CxnC. This corresponds to the FeS binding motif of vertebrate adrenodoxins (Fukuyama 2004), which are typically part of the mitochondrial metabolism (Ewen et al. 2012). Due to the fact that the FDX10 peptide sequence reported in Yang et al. (2015) does not contain either of these binding motifs, we discarded it from our list of FDX isoforms and instead renamed MFDX to FDX10, which leaves a total of 12 FDX paralogs (PETF and FDX2-12, Supplementary Table S1 and Fig. 1b). The phylogenetic analysis of a multiple sequence alignment comprising all 12 FDX isoforms of C. reinhardtii and 69 orthologs from the entire spectrum of oxygenic phototrophs, including sequences from cyanobacteria, Chlorophyta, Bryophyta and higher plants, enabled us to construct a phylogenetic tree, containing all ferredoxin isotypes (Fig. 1a).
Multiple sequence alignment, evolutionary relationships and phylogeny of plant-type ferredoxin isoforms of Chlamydomonas reinhardtii. a A phylogenetic tree comprising all plant-type ferredoxin isotypes present in C. reinhardtii (PETF and FDX2-10) was constructed using the minimum evolution (ME) method (Rzhetsky and Nei 1992). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. The ME tree was searched using the close-neighbour-Interchange (CNI) algorithm (Nei Mak 2000) at a search level of 1. The neighbour-joining algorithm (Saitou and Nei 1987) was used to generate the initial tree. The analysis included 81 amino acid sequences from cyanobacteria (Syn: Synechocystis sp. PCC 6803; Nostoc: Nostoc sp. PCC 7524), Chlorophyta (Cr: C. reinhardtii, Vc: Volvox carteri nagariensis, Gp: Gonium pectorale, Cv: Chlorella variabilis NC64A, Mc Micromonas commoda, Os: Ostreococcus sp. RCC 809, Ol: Ostreococcus lucimarinus), Bryophyta (Pp: Physcomitrella patens) and Planta (Ara: Arabidopsis thaliana, Rs: Raphanus sativus, Gm: Glycine max). All positions containing gaps and missing data were eliminated. There were a total of 45 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 (Tamura et al. 2013). b Polypeptide alignment of plant-type ferredoxins PETF (marked in red) and FDX2-10 from C. reinhardtii and C. variabilis NC64A. Cysteine residues which coordinate the [2Fe2S]-cluster are marked with an asterisk. Amino acid positions of PETF for which experimental data indicate their participation in protein–protein interactions during complex formation with HYDA1 are indicated by arrows. Position labels with coloured backgrounds indicate residues, which, when exchanged, had the strongest impact on PETF-dependent HYDA1 activity. c Phylogenetic development of the different PETF/FDX paralogs identified in C. reinhardtii during the evolution of oxygenic phototrophic organisms, comprising cyanobacteria, Chlorophyta (Prasinophyceae, Trebouxiophyceae and Chlorophyceae), Bryophyta and higher plants. Species abbreviations representing the respective taxa are shown in blue
With the exception of FDX2, FDX9 and FDX12, all C. reinhardtii FDX paralogs are located in distinct monophyletic clades. They cluster together with orthologs of other chlorophycean, trebouxiophycean or prasinophycean algal species. For example, the sub-trees of isoforms FDX1, FDX4, FDX5, FDX6, FDX10 and FDX11 also include orthologs of higher plants and/or bryophytes, indicating that these FDX isoforms played a vital role as part of the genetic heritage for the evolution of higher photosynthetic organisms (Fig. 1c). Meanwhile, the sub-trees of paralogs FDX1 (PETF), FDX7 and FDX8 contain homologs of cyanobacterial origin such as from Synechocystis or Nostoc species, advocating for differential evolutionary pathways that might even date back to the endosymbiotic origin of chloroplasts from endocytosed cyanobacteria (Okazaki et al. 2010; Pedroza-Garcia et al. 2016; Rodriguez-Ezpeleta and Philippe 2006). Paralogs FDX3, FDX4, FDX6 and FDX10 appear to have evolved in the common ancestral lineage of the three Chlorophyta classes, while homologs of FDX2 can only be found in the chlorophycean and trebouxiophycean branches. Among the three algal classes, isoforms related to FDX5, FDX9, FDX11 and FDX12 are restricted to chlorophycean species and thus presumably differentiated from other isoforms rather late in C. reinhardtii evolution. The high bootstrap value of 88 suggests a common ancestor for FXD8, FDX10 and FDX11, which likely corresponds to the cyanobacterial origin of FDX8, represented here by Nostoc FDX8 (Fig. 1a). While FDX9 and FDX12 seem to be unique for C. reinhardtii, orthologs of FDX5 and FDX11 appear to have evolved together with subtypes FDX4, FDX6 and FDX10, as they can be found in higher photosynthetic organisms such as vascular plants and/or mosses. For FDX4 and FDX6, this corresponds to earlier data from Terauchi et al. (2009). A remaining question is the conservation of ferredoxin function. Once the individual chlorophyta isotype functions have been determined, the conservation of their functions during the course of evolution up to the orthologs of higher photosynthetic organisms can be examined.
In higher plants, different ferredoxins are located in different tissues. In C. reinhardtii, most of the previously examined ferredoxin paralogs (FDX1-3, FDX5 and FDX6) have been predicted or experimentally demonstrated to be localised in the chloroplast (Jacobs et al. 2009; Terauchi et al. 2009; Yang et al. 2015). According to PredAlgo (https://giavap-genomes.ibpc.fr/cgi-bin/predalgodb.perl?page=main) predictions, isoforms FDX8-9 are also located in the chloroplast, while the adrenodoxin-like isoform FDX10 is strongly predicted to be localised within the mitochondria. For FDX paralogs 7, 11 and 12, PredAlgo fails to predict a definite sub-cellular localisation, although the FDX7 ortholog of the closely related Chlorophycean species Volvox carteri, has a solid c-score value (Supplementary Table S2), suggesting its localisation within the chloroplast.
An earlier phylogenetic analysis based on 30 FDX sequences from algae, moss and higher plants suggested that PETF and FDX5 are most closely related to photosynthetic leaf-type FDXs, while FDX2 instead groups with non-photosynthetic root-type FDXs (Terauchi et al. 2009). The low bootstrap values in the earlier branching points do not allow any conclusions to be drawn on that matter from the larger scaled phylogram in Fig. 1a.
It is still not quite clear why such a multitude of different ferredoxin isotypes are required in a unicellular green alga. However, emerging evidence suggests that the presence of different isoforms allows exceptionally flexible algal species such as C. reinhardtii to quickly respond and adapt to changing environmental conditions (Peden et al. 2013; Terauchi et al. 2009; Yang et al. 2015). For example, each of the Fdx genes is differentially regulated in response to changes in nutrient supply (Fig. 2). PETF is the predominant isoform during photoautotrophic growth, comprising 98% of the total ferredoxin transcript pool in cells grown in Tris-acetate-phosphate medium (Terauchi et al. 2009). Fdx2 is induced in nitrate-grown cells, with its transcript abundance 300 to 400-fold higher with nitrate as the nitrogen source instead of ammonia (Terauchi et al. 2009), while Fdx5 is induced under copper or sulphur deprivation (Jacobs et al. 2009; Terauchi et al. 2009), with transcript abundance reaching almost the same level as Petf under copper deficiency. Indeed, each ferredoxin appears to play a specific role under specific conditions. FDX2 is assumed to participate in state transitions, a photoprotective mechanism where excitation energy is balanced between PSII and PSI by the reversible dissociation of light harvesting complex II (LHCII) proteins from PSII, and dark anoxia. FDX3 appears to be involved in N assimilation, while FDX4 possibly functions in glycolysis and ROS responses. FDX5 has been described as being important for hydrogenase maturation and furthermore appears to play a role in fatty acid desaturation and in maintaining the composition and functionality of the thylakoid membrane, especially during growth in the dark (Lambertz et al. 2010; Mus et al. 2007; Peden et al. 2013; Terauchi et al. 2009; Yang et al. 2015).
Schematic of putative ferredoxin interactions and functions in Chlamydomonas reinhardtii. Six out of 12 ferredoxin isoforms have been characterised in C. reinhardtii: Ferredoxin 1 (FDX1/PETF), FDX2, FDX3, FDX4, FDX5 and FDX6. PETF is the main electron donor of the hydrogenase HYDA, providing electrons (black lines) received from photosystem I (PSI) via the photosynthetic electron transfer chain, which additionally involves photosystem II (PSII), plastoquinone (PQ), cytochrome b6f (Cytb6f) and plastocyanin (PC), but also donates electrons to other redox enzymes, above all others to the ferredoxin-NADP+ oxidoreductase (FNR). LHC light harvesting complex, ATPase ATP synthase
C. reinhardtii ferredoxin interaction partners
The C. reinhardtii ferredoxins are not only regulated by different stimuli, but also have a wide range of interaction partners. In a recent yeast two-hybrid study, 216 putative ferredoxin interacting proteins were identified (Peden et al. 2013). PETF, in addition to interacting with FNR and HYDA1, also interacts with HYDA2 (Chang et al. 2007), as well as with sulphite and nitrite reductases, which are involved in nitrate and sulphate assimilation; ferredoxin thioredoxin reductase, which is part of the thiol signalling pathway; the glutamine-oxoglutarate amidotransferase, which functions in amino acid synthesis; stearoyl-ACP Δ9-desaturase, which functions in fatty acid desaturation; and phycocyanobilin ferredoxin oxidoreductase, which is involved in the synthesis of phytochromobilin (Hemschemeier and Happe 2011; Peden et al. 2013). FDX2, FDX3, FDX4 and FDX5 were also found to interact with HYDA1, although with lower affinities than for PETF (FDX2 displayed a moderate interaction and was capable of driving NADPH and H2 production, albeit at half the rate of PETF, while FDX3, FDX4 and FDX5 only displayed very weak interactions). The moderate affinity of FDX2 for HYDA1 is not surprising, as FDX2 is structurally very similar to PETF, with a recent finding that differences at only two amino acid positions contribute to major functional differences between the two proteins (Boehm et al. 2016). In addition to interacting with HYDA1, FDX3 was also found to interact with HYDA2, although the interaction was not as strong as that of PETF and HYDA2. FDX2, FDX4 and FDX5 also appear to interact with HYDEF, with FDX5 displaying the strongest interaction (note that due to problems with the promoter system, PETF, FDX3 and FDX6 were not able to be screened) (Peden et al. 2013).
The first appearance of the FDX5 isotype within the chlorophycean branch interestingly coincides with the evolution of small M1 type [FeFe]-hydrogenases (Meyer 2007) in the same algal class, which appears to support the connection between FDX5 and [FeFe]-hydrogenase maturation as proposed by Peden et al. (2013). However, the trebouxiophycean species C. variabilis NC64A also encodes an [FeFe]-hydrogenase of the more complex M3 structure type (Meyer 2007), which more closely resembles the presumptive bacterial type ancestor. Additionally, [FeFe]-hydrogenase activity has been reported for prasinophycean species such as Tetraselmis subcordiformis (D’Adamo et al. 2014; Guo et al. 2016). As members of neither of these two classes contain an FDX5 ortholog, hydrogenase maturation presumably depends on alternative FDX isoforms.
Transient protein interactions—mechanisms
The interaction between ferredoxins and their partners, like that of all protein electron transfer complexes, is transient, and thus displays characteristics typical of these interactions. These include a low binding affinity, with K d’s in the micromolar to millimolar range, but high complex turnover rates, which enable a continuous electron current (Bashir et al. 2011; Crowley and Ubbink 2003). The initial binding state, also known as the encounter state, which consists of an ensemble of orientations, plays an important role in complex formation (Bashir et al. 2011). The encounter state has a low specificity, with molecules held together by electrostatic interactions just long enough to increase their chance of finding a more favourable configuration with complementary electrostatic and hydrophobic interactions (Berg and Vonhippel 1985; Suh et al. 2007). This more stable configuration is then likely supported by van der Waals forces (Crowley and Ubbink 2003). Electron transfer complexes usually have small complex interfaces (Crowley and Ubbink 2003) which allow a rapid detachment after electron transfer.
In the chloroplast, changes in the affinity between PETF and HYDA1 appear to be regulated by the stromal pH. Unlike PETF-FNR complex formation, whose rate constant was found to be unaffected by pH variations across a wide range of physiological values, the modelled rate constant of the PETF–HYDA1 interaction was interestingly found to depend on the pH (Diakonova et al. 2016), with an increase in pH by two units from 7 to 9, resulting in a threefold increase in rate constant. The authors explain this effect by pH dependent changes in the surface charge distribution, which shows an increasingly polar distribution with a confined focus of positive net charge at the surface region covering the [4Fe4S]-cluster at higher pH values (>pH 8). This behaviour corresponds to pH changes in the stroma under illumination where in the dark a pH of 6 can be measured. Under full photosynthetic activity in the light, the pH increases to pH 8.5, and thus amplifies the rate constant of complex formation with HYDA1 in contrast to FNR (Diakonova et al. 2016). In this way, the emergency valve function of HYDA1 to deplete excessive reducing power in photosynthetic electron transport only sets in when actually needed, under high photo-fermentative activity (Hemschemeier and Happe 2011).
PETF–HYDA1 interaction
The PETF–HYDA1 binding interface has been extensively examined through molecular dynamics and modelling studies, nuclear magnetic resonance (NMR) titration analysis and site-directed mutagenesis. In an initial study by Long et al. (2008), Brownian and molecular dynamics simulations were used to model PETF–HYDA1 complex formation. It was found that PETF binds to HYDA1 mainly via electrostatic interactions between the binding surfaces of the two proteins and that the binding resembled that of PETF with other binding partners. In the case of HYDA2, Brownian dynamics (BD) and all-atom molecular dynamics (MD) simulations suggest that only one binding surface exists for PETF. PETF is however able to bind to HYDA2 in multiple orientations, with several encounter complexes identified (Long et al. 2008). According to the similarity of the PETF-binding regions on the surfaces of HYDA1 and HYDA2, the same can be assumed for HYDA1.
The PETF–HYDA1 complex was further probed in site-directed mutagenesis studies. Winkler et al. (2009) identified a number of amino acids in both, HYDA1 and PETF, to be essential for binding. In HYDA1, K396 appeared to be the most essential residue for complex formation (Fig. 3), with a K396Q variant displaying only 18% the activity of the wild type. The residues K179, K262, K397 and K433 were also found to be important, with substitutions of these residues resulting in moderate losses in activity. In PETF, D64, D90 and D95 were found to play a role in intermolecular attraction and orientation in the initial stages of complex formation, while D56, E122 and F93 were proposed to be essential for stabilising the final complex orientation. An electron transfer complex model suggested that electrostatic interactions between HYDA1 residue K396 and PETF C-terminal residue Y126 and PETF E122 and HYDA1 N-terminal A57 play a central role in complex formation (Winkler et al. 2010, 2009). Boehm et al. (2016) confirmed that positions F93 and Y126 are crucial for the interaction of PETF with HYDA1 by demonstrating that amino acid substitutions at the corresponding positions F62 and Y95 in FDX2 lowered its midpoint redox potential to values closer to those of PETF, which consequently increased H2 production threefold. Finally, a second site-directed mutagenesis study by Sybirna and Bottin (2013) additionally identified R227 in HYDA1 as being essential for complex formation with PETF.
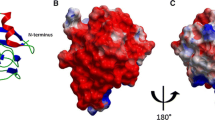
PETF–HYDA1 structure. a Surface charge distribution of HYDA1 and PETF and the PETF–HYDA1 complex. b Putative salt bridge contacts between HYDA1 and PETF in one of the best final complex solutions published in Rumpel et al. (2015), with a cluster to cluster distance of 11 Å. The labels with coloured backgrounds indicate residues which when were exchanged had the strongest impact on PETF-dependent HYDA1 activity
An NMR titration study by Rumpel et al. (2014) verified the findings of the above mentioned site-directed mutagenesis analysis studies. The authors compared the PETF–HYDA1 binding interface with that of PETF-FNR by adding increasing amounts of HYDA1 or FNR to 15N-labelled PETF and monitoring changes in chemical shifts of the backbone amide resonances of PETF. Upon addition of either HYDA1 or FNR, the largest changes in chemical shifts were seen for the backbone amides of PETF residues 55–60 (23–28 in the mature protein) in α helix 1, 90–99 (58–67) in α helix 2 and the proceeding loop and 121–126 (89–94) of the C-terminal helix, indicating a common binding interface. These residues are located in flexible regions surrounding the residues coordinating the [2Fe2S]-cluster on the surface of PETF (Rumpel et al. 2014). Interestingly, upon addition of the FNR interaction partner, selective changes in chemical shift were only seen for positions D51 and D90 (D19 and D58), indicating that these residues are important for PETF-FNR binding, but not for PETF–HYDA1 binding. Substitution of these residues resulted in an increased H2 production when assaying PETF-dependent H2 production under competitive conditions in the presence of FNR, confirming that D19 and D58 are indeed selective for complex formation with FNR (Rumpel et al. 2014). In contrast to these results, Winkler et al. (2009) determined that D90 does affect the kinetics of PETF-dependent HYDA1 activity by doubling the Km value. However, this comparatively mild effect can obviously be overlooked when evaluating the significant net increase of H2 production under competitive conditions.
In another study by Rumpel et al. (2015), an NMR solution structure of gallium(III)-substituted PETF was used in titrations where chemical shift data of residues coordinating and surrounding the [2Fe2S]-cluster were accessible for the first time. Upon addition of HYDA1, PETF residues 67–78 (35–46), which are located in the now visible long [2Fe2S]-cluster-coordinating loop, showed the strongest chemical shift perturbations. This would support the assumption that complex formation triggers a flip in the peptide bond orientation between positions C74 and S75, which through a slight displacement of the corresponding cluster binding thiol group of C74, affects the redox potential of the [2Fe2S]-cluster, thus promoting electron transfer to the respective redox partner (Morales et al. 1999; Winkler et al. 2009). Rumpel et al. also generated a docking model of the PETF NMR solution structure and the X-ray crystal structure of non-mature HYDA1 (as of yet no 3D structure has been solved for mature HYDA1). In this model, HYDA1 residue R227 (R187 in the processed protein), which was previously shown by Sybirna and Bottin (2013) to be essential for complex formation, was found to potentially form several H-bond and salt bridge interactions with PETF. On the other side of the interface, PETF residue E122 (E90), which was previously identified by Winkler et al. (2009) to play a key role in complex formation, was in close proximity to R227 and displayed the largest number of possible H-bonds and salt bridges to HYDA1. As indicated above, substitutions of E122 led to a severely diminished H2 production activity. Additionally, several HYDA1 residues with hydrophobic and polar side chains near the [4Fe4S]-cluster-coordinating cysteines (C90, C145, C337 and C341) were proposed to be essential for PETF–HYDA1 complex formation (Rumpel et al. 2015; Winkler et al. 2009).
In summary, acidic PETF side chains and basic HYDA1 side chains are important for complex formation, particularly E122 of PETF and residues R227 and K396 in HYDA1, as well as residues with hydrophobic and polar side chains in HYDA1 close to the [4Fe4S]-cluster coordinating cysteines and in the long [2Fe2S]-cluster-coordinating loop of PETF (Fig. 3). This information can be used to guide genetic engineering approaches to improve H2 production through optimising the kinetics of complex formation.
PETF–HYDA2 interaction
While several MD simulations have analysed the PETF–HYDA2 interaction process on a theoretical level, the findings from these studies are yet to be verified by experimental data. Although HYDA1 and HYDA2 exhibit a similar number and pattern of basic residues close to the electron accepting [4Fe4S]-cluster, in silico docking between PETF and HYDA2 performed by Chang et al. (2007) resulted in two differently orientated docking complexes with a 6 and 8.5 Å cluster to cluster distance. While the closer distance compared to the PETF–HYDA1 complex of Rumpel et al. (2015) (11 Å) would clearly be beneficial for fast electron tunnelling, the pattern of proposed salt bridge contacts between both interaction partners is less convincing, especially given the significant role of R227 for complex formation with HYDA1. R227 is highly but exclusively conserved among the group of small algal [FeFe]-hydrogenases of the M1-structure type (Winkler et al. 2013) which suggests an essential role for this Arg residue for all M1-type [FeFe]-hydrogenases, including HYDA2. In one of the two PETF-HYDA2 complexes suggested by Chang et al. (2007), the corresponding residue R230 of HYDA2 would not be involved in the final complex configuration, and would instead end up as an uncompensated charge covering the [4Fe4S]-cluster, which appears unlikely. In the second complex model, R230 does interact with an Asp residue of PETF (D90), which was demonstrated by Rumpel et al. (2014) to be dispensable for electron transfer to HYDA1. MD simulations, such as that of Long et al. (2008), are valuable for our general understanding of the process of complex formation, but should be complemented and verified by experimental data such as mutagenesis studies or NMR titrations. At this point, the investigations on PETF complexes with both HYDA isoforms are incomplete. For the HYDA1–PETF interaction, simulations are missing, while for the HYDA2–PETF interaction, experimental data are required to examine electron transfer between HYDA2 and PETF.
Other potential HYDA1 interaction partners
At this point, it cannot be ruled out that apart from the already examined isoforms FDX1-6, one of the more recently discovered plastidial FDX paralogs might also turn out to be a native electron donor for HYDA1. However, the characteristic contact pattern for PETF–HYDA1 complex formation described above would not be applicable, as some of the most important positions discussed here and indicated in the multiple sequence alignment of Fig. 1b are absent in most of the new ferredoxin sequences.
Biotechnological applications—improving electron transfer and therefore hydrogen production by altering the PETF–HYDA1 interaction
Photobiological H2 production is of enormous interest due to its potential to be renewable, as well as carbon–neutral or even carbon-negative (Dubini and Ghirardi 2015; Eroglu and Melis 2016; Oey et al. 2016). However, significantly higher H2 production efficiencies are required before biological H2 production can become commercially viable. Microalgal systems currently only have photon conversion efficiencies of 3%, while the theoretically achievable maximum is ~12–14% (Scoma et al. 2012; Volgusheva et al. 2013). The highest efficiency strains will therefore require significant re-engineering of the H2 production process. One of the major factors limiting sustained H2 production is electron supply to the hydrogenase. However, genetic engineering approaches based on in vitro-tested results have the potential to overcome this problem.
A few different strategies can be followed. One approach is the replacement of the native PETF and/or HYDA1 with protein variants which either have higher affinities to each other, or in the case of PETF, a significantly lower affinity for FNR. One of these variants which could be expressed in algae is the D19A/D58A double variant of PETF, which according to Rumpel et al. (2014) resulted in a higher in vitro H2 production activity due to a decreased affinity to the competitor FNR compared to wild-type ferredoxin. To further manipulate the PETF-binding interface selectively, either in favour of HYDA1 or disfavour of FNR, the substitution of other amino acid residues of 15N-labelled PETF, which displayed differing chemical shift perturbations in FNR- and HYDA1-dependent 2D 1H-15N-TROSY-HSQC experiments, could also be tested (Rumpel et al. 2014). However, the possible number of expedient changes on the surface of PETF is limited due to the large overlap between the binding interfaces for FNR and HYDA1. Furthermore, manipulation of PETF can also affect complex formation with the stromal subunits of PSI, as reported by Rumpel et al.
Instead of introducing further amino acid exchanges into PETF, it might be more favourable for future researchers to target HYDA1. To improve its binding affinity for PETF into a range which renders HYDA1 as the main electron acceptor in the presence of competitors such as FNR, is however a challenging task, which likely requires a tailored directed evolution strategy rather than a rational site-directed mutagenesis approach. The severe side effects of manipulating R227 (R189) demonstrate that single exchanges close to the active centre of redox enzymes such as HYDA1 can affect multiple enzymatic features. Applied research concepts targeting an improved electron transfer to HYDA1 may profit from refreshingly alternative approaches and perspectives, such as those described by Diakonova et al., which bring into focus formerly neglected aspects of transient complex formation (Diakonova et al. 2016).
Unfortunately, targeted nuclear genome engineering is difficult in microalgae. Techniques utilising homologous recombination (HR), the process required for site-directed genomic manipulation, such as zinc-finger nucleases, TALENs, or CRISPR/Cas9 (Jinkerson and Jonikas 2015), are still not fully established for targeting the C. reinhardtii nuclear genome (where both PETF and HYDA1 are encoded). However, progress is being made, with the CRISPR/Cas9 system recently showing promising results (Baek et al. 2016; Jiang et al. 2014; Shin et al. 2016). While a C. reinhardtii hydA1 hydA2 double-knockout strain is available (Meuser et al. 2012), and thus can be used for the ectopic integration of an engineered HydA1 variant, a corresponding knockout mutant for the Petf locus is yet to be identified and would likely be lethal. A possible approach to circumvent this problem could be to first integrate another copy of the Petf gene into the chloroplast genome, which in contrast to the nuclear genome is amenable to homologous recombination, before screening for nuclear ΔPetf knockout mutants.
Another idea being tested is the peptide linker-based fusion of HYDA1 and PETF. This has been shown to lead to improved levels of H2 production both, in vitro and in vivo. C. reinhardtii hydA1-1 hydA2-1 transformants expressing the fusion protein exhibited a 4.5-fold increased photosynthetic H2 production rate standardised for hydrogenase amount (PHPRH) compared to native HYDA1. According to Eilenberg et al. (2016), this mainly results from a close tethering of HYDA1 to PSI via the PETF domain of the fusion protein. The HYDA1–PETF fusion protein furthermore shows a slightly decreased sensitivity of HYDA1 towards O2 (Eilenberg et al. 2016; Yacoby et al. 2011), which might be explained by additional electrons provided by the [2Fe2S]-redox centre of PETF promoting a fast reductive turnover of O2 to H2O at the H-cluster of HYDA1.
Concluding remarks
In conclusion, microalgae provide an unusual wealth of ferredoxin isoforms which appear to play important roles in a number of different pathways in response to various stimuli. The C. reinhardtii ferredoxins represent the most widespread subgroup of ferredoxins from cyanobacteria to higher plants. However, the differential functions of most of the C. reinhardtii ferredoxins have either not been examined or not sufficiently supported on the experimental level. PETF on the other hand has been studied extensively due to its function as an electron donor for a number of important redox enzymes, including HYDA1. The complete characterisation of the complex interface between HYDA1 and PETF has already enabled engineered improvements in photohydrogen production, a process currently limited by the presence of major competitors such as FNR, and the utilisation of strategies such as directed evolution is likely to lead to further improvements in the future.
References
Baek K, Kim DH, Jeong J, Sim SJ, Melis A, Kim JS, Jin E, Bae S (2016) DNA-free two-gene knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 ribonucleoproteins. Sci Rep. doi:10.1038/srep30620
Bashir Q, Scanu S, Ubbink M (2011) Dynamics in electron transfer protein complexes. FEBS J 278:1391–1400. doi:10.1111/j.1742-4658.2011.08062.x
Berg OG, Vonhippel PH (1985) Diffusion-controlled macromolecular interactions. Annu Rev Biophys Biol 14:131–160. doi:10.1146/annurev.biophys.14.1.131
Boehm M, Alahuhta M, Mulder DW, Peden EA, Long H, Brunecky R, Lunin VV, King PW, Ghirardi ML, Dubini A (2016) Crystal structure and biochemical characterization of Chlamydomonas FDX2 reveal two residues that, when mutated, partially confer FDX2 the redox potential and catalytic properties of FDX1. Photosynth Res 128:45–57. doi:10.1007/s11120-015-0198-6
Chang CH, King PW, Ghirardi ML, Kim K (2007) Atomic resolution modeling of the ferredoxin: [FeFe] hydrogenase complex from Chlamydomonas reinhardtii. Biophys J 93:3034–3045. doi:10.1529/biophysj.107.108589
Crowley PB, Ubbink M (2003) Close encounters of the transient kind: Protein interactions in the photosynthetic redox chain investigated by NMR spectroscopy. Accounts Chem Res 36:723–730. doi:10.1021/ar0200955
D’Adamo S, Jinkerson RE, Boyd ES, Brown SL, Baxter BK, Peters JW, Posewitz MC (2014) Evolutionary and biotechnological implications of robust hydrogenase activity in halophilic strains of tetraselmis. PLoS ONE. doi:10.1371/journal.pone.0085812
Diakonova AN, Khrushchev SS, Kovalenko IB, Riznichenko GY, Rubin AB (2016) Influence of pH and ionic strength on electrostatic properties of ferredoxin, FNR, and hydrogenase and the rate constants of their interaction. Phys Biol 13:056004. doi:10.1088/1478-3975/13/5/056004
Dubini A, Ghirardi ML (2015) Engineering photosynthetic organisms for the production of biohydrogen. Photosynth Res 123:241–253. doi:10.1007/s11120-014-9991-x
Eilenberg H, Weiner I, Ben-Zvi O, Pundak C, Marmari A, Liran O, Wecker MS, Milrad Y, Yacoby I (2016) The dual effect of a ferredoxin-hydrogenase fusion protein in vivo: successful divergence of the photosynthetic electron flux towards hydrogen production and elevated oxygen tolerance. Biotechnol Biofuels. doi:10.1186/s13068-016-0601-3
Eroglu E, Melis A (2016) Microalgal hydrogen production research. Int J Hydrogen Energ 41:12772–12798. doi:10.1016/j.ijhydene.2016.05.115
Ewen KM, Ringle M, Bernhardt R (2012) Adrenodoxin: a versatile ferredoxin. IUBMB Life 64:506–512. doi:10.1002/iub.1029
Fukuyama K (2004) Structure and function of plant-type ferredoxins. Photosynth Res 81:289–301. doi:10.1023/B:PRES.0000036882.19322.0a
Guo Z, Li Y, Guo HY (2016) Characterization of H2 photoproduction by marine green alga Tetraselmis subcordiformis integrated with an alkaline fuel cell. Biotechnol Lett 38:435–440. doi:10.1007/s10529-015-2008-9
Hanke G, Mulo P (2013) Plant type ferredoxins and ferredoxin-dependent metabolism. Plant Cell Environ 36:1071–1084. doi:10.1111/pce.12046
Happe T, Naber JD (1993) Isolation, characterization and N-terminal amino acid sequence of hydrogenase from the green alga Chlamydomonas reinhardtii. Eur J Biochem 214:475–481. doi:10.1111/j.1432-1033.1993.tb17944.x
Happe T, Mosler B, Naber JD (1994) Induction, localization and metal content of hydrogenase in the green alga Chlamydomonas reinhardtii. Eur J Biochem 222:769–774. doi:10.1111/j.1432-1033.1994.tb18923.x
Hemschemeier A, Happe T (2011) Alternative photosynthetic electron transport pathways during anaerobiosis in the green alga Chlamydomonas reinhardtii. BBA 1807:919–926. doi:10.1016/j.bbabio.2011.02.010
Jacobs J, Pudollek S, Hemschemeier A, Happe T (2009) A novel, anaerobically induced ferredoxin in Chlamydomonas reinhardtii. FEBS Lett 583:325–329. doi:10.1016/j.febslet.2008.12.018
Jiang WZ, Brueggeman AJ, Horken KM, Plucinak TM, Weeks DP (2014) Successful transient expression of Cas9 and single guide RNA genes in Chlamydomonas reinhardtii. Eukaryot Cell 13:1465–1469. doi:10.1128/ec.00213-14
Jinkerson RE, Jonikas MC (2015) Molecular techniques to interrogate and edit the Chlamydomonas nuclear genome. Plant J 82:393–412. doi:10.1111/tpj.12801
Lambertz C, Hemschemeier A, Happe T (2010) Anaerobic expression of the ferredoxin-encoding FDX5 gene of Chlamydomonas reinhardtii is regulated by the Crr1 transcription factor. Eukaryot Cell 9:1747–1754. doi:10.1128/EC.00127-10
Long H, Chang CH, King PW, Ghirardi ML, Kim K (2008) Brownian dynamics and molecular dynamics study of the association between hydrogenase and ferredoxin from Chlamydomonas reinhardtii. Biophys J 95:3753–3766. doi:10.1529/biophysj.107.127548
Meuser JE, D’Adamo S, Jinkerson RE, Mus F, Yang WQ, Ghirardi ML, Seibert M, Grossman AR, Posewitz MC (2012) Genetic disruption of both Chlamydomonas reinhardtii [FeFe]-hydrogenases: insight into the role of HYDA2 in H2 production. Biochem Biophys Res Commun 417:704–709. doi:10.1016/j.bbrc.2011.12.002
Meyer J (2007) [FeFe] hydrogenases and their evolution: a genomic perspective. Cell Mol Life Sci 64:1063–1084. doi:10.1007/s00018-007-6477-4
Morales R, Chron MH, Hudry-Clergeon G, Petillot Y, Norager S, Medina M, Frey M (1999) Refined X-ray structures of the oxidized, at 1.3 angstrom, and reduced, at 1.17 angstrom, [2Fe-2S] ferredoxin from the cyanobacterium Anabaena PCC7119 show redox-linked conformational changes. BioChemistry 38:15764–15773. doi:10.1021/bi991578s
Mus F, Dubini A, Seibert M, Posewitz MC, Grossman AR (2007) Anaerobic acclimation in Chlamydomonas reinhardtii: anoxic gene expression, hydrogenase induction, and metabolic pathways. J Biol Chem 282:25475–25486. doi:10.1074/jbc.M701415200
Mussgnug JH (2015) Genetic tools and techniques for Chlamydomonas reinhardtii. Appl Microbiol Biotechnol 99:5407–5418. doi:10.1007/s00253-015-6698-7
Nei MaK S (2000) Molecular evolution and phylogenetics. Oxford University Press, New York
Oey M, Sawyer AL, Ross IL, Hankamer B (2016) Challenges and opportunities for hydrogen production from microalgae. Plant Biotechnol J. doi:10.1111/pbi.12516
Okazaki K, Kabeya Y, Miyagishima SY (2010) The evolution of the regulatory mechanism of chloroplast division. Plant Signal Behav 5:164–167 pii]
Peden EA, Boehm M, Mulder DW, Davis R, Old WM, King PW, Ghirardi ML, Dubini A (2013) Identification of global ferredoxin interaction networks in Chlamydomonas reinhardtii. J Biol Chem 288:35192–35209. doi:10.1074/jbc.M113.483727
Pedroza-Garcia JA, Domenichini S, Bergounioux C, Benhamed M, Raynaud C (2016) Chloroplasts around the plant cell cycle. Curr Opin Plant Biol 34:107–113
Rodriguez-Ezpeleta N, Philippe H (2006) Plastid origin: replaying the tape. Curr Biol 16:R53–R56
Rumpel S, Siebel JF, Fares C, Duan J, Reijerse E, Happe T, Lubitz W, Winkler M (2014) Enhancing hydrogen production of microalgae by redirecting electrons from photosystem I to hydrogenase. Energy Environ Sci 7:3296–3301. doi:10.1039/c4ee01444h
Rumpel S, Siebel JF, Diallo M, Fares C, Reijerse EJ, Lubitz W (2015) Structural insight into the complex of ferredoxin and [FeFe] hydrogenase from Chlamydomonas reinhardtii. Chembiochem 16:1663–1669. doi:10.1002/cbic.201500130
Rzhetsky A, Nei M (1992) A simple method for estimating and testing minimum-evolution trees. Mol Biol Evol 9:945–967
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Scoma A, Krawietz D, Faraloni C, Giannelli L, Happe T, Torzillo G (2012) Sustained H2 production in a Chlamydomonas reinhardtii D1 protein mutant. J Biotechnol 157:613–619. doi:10.1016/j.jbiotec.2011.06.019
Shin M (2004) How is ferredoxin-NADP reductase involved in the NADP photoreduction of chloroplasts? Photosynth Res 80:307–313. doi:10.1023/B:PRES.0000030456.96329.f9
Shin SE, Lim JM, Koh HG, Kim EK, Kang NK, Jeon S, Kwon S, Shin WS, Lee B, Hwangbo K, Kim J, Ye SH, Yun JY, Seo H, Oh HM, Kim KJ, Kim JS, Jeong WJ, Chang YK, Jeong BR (2016) CRISPR/Cas9-induced knockout and knock-in mutations in Chlamydomonas reinhardtii. Sci Rep. doi:10.1038/srep27810
Suh J-Y, Tang C, Clore GM (2007) Role of electrostatic interactions in transient encounter complexes in protein-protein association investigated by paramagnetic relaxation enhancement. J Am Chem Soc 129:12954. doi:10.1021/ja0760978
Sybirna K, Bottin H (2013) New insights into the systems for heterologous synthesis and maturation of hydrogenases, the most efficient biohydrogen producers. Int J Hydrog Energ 38:13164–13171. doi:10.1016/j.ijhydene.2013.07.106
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Terauchi AM, Lu SF, Zaffagnini M, Tappa S, Hirasawa M, Tripathy JN, Knaff DB, Farmer PJ, Lemaire SD, Hase T, Merchant SS (2009) Pattern of expression and substrate specificity of chloroplast ferredoxins from Chlamydomonas reinhardtii. J Biol Chem 284:25867–25878. doi:10.1074/jbc.M109.023622
Volgusheva A, Styring S, Mamedov F (2013) Increased photosystem II stability promotes H2 production in sulfur-deprived Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 110:7223–7228. doi:10.1073/pnas.1220645110
Winkler M, Kuhlgert S, Hippler M, Happe T (2009) Characterization of the key step for light-driven hydrogen evolution in green algae. J Biol Chem 284:36620–36627. doi:10.1074/jbc.M109.053496
Winkler M, Hemschemeier A, Jacobs J, Stripp S, Happe T (2010) Multiple ferredoxin isoforms in Chlamydomonas reinhardtii: their role under stress conditions and biotechnological implications. Eur J Cell Biol 89:998–1004. doi:10.1016/j.ejcb.2010.06.018
Winkler M, Esselborn J, Happe T (2013) Molecular basis of [FeFe]-hydrogenase function an insight into the complex interplay between protein and catalytic cofactor. BBA 1827: 974–985. doi:10.1016/j.bbabio.2013.03.004
Yacoby I, Pochekailov S, Toporik H, Ghirardi ML, King PW, Zhang SG (2011) Photosynthetic electron partitioning between [FeFe]-hydrogenase and ferredoxin:NADP+-oxidoreductase (FNR) enzymes in vitro. Proc Natl Acad Sci USA 108:9396–9401. doi:10.1073/pnas.1103659108
Yang WQ, Wittkopp TM, Li XB, Warakanont J, Dubini A, Catalanotti C, Kim RG, Nowack ECM, Mackinder LCM, Aksoy M, Page MD, D’Adamo S, Saroussi S, Heinnickel M, Johnson X, Richaud P, Alric J, Boehm M, Jonikas MC, Benning C, Merchant SS, Posewitz MC, Grossman AR (2015) Critical role of Chlamydomonas reinhardtii ferredoxin-5 in maintaining membrane structure and dark metabolism. Proc Natl Acad Sci USA 112:14978–14983. doi:10.1073/pnas.1515240112
Acknowledgements
This research was supported by the Federal Ministry of Education and Research, Germany (ERASynBio consortium “Sun2Chem”) and the Volkswagen Foundation (LigH2t).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sawyer, A., Winkler, M. Evolution of Chlamydomonas reinhardtii ferredoxins and their interactions with [FeFe]-hydrogenases. Photosynth Res 134, 307–316 (2017). https://doi.org/10.1007/s11120-017-0409-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11120-017-0409-4