Abstract
Fucoxanthin chlorophyll a/c-binding protein (FCP) is a unique light-harvesting apparatus in diatoms. Several biochemical characteristics of FCP oligomer and trimer from different diatom species have been reported previously. However, the integration of information about molecular organizations and polypeptides of FCP through a comparison among diatoms has not been published. In this study, we used two-dimensional clear-native/SDS-PAGE to compare the oligomeric states and polypeptide compositions of FCP complexes from four diatoms: Chaetoceros gracilis, Thalassiosira pseudonana, Cyclotella meneghiniana, and Phaeodactylum tricornutum. FCP oligomer was found in C. gracilis, T. pseudonana, and C. meneghiniana, but not in P. tricornutum. The oligomerization varied among the three diatoms, although a predominant subunit having similar molecular weight was recovered in each FCP oligomer. These results suggest that the predominant subunit is involved in the formation of high FCP oligomerization in each diatom. In contrast, FCP trimer was found in all the diatoms. The trimerizations were quite similar, whereas the polypeptide compositions were markedly different. On the basis of this information and that from mass spectrometric analyses, the gene products in each FCP complex were identified in T. pseudonana and P. tricornutum. Based on these results, we discuss the role of FCP oligomer and trimer from the four diatoms.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Diatoms are unicellular photosynthetic eukaryotes found throughout the world’s oceans and freshwater environments (Field et al. 1998). They are ecologically important for their high abundance in aquatic environments, accounting for up to one quarter of global primary productivity (Falciatore and Bowler 2002). Their successful prosperity is partly because of their light-harvesting strategies. Diatoms have a unique light-harvesting apparatus, fucoxanthin chlorophyll (Chl) a/c-binding protein (FCP). FCP shows sequence similarity with light-harvesting Chl a/b-binding protein (LHC) of green plants, but pigment compositions differ significantly (Green and Pichersky 1994). Briefly, FCP possesses Chl c as antennae Chl. Fucoxanthin is a major light-harvesting carotenoid. Diadinoxanthin and diatoxanthin are minor carotenoids involved in diadinoxanthin cycle, which is a type of xanthophyll cycle (Lohr and Wilhelm 1999). These pigments can absorb blue-green spectral components (450–580 nm), which remain even in deep water layers (Depauw et al. 2012). Since diatoms have developed such an excellent light harvesting system, their evolution resulted in successfully occupying ecological niches in aquatic environments.
Molecular organization and polypeptide compositions of FCP complexes have been characterized in four diatom species. Cyclotella meneghiniana is a freshwater centric diatom whose FCPs have been well characterized in recent years. C. meneghiniana contains two FCP complexes, namely the trimeric FCPa and oligomeric FCPb. The two FCP complexes are composed of different polypeptides but are spectroscopically rather similar (Büchel 2003; Beer et al. 2006; Gildenhoff et al. 2010). Thalassiosira pseudonana and Phaeodactylum tricornutum are marine centric and pennate diatoms, respectively. Whole genome sequences are available for these two diatoms (Armbrust et al. 2004; Bowler et al. 2008). T. pseudonana contains oligomeric and trimeric FCP complexes, whereas P. tricornutum contains only a trimeric FCP complex (Grouneva et al. 2011). Chaetoceros gracilis is a marine centric diatom and is a good model for study on the diatom photosystem II (PSII) (Nagao et al. 2007, 2010a, b, 2013a; Okumura et al. 2008). We recently reported the isolation of oligomeric FCP-A and trimeric FCP-B/C complexes from C. gracilis (Nagao et al. 2012). The two complexes were composed of different polypeptides and showed different spectroscopic properties (Nagao et al. 2012, 2013b). Although the biochemical characterization of FCPs has been established for each diatom, it remains unclear whether the oligomeric states and polypeptide compositions of FCP complexes are similar among the diatom species.
In the present study, we compared oligomeric states and polypeptide compositions of FCP complexes among the four diatoms: C. gracilis, T. pseudonana, C. meneghiniana, and P. tricornutum. FCP complexes were isolated by clear-native PAGE (CN-PAGE) after solubilizing thylakoid membranes from each diatom. The oligomerization and polypeptides of FCPs differed markedly. The information regarding diatom FCPs was unified for the first time in this report.
Materials and methods
Cultures
Cells of the marine centric diatom C. gracilis Schütt (UTEX LB 2658; termed Chaetoceros in the text) were grown in artificial seawater at 298 K under continuous illumination at 30 μmol photons m−2 s−1 with air bubbling, as described previously (Nagao et al. 2007, 2010b). Cells of the marine centric diatom T. pseudonana (CCMP 1335; termed Thalassiosira in the text) and the pennate diatom P. tricornutum (CCMP2561; termed Phaeodactylum in the text) were grown in the same seawater with Chaetoceros at 291 K under continuous illumination at 30 μmol photons m−2 s−1 with stirring and air bubbling for 10–11 days. Cells of the freshwater centric diatom C. meneghiniana (SAG strain 1020-1a; termed Cyclotella in the text) were grown in CSi medium at 291 K under continuous illumination at 30 μmol photons m−2 s−1 with stirring and air bubbling for 10–11 days (Ichimura 1971). The grown cells were harvested by filtration using a Pellicon system (Millipore, USA) and collected by centrifugation at 8,000×g for 10 min. The cells were suspended in a medium containing 1 M betaine and 50 mM MES–NaOH (pH 6.5) (buffer A) and then immediately frozen by liquid nitrogen and stored at 193 K.
Isolation of thylakoid membranes
Isolation was performed at 277 K unless otherwise indicated. Thylakoid membranes of Chaetoceros were isolated as described previously (Nagao et al. 2007, 2010b). The cells of Thalassiosira, Phaeodactylum, and Cyclotella were loaded into a pre-chilled Bead-Beater chamber (Bio-Spec Products) and glass beads (100-μm diameter) were added to give a 1:10 ratio (w/w) of glass beads to cell suspension. The cells were then broken with 19 break cycles, each cycle consisting of 10 s of homogenization followed by 3 min of cooling. After centrifugation at 8,000×g for 10 min to discard unbroken materials, the thylakoid membranes were collected by centrifugation at 40,000×g for 10 min. The membranes were suspended in buffer A and Chl concentrations (Chls a and c) were determined in 90 % acetone using the equation of Jeffrey and Humphrey (1975). The thylakoids were immediately frozen by liquid nitrogen and stored at 193 K.
Two-dimensional CN/SDS-PAGE
CN-PAGE was performed as described (Nagao et al. 2012) with some modifications. Thylakoid membranes were solubilized with 4 % n-dodecyl-β-d-maltoside at 1.0 mg Chl ml−1 for 10 min on ice in the dark. After centrifugation at 40,000×g for 10 min, the supernatant was applied to a 4–18 % polyacrylamide gel. Each sample containing 5 μg Chl was loaded in a separate lane. A standard molecular marker (NativeMark™; Invitrogen, USA) was used. Electrophoresis was performed at 277 K in electrophoresis buffers containing 25 mM imidazole (pH 7.0) as the anode buffer, and 50 mM Tricine, 7.5 mM imidazole, 0.02 % sodium deoxycholate, and 0.02 % (w/v) Triton X-100 as the cathode buffer.
For two-dimensional CN/SDS-PAGE, CN-PAGE lanes or bands were cut out and denatured using 2 % lithium lauryl sulfate and 2 % 2-mercaptoethanol in buffer A at 298 K for 30 min, and then subjected to SDS-PAGE using 19 % acrylamide and 7.5 M urea. The two-dimensional gel was stained with silver (Aro et al. 2005) or Coomassie Brilliant Blue R-250. A standard molecular marker (LMW Marker; GE Healthcare, UK) was used in SDS-PAGE.
Mass spectrometric analysis
A protein band of FCP obtained by SDS-PAGE was reduced by incubation with 50 mM dithiothreitol for 2 h, then alkylated by 110 mM acrylamide for 30 min at 298 K. After washing the gel, the protein band was digested using sequencing grade modified trypsin (Promega, USA) at 310 K overnight. An aliquot of the digest was analyzed by nano LC–MS/MS using LCQ Deca XP (Finnigan, USA). The peptides were separated using a nano ESI spray column (100 μm i.d. × 375 μm o.d.) packed with a reversed-phase material (Inertsil ODS-3, 3 μm, GL Sciences, Japan) using a gradient of 1–60 % acetonitrile gradient containing 0.75 % formic acid at a flow rate 400 nl min−1. The mass spectrometer was operated in the positive ion mode and the spectra were acquired in a data-dependent MS/MS mode. The MS/MS spectra were searched against an in-house database containing T. pseudonana and P. tricornutum protein sequences using an in-house Mascot server (version: 2.3, Matrix Sciences, UK). The parameters used for database searching of the MS/MS spectra were as follows: type of search, MS/MS ion search; enzyme, trypsin; fixed modifications, propionamide (C); variable modifications, oxidation (M), Gln- > pyro-Glu (N-term Q), acetyl (protein N-term), formyl (protein N-term); mass values, monoisotopic; peptide mass tolerance, ±2 Da; fragment mass tolerance, ±0.8 Da; max missed cleavages, 2; instrument type, ESI-Trap. If peptide assignments passed this scoring filter, the corresponding MS/MS spectra were manually evaluated. Manual data assignment is effective for identifying unexpected fragmentation peptides.
Results and discussion
Distribution of pigment–protein complexes
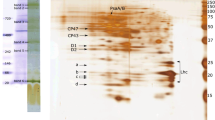
Pigment–protein complexes were separated by CN-PAGE from the four diatoms—Chaetoceros, Thalassiosira, Cyclotella, and Phaeodactylum (Fig. 1), and polypeptide compositions were resolved by two-dimensional CN/SDS-PAGE (Fig. 2). In Chaetoceros, the distribution of protein complexes was consistent with the previous results (Nagao et al. 2012). In the present study, the complexes were renamed as PSI, PSII dimer (PSIID), PSII monomer (PSIIM), FCP oligomer (FCPO), and FCP trimer (FCPT). On the basis of the results of Chaetoceros, we identified the pigment–protein complexes in the other three diatoms. The five protein complexes were also detected in Thalassiosira and Cyclotella, whereas only four protein complexes, namely PSI, PSIID, PSIIM, and FCPT, were detected in Phaeodactylum. The assignment of PSI and PSII were done from their mobility on electrophoresis, and it was almost same as the previous report (Grouneva et al. 2011). The assignment of PSII complexes was also confirmed by fluorescence analysis (data not shown), although a small amount of PSII monomer can be seen in Chaetoceros.
FCPT was found in all the four diatoms. Their molecular weights and brown colors resembled to some extent, although the FCPT bands became deformed from the effect of the detergent on the bottom of the gel. This suggests that trimeric FCP complexes are conserved in diatoms.
In contrast, FCPO was found in Chaetoceros, Thalassiosira, and Cyclotella, but it was not found in Phaeodactylum (Fig. 1). The molecular weight of FCPO of Chaetoceros was slightly higher than those of Thalassiosira and Cyclotella. The molecular weight of FCPO of Thalassiosira was very similar to that of Cyclotella. These results suggest that oligomeric FCP complexes have a wide variety of its oligomerization. The molecular weight of subunit in FCPO was higher than that in FCPT (Fig. 2). However, such a higher molecular band, corresponding to the subunit in FCPO, could not be detected from Phaeodactylum in the two-dimensional PAGE, indicating that the high molecular weight FCP subunit is originally absent in Phaeodactylum, regardless of the effect of strict detergent and the condition of electrophoresis.
In the other complexes, the molecular weights and green colors of PSIID and PSIIM were similar among the four diatoms, but those of PSI differed remarkably. Since it is known that PSI complexes are monomeric in diatoms (Veith and Büchel 2007), the binding patterns of FCPs to the monomeric PSI complexes appear to be different among the diatoms. Actually, the polypeptide compositions in the PSI fraction were distinctly different among Chaetoceros, Thalassiosira, Cyclotella, and Phaeodactylum (Fig. 2). This result was consistent with those of the previous studies in Chaetoceros (Nagao et al. 2007; Ikeda et al. 2008), Thalassiosira (Grouneva et al. 2011), and Cyclotella (Veith et al. 2009). In Phaeodactylum, however, PSI was detected near PSIIM in CN-PAGE (Fig. 1); FCP subunits were hardly found in the PSI fraction by two-dimensional PAGE (Fig. 2). Since the monomeric PSI complex is near to the position of the monomeric PSII complex in native PAGE (Watanabe et al. 2011), FCPs may be dissociated from PSI complexes in Phaeodactylum. This is in agreement with the previous study (Berkaloff et al. 1990), whereas it is inconsistent with the other studies (Veith and Büchel 2007; Grouneva et al. 2011). The differences seem to occur by strict detergent concentration in addition to other biochemical condition. If high concentrated detergent is used for the solubilization of thylakoids or the treatment of PSI fraction, the tightly bounded FCP is likely removed from PSI. This is in good agreement with the recent biochemical study that showed the effect of excess detergent on the removal of FCP from PSI (Ikeda et al. 2013). These findings are indicative of a species-dependent variation in the distribution of FCP complexes in diatoms.
Comparison of polypeptide compositions of FCP complexes
In this study, we focused on the polypeptide compositions of FCPs in the four diatoms. Figure 3 shows the comparison of FCP subunits in each FCP complex, with addition of sample numbers (Nos. 1–10). FCPO of Chaetoceros was mainly composed of FCP-A, which corresponded with our previous results (Nagao et al. 2012). FCPO of Thalassiosira was composed of one subunit, which had a similar molecular weight with FCP-A. The subunit (No. 1) was identified as Lhcf8 by MS analysis (Table 1). FCPO of Cyclotella was also composed of one subunit (No. 5), which had a similar molecular weight with FCP-A and Lhcf8. The subunit in FCPO of Cyclotella appears to be a product of the fcp5 gene (Beer et al. 2006). These results suggest that similar predominant subunits are involved in the formation of high oligomerization of FCP in each diatom.
FCPT of Chaetoceros was mainly composed of FCP-B/C, which corresponded with our previous results (Nagao et al. 2012). FCPT of Thalassiosira was composed of three bands: the first (No. 2) had a similar molecular weight with FCP-A, the second (No. 3) had a slightly higher molecular weight than FCP-B, and the third (No. 4) had a slightly higher molecular weight than FCP-C but it was a minor component. The first band was identified as Lhcf1 and Lhcf8; the second as Lhcf1, Lhcf3, Lhcf4, Lhcf5, Lhcf8/9, and Lhcx6_1; and the third as Lhcf1, Lhcf3, Lhcf6, Lhcf7, Lhcf11, and two FCP-related proteins (Table 1). In Cyclotella, the polypeptide compositions of FCPT were very similar to those in Thalassiosira. The first band (No. 6) appeared to be a product of the fcp6/7 genes and the second (No. 7) appeared to be a product of the fcp1-3 genes (Beer et al. 2006), although a possible candidate of the third (No. 8) was not found. FCPT of Phaeodactylum was composed of a major subunit (No. 9) and a minor subunit (No. 10). The major subunit had a similar molecular weight with FCP-B and was identified as Lhcf1, Lhcf2, Lhcf3/4, Lhcf4, Lhcf5, Lhcf8, Lhcf10, Lhcf11, Lhcr2, Lhcr4, Lhcr12, and two FCP-related proteins (Table 2). The minor subunit had a similar molecular weight with FCP-C and was identified as Lhcf1, Lhcf2, Lhcf4, Lhcf5, Lhcf8, Lhcf11, Lhcf14, Lhcf16, Lhcr1, Lhcr3, Lhcr13, and one FCP-related protein (Table 2). These results suggest that trimeric FCP complexes have a variety of polypeptide compositions, although the trimerizations were quite similar.
Three main groups of FCPs can be distinguished in diatoms: Lhcf, Lhcr, and Lhcx. The Lhcf family consists of major light-harvesting proteins, the Lhcr family is thought to function as PSI antennae, and the Lhcx family is involved in non-photochemical quenching of Chl excitation, with high similarity to the LI818 or LhcSR proteins of the green alga Chlamydomonas reinhardtii. Since whole genome sequences have been elucidated in Thalassiosira and Phaeodactylum (Armbrust et al. 2004; Bowler et al. 2008), we could identify the gene products in the FCP complexes of the two diatoms. In Thalassiosira, Lhcf8 was detected in the predominant subunit of the FCP oligomer. This result corresponded with that of a previous study (Grouneva et al. 2011). No Lhcr proteins were detected in the FCP oligomer and trimer, as before. However, the gene products of Lhcf and Lhcx in the FCP trimer were markedly different from those found in the previous study: Lhcf3 and Lhcf11 were found in the present study, whereas no Lhcf2, Lhcx1, Lhcx2, and Lhcx4 were found. FCP gene expressions are required for light irradiation (Leblanc et al. 1999; Nagao et al. 2013a), and the patterns of protein expression vary easily in response to light intensities (Gundermann and Büchel 2012; Gundermann et al. 2013). Therefore, the growth-light condition of diatom cells is of importance for the transcription and translation of FCPs. In the present study, we used continuous low light for cell growth, whereas Grouneva et al. (2011) used a light/dark cycle. These findings suggest that the different protein expressions of Lhcf and Lhcx in the FCP trimers are due to the different growth-light conditions between the two studies, although we cannot exclude the possibility of the differences in FCP preparation and MS analysis. This is in agreement with the previous study that showed the variability of polypeptide compositions in the FCP trimer of Phaeodactylum (Gundermann et al. 2013).
In Phaeodactylum, Lhcf and Lhcr proteins were detected in the predominant subunits of the FCP trimer. Some reports have described the proteomic analysis of FCPT of Phaeodactylum (Lepetit et al. 2007, 2010; Grouneva et al. 2011; Gundermann et al. 2013). No Lhcr proteins were detected in any of the studies. However, the studies showed different Lhcf proteins compared with our results. The differences seem to be partly caused by the different growth-light conditions, as described above. Intriguingly, Grouneva et al. (2011) have described that Lhcr1, Lhcr2, Lhcr3, Lhcr4, Lhcr12, and Lhcr13 have been identified in the PSI fraction but not the FCPT fraction by blue-native PAGE, supporting the idea that some FCP subunits are dissociated from the PSI complex by the effect of strict detergent concentration.
Here again, we highlight the comparison of FCP complexes from the four diatoms. We were particularly interested in the biochemical properties of the FCP oligomer rather than the FCP trimer. The oligomerization of FCP varied among the three diatoms and was absent in Phaeodactylum; however, the polypeptide composition of FCPO was quite similar in the three diatoms. The major subunit in FCPO of Thalassiosira, Lhcf8, belongs to the Lhcf family and is most probably involved in light harvesting. Since Fcp5 in FCPO of Cyclotella has homology to Lhcf8 (Gundermann and Büchel 2012), it appears to be involved in light harvesting. Gundermann and Büchel (2008, 2012) have proposed that the oligomeric FCP complex (termed FCPb) in Cyclotella lacks the photoprotection mechanism called non-photochemical quenching and is responsible for pure light harvesting. However, we revealed that the FCP complexes in Chaetoceros, such as FCP oligomer and trimer, have a high excitation energy quenching system in addition to light-harvesting system (Nagao et al. 2013b). The quenching of FCP complexes was greater than that of the LHC associated with PSII (LHCII) (Nagao et al. 2013b). These findings suggest that FCP oligomer plays a different role in light harvesting and excitation energy quenching among diatom species.
Diatoms are roughly classified into two groups: centric and pennate. Chaetoceros, Thalassiosira, and Cyclotella are centric diatoms, whereas Phaeodactylum is a pennate diatom. This may suggest that FCP oligomer is not present in this pennate diatom. Gundermann et al. (2013) also showed this possibility. If so, the light harvesting and excitation energy quenching strategies would be different between centric and this pennate diatoms. To prove this hypothesis, we designed the spectroscopic analyses using the CN-PAGE gel. We are currently conducting studies to elucidate the differences in light harvesting and excitation energy quenching among the FCP complexes of the four diatom species by time-resolved spectroscopic analysis.
Abbreviations
- Chl:
-
Chlorophyll
- Cg:
-
Chaetoceros gracilis
- Cm:
-
Cyclotella meneghiniana
- FCP:
-
Fucoxanthin chlorophyll a/c-binding protein
- MS:
-
Mass spectrometric
- Pt:
-
Phaeodactylum tricornutum
- PS:
-
Photosystem
- Tp:
-
Thalassiosira pseudonana
References
Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou S, Allen AE, Apt KE, Bechner M, Brzezinski MA, Chaal BK, Chiovitti A, Davis AK, Demarest MS, Detter JC, Glavina T, Goodstein D, Hadi MZ, Hellsten U, Hildebrand M, Jenkins BD, Jurka J, Kapitonov VV, Kröger N, Lau WW, Lane TW, Larimer FW, Lippmeier JC, Lucas S, Medina M, Montsant A, Obornik M, Parker MS, Palenik B, Pazour GJ, Richardson PM, Rynearson TA, Saito MA, Schwartz DC, Thamatrakoln K, Valentin K, Vardi A, Wilkerson FP, Rokhsar DS (2004) The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306:79–86
Aro E-M, Suorsa M, Rokka A, Allahverdiyeva Y, Paakkarinen V, Saleem A, Battchikova N, Rintamäki E (2005) Dynamics of photosystem II: a proteomic approach to thylakoid protein complexes. J Exp Bot 56:347–356
Beer A, Gundermann K, Beckmann J, Büchel C (2006) Subunit composition and pigmentation of fucoxanthin-chlorophyll proteins in diatoms: evidence for a subunit involved in diadinoxanthin and diatoxanthin binding. Biochemistry 45:13046–13053
Berkaloff C, Caron L, Rousseau B (1990) Subunit organization of PSI particles from brown algae and diatoms: polypeptide and pigment analysis. Photosynth Res 23:181–193
Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, Maheswari U, Martens C, Maumus F, Otillar RP, Rayko E, Salamov A, Vandepoele K, Beszteri B, Gruber A, Heijde M, Katinka M, Mock T, Valentin K, Verret F, Berges JA, Brownlee C, Cadoret JP, Chiovitti A, Choi CJ, Coesel S, De Martino A, Detter JC, Durkin C, Falciatore A, Fournet J, Haruta M, Huysman MJ, Jenkins BD, Jiroutova K, Jorgensen RE, Joubert Y, Kaplan A, Kröger N, Kroth PG, La Roche J, Lindquist E, Lommer M, Martin-Jézéquel V, Lopez PJ, Lucas S, Mangogna M, McGinnis K, Medlin LK, Montsant A, Oudot-Le Secq MP, Napoli C, Obornik M, Parker MS, Petit JL, Porcel BM, Poulsen N, Robison M, Rychlewski L, Rynearson TA, Schmutz J, Shapiro H, Siaut M, Stanley M, Sussman MR, Taylor AR, Vardi A, von Dassow P, Vyverman W, Willis A, Wyrwicz LS, Rokhsar DS, Weissenbach J, Armbrust EV, Green BR, Van de Peer Y, Grigoriev IV (2008) The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456:239–244
Büchel C (2003) Fucoxanthin-chlorophyll proteins in diatoms: 18 and 19 kDa subunits assemble into different oligomeric states. Biochemistry 42:13027–13034
Depauw FA, Rogato A, Ribera d’Alcalá M, Falciatore A (2012) Exploring the molecular basis of responses to light in marine diatoms. J Exp Bot 63:1575–1591
Falciatore A, Bowler C (2002) Revealing the molecular secrets of marine diatoms. Annu Rev Plant Biol 53:109–130
Field CB, Behrenfeld MJ, Randerson JT, Falkowski P (1998) Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281:237–240
Gildenhoff N, Amarie S, Gundermann K, Beer A, Büchel C, Wachtveitl J (2010) Oligomerization and pigmentation dependent excitation energy transfer in fucoxanthin-chlorophyll proteins. Biochim Biophys Acta 1797:543–549
Green BR, Pichersky E (1994) Hypothesis for the evolution of three-helix Chl a/b and Chl a/c light-harvesting antenna proteins from two-helix and four-helix ancestors. Photosynth Res 39:149–162
Grouneva I, Rokka A, Aro E-M (2011) The thylakoid membrane proteome of two marine diatoms outlines both diatom-specific and species-specific features of the photosynthetic machinery. J Proteome Res 10:5338–5353
Gundermann K, Büchel C (2008) The fluorescence yield of the trimeric fucoxanthin-chlorophyll-protein FCPa in the diatom Cyclotella meneghiniana is dependent on the amount of bound diatoxanthin. Photosynth Res 95:229–235
Gundermann K, Büchel C (2012) Factors determining the fluorescence yield of fucoxanthin-chlorophyll complexes (FCP) involved in non-photochemical quenching in diatoms. Biochim Biophys Acta 1817:1044–1052
Gundermann K, Schmidt M, Weisheit W, Mittag M, Büchel C (2013) Identification of several sub-populations in the pool of light harvesting proteins in the pennate diatom Phaeodactylum tricornutum. Biochim Biophys Acta 1827:303–310
Ichimura T (1971) Sexual cell division and conjugation-papilla formation in sexual reproduction of Closterium strigosum. In: Nishizawa K (ed) Proceedings of the 7th international seaweed symposium, University of Tokyo Press, Tokyo, pp 208–214
Ikeda Y, Komura M, Watanabe M, Minami C, Koike H, Itoh S, Kashino Y, Satoh K (2008) Photosystem I complexes associated with fucoxanthin-chlorophyll-binding proteins from a marine centric diatom, Chaetoceros gracilis. Biochim Biophys Acta 1777:351–361
Ikeda Y, Yamagishi A, Komura M, Suzuki T, Dohmae N, Shibata Y, Itoh S, Koike H, Satoh K (2013) Two types of fucoxanthin-chlorophyll-binding proteins I tightly bound to the photosystem I core complex in marine centric diatoms. Biochim Biophys Acta 1827:529–539
Jeffrey SW, Humphrey GF (1975) New spectrophotometric equations for determining chlorophylls a, b, c 1 and c 2 in higher plants, algae and natural phytoplankton. Biochem Physiol Pflanzen 167:191–194
Leblanc C, Falciatore A, Watanabe M, Bowler C (1999) Semi-quantitative RT-PCR analysis of photoregulated gene expression in marine diatoms. Plant Mol Biol 40:1031–1044
Lepetit B, Volke D, Szabó M, Hoffmann R, Garab G, Wilhelm C, Goss R (2007) Spectroscopic and molecular characterization of the oligomeric antenna of the diatom Phaeodactylum tricornutum. Biochemistry 46:9813–9822
Lepetit B, Volke D, Gilbert M, Wilhelm C, Goss R (2010) Evidence for the existence of one antenna-associated, lipid-dissolved and two protein-bound pools of diadinoxanthin cycle pigments in diatoms. Plant Physiol 154:1905–1920
Lohr M, Wilhelm C (1999) Algae displaying the diadinoxanthin cycle also possess the violaxanthin cycle. Proc Natl Acad Sci USA 96:8784–8789
Nagao R, Ishii A, Tada O, Suzuki T, Dohmae N, Okumura A, Iwai M, Takahashi T, Kashino Y, Enami I (2007) Isolation and characterization of oxygen-evolving thylakoid membranes and photosystem II particles from a marine diatom Chaetoceros gracilis. Biochim Biophys Acta 1767:1353–1362
Nagao R, Moriguchi A, Tomo T, Niikura A, Nakajima S, Suzuki T, Okumura A, Iwai M, Shen J-R, Ikeuchi M, Enami I (2010a) Binding and functional properties of five extrinsic proteins in oxygen-evolving photosystem II from a marine centric diatom, Chaetoceros gracilis. J Biol Chem 285:29191–29199
Nagao R, Tomo T, Noguchi E, Nakajima S, Suzuki T, Okumura A, Kashino Y, Mimuro M, Ikeuchi M, Enami I (2010b) Purification and characterization of a stable oxygen-evolving photosystem II complex from a marine centric diatom, Chaetoceros gracilis. Biochim Biophys Acta 1797:160–166
Nagao R, Tomo T, Noguchi E, Suzuki T, Okumura A, Narikawa R, Enami I, Ikeuchi M (2012) Proteases are associated with a minor fucoxanthin chlorophyll a/c-binding protein from the diatom, Chaetoceros gracilis. Biochim Biophys Acta 1817:2110–2117
Nagao R, Tomo T, Nariakwa R, Enami I, Ikeuchi M (2013a) Light-independent biosynthesis and assembly of the photosystem II complex in the diatom Chaetoceros gracilis. FEBS Lett 587:1340–1345
Nagao R, Yokono M, Akimoto S, Tomo T (2013b) High excitation energy quenching in fucoxanthin chlorophyll a/c-binding protein complexes from the diatom Chaetoceros gracilis. J Phys Chem B 117:6888–6895
Okumura A, Nagao R, Suzuki T, Yamagoe S, Iwai M, Nakazato K, Enami I (2008) A novel protein in photosystem II of a diatom Chaetoceros gracilis is one of the extrinsic proteins located on lumenal side and directly associates with PSII core components. Biochim Biophys Acta 1777:1545–1551
Veith T, Büchel C (2007) The monomeric photosystem I-complex of the diatom Phaeodactylum tricornutum binds specific fucoxanthin chlorophyll proteins (FCPs) as light-harvesting complexes. Biochim Biophys Acta 1767:1428–1435
Veith T, Brauns J, Weisheit W, Mittag M, Büchel C (2009) Identification of a specific fucoxanthin-chlorophyll protein in the light harvesting complex of photosystem I in the diatom Cyclotella meneghiniana. Biochim Biophys Acta 1787:905–912
Watanabe M, Kubota H, Wada H, Narikawa R, Ikeuchi M (2011) Novel supercomplex organization of photosystem I in Anabaena and Cyanophora paradoxa. Plant Cell Physiol 52:162–168
Acknowledgments
This work was supported by Grant-in-Aids for Scientific Research from the Ministry of Education of Japan (No. 22370017, 24370025 to T.T.), a grant from JST PRESTO (T.T.), and a grant from the Australian Research Council’s Discovery Projects funding scheme (project number DP12101360) to T.T.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nagao, R., Takahashi, S., Suzuki, T. et al. Comparison of oligomeric states and polypeptide compositions of fucoxanthin chlorophyll a/c-binding protein complexes among various diatom species. Photosynth Res 117, 281–288 (2013). https://doi.org/10.1007/s11120-013-9903-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11120-013-9903-5