Abstract
A practical approach to increasing crop yields is to extend the duration of active photosynthesis. Stay-green is a term that is used to describe mutant and transgenic plants or cultivars with the trait of maintaining their leaves for a longer period of time than the wild-type or crosses from which they are derived. Analyzing stay-green genotypes contributes to our understanding of the molecular mechanism regulating leaf senescence which may allow us to extend the duration of active photosynthesis in crop plants. This article summarizes recent studies on stay-green plants and the insights they provide on the mechanism of leaf senescence. Briefly, mutations suppressing ethylene, abscisic acid, brassinosteroid, and strigolactone signal transduction or those activating cytokinin signaling often lead to stay-green phenotypes indicating a complex signaling network regulating leaf senescence. Developmentally regulated transcription factors, including NAC or WRKY family members, play key roles in the induction of leaf senescence and thus alteration in the activity of these transcription factors also result in stay-green phenotypes. Impairment in the enzymatic steps responsible for chlorophyll breakdown also leads to stay-green phenotypes. Some of these genotypes die in the middle of the process of chlorophyll breakdown due to the accumulation of toxic intermediates, while others appear to stay-green but their photosynthetic activity declines in a manner similar to wild-type plants. Alterations in certain metabolic pathways in chloroplasts (e.g., photosynthesis) can lead to a delayed onset of leaf senescence with maintenance of photosynthetic activity longer than wild-type plants, indicating that chloroplast metabolism can also affect the regulatory mechanism of leaf senescence.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Stay-green mutants and the molecular analysis of leaf senescence
Increased crop yield has been one of the underlying goals of photosynthesis research and could be achieved by multiple means, including improving the efficiency of photosynthesis or carbon assimilation (Zhu et al. 2010). Another approach to achieve this objective would be to delay senescence and maintain the photosynthetic activity of plants for a longer duration of time (Dohleman and Long 2009). To achieve this goal in a variety of plant species, it is essential to understand the mechanism of leaf senescence at the molecular level.
Leaf senescence is a complex process in which multiple cellular events proceed in parallel or sequentially (Lim et al. 2007). It typically initiates with changes in nuclear gene expression profiles which are induced at certain developmental stages or by environmental stimuli. Although such changes are not visibly obvious at the early stages of leaf senescence, they are soon easily recognizable as the breakdown of photosynthetic pigments (Buchanan-Wollaston et al. 2003). These events are typically followed by the breakdown of plastids, mitochondria, vacuoles and nuclei, leading to ultimate cell death (see review, Buchanan-Wollaston 1997). Although leaf senescence is a degradatory process, it is a highly coordinated process involving the activity and function of numerous specific genes and proteins. While researchers have identified many of these genes and proteins, our understanding of the complete picture of leaf senescence is still limited.
One major approach to dissect the process of leaf senescence is to analyze mutants or transgenic plants which show altered leaf senescence phenotypes (Fig. 1). The advantage of such an approach is that it enables one to investigate each step of leaf senescence independently from other events of senescence. In other words, like other genetic studies, the analysis of particular stay-green mutants allows one to understand the influence of each gene (or protein). The objective of the present review is to provide a summary of leaf senescence studies emphasizing those that have utilized stay-green plants. Stay-green refers to mutants, varieties, or transgenic lines of plants that retain their green color longer than wild-type plants or standard varieties. The use of stay-green plants not only provides insights into the molecular mechanism of leaf senescence but may also represent a practical approach to improve crop yields. The classification of stay-green plants into five categories, as proposed by Thomas and Howarth (2000) will be utilized in this review (Table 1). When initiation of senescence is delayed compared to wild-type plants, these plants are classified as type A stay-greens. In type B stay-green plants, progression of senescence is delayed. In both type A and B stay-green plants, the duration that plants are photosynthetically active is prolonged, thus these plants are termed functional stay-greens (Hörtensteiner 2009). It is possible for chlorophyll breakdown to be impaired while the rest of senescence-related events proceed at a comparable rate to the wild type. In such a case, plants are classified as type C and sometimes described as cosmetic stay-greens (Hörtensteiner 2009). Type D is described as pseudo-stay-green because the plant dies before or in the middle of senescence, and as a consequence the plant appears green. Type E stay-greens accumulate higher levels of chlorophyll than wild-type plants, thus it takes longer to degrade chlorophyll during senescence. Although mutant plants that retain higher chlorophyll contents have been reported (e.g., Wang et al. 2008), to the best of our knowledge, the progress of leaf senescence in these mutants has not been described. Therefore, there will be only a limited discussion of type E stay-greens in this review. In the future, pigment metabolism of high-chlorophyll mutants should be further investigated in order to gain more insight into the relationship between pigment metabolism and leaf senescence.
Representative stay-green plants. a Mendel’s green cotyledon mutant shows a cosmetic stay-green phonotype. Left: Mature seeds in a pea pod from a plant that is heterozygous for the I locus (i homozygote has green seed color). Right: Dark-incubated pea leaves with the II (upper) and ii (lower) genotypes. b Arabidopsis plants expressing truncated CAO protein show a functional stay-green phenotype. Left: 7-week-old wild-type Arabidopsis plants. Right: 7-week-old BCG plants
Initiation and progression of leaf senescence is regulated by various phytohormones. Therefore, it is not surprising that many mutants in which phytohormone metabolism or signaling is modified show type A or type B stay-green phenotypes. The analysis of these mutants provides insight into the link between these hormones and senescence (or programmed cell death). The second half of this review describes stay-green plant studies related to phytohormone metabolism and signaling. The majority of such signaling pathways are controlled and/or mediated by nuclear gene expression. Several mutants impaired in specific chloroplast functions also exhibit type A or B type stay-green phenotypes indicating that functions of the chloroplast also contribute to the signaling governing leaf senescence. Various mutants, specifically impaired in chlorophyll breakdown, are type C or D stay-greens. Analysis of these mutants provides insight into how chlorophyll breakdown is linked to the other events of leaf senescence (e.g., the breakdown of chloroplast membranes).
For more information, readers may refer to other relevant review articles including Hörtensteiner and Kräutler (2011) and Hörtensteiner (2006) for the chlorophyll breakdown pathway, Thomas et al. (2002) and Thomas (2013) for more physiological aspects of plant senescence, and Buchanan-Wollaston et al. (2003) and Lim et al. (2007) for a comprehensive overview of the molecular analysis of leaf senescence.
Screening and isolation of stay-green mutants
Stay-green plants are usually screened by a visible assessment of greenness, as Gregor Mendel must have done when he isolated his famous stay-green pea variety (Mendel 1866; see Fig. 1a). It is also practical to use a portable photometric device, such as SPAD-502 chlorophyll meter (Konica-Minolta, Co. Ltd, Tokyo, Japan), to estimate leaf chlorophyll contents in a non-destructive manner (Monje and Bugbee 1992). For the isolation of stay-green mutants, more accurate, time-consuming techniques (such as the spectrophotometric determination of chlorophyll in organic solvents) is not necessary since only obvious stay-green phenotypes can be detected due to substantial variation in chlorophyll content among leaves.
Increased accuracy of techniques is necessary when isolated mutants are analyzed in detail. These techniques include the extraction of pigments with organic solvents that is followed by spectrophotometric determination of pigment concentrations and chlorophyll a to b ratios (Porra et al. 1989). In some instances, these techniques are coupled with the separation with high-performance liquid chromatography (HPLC) for the simultaneous determination of various photosynthetic pigments (Zapata et al. 2000). For the latter method, extra care must be taken for the extraction of chlorophyll, since HPLC separation is more susceptible to artifactual modification during pigment extraction as compared to conventional spectrophotometric methods. Extraction at sub-zero temperatures is recommended for suppression of such artifactual modification.
Stay-green signaling in chloroplasts
Loss of green color is a hallmark of leaf senescence. However, it is not necessarily associated with the rest of senescence-related events. Analysis of various mutants or transgenic plants in which an enzymatic step of the chlorophyll breakdown pathway is impaired provides insight into how this pathway is integrated in the entire process of senescence. This section provides an overview of the chlorophyll breakdown pathway and subsequently describes mutants or the transgenic plants associated with each step of the pathway. A, B, or C stay-green plants with defects in the function of chloroplast proteins other than those involved in chlorophyll breakdown are then described and discussed. These stay-green plants demonstrate that chloroplast activities play an integral role in the regulatory mechanism of leaf senescence.
The chlorophyll breakdown pathway
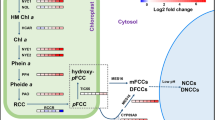
In order to describe the chlorophyll breakdown pathway, it is necessary to describe the two chlorophyll species synthesized by plants and how they are associated with the photosynthetic apparatus. All land plants synthesize chlorophyll a and chlorophyll b. The former contains a methyl group at position 7 on the tetrapyrrole macrocycle, while the latter contains a formyl group at the same position (Fig. 2). Chlorophyll a resides both in the core antenna complexes of photosystem (PS) 1 and 2, and the peripheral antenna complexes, which are composed of light-harvesting chlorophyll-binding proteins (LHC proteins: Nelson and Yocum 2006). Chlorophyll a not only harvests and transfers light energy, but also plays a role in photochemistry and electron transfer. In contrast, chlorophyll b is exclusively associated with the peripheral antenna complexes in plants. The major function of chlorophyll b is harvesting and transferring light energy but this function is also a property of chlorophyll a. Thus, chlorophyll b is not essential to plant photosynthesis. Nevertheless, plants need chlorophyll b to stabilize LHC (see review, Tanaka and Tanaka 2011).
Chlorophyll breakdown pathway. In the structure of Arabidopsis FCC, R1 denotes ethyl, while R2 can be either OH or H, and R3 can be either H or a methyl group (Hörtensteiner and Kräutler 2011). In the structure of Arabidopsis NCC, R1 is ethyl, R2 is H, OH or O-glucosyl, and R3 is either H or a methyl group (Hörtensteiner and Kräutler 2011). The side chains of FCC and NCC vary in other species (Hörtensteiner and Kräutler 2011). A methylesterase, MES16, was identified to be involved in the conversion of FCC to NCC (Christ et al. 2012). The other enzymes that are involved in this conversion have not been identified yet. Corresponding stay-green phenotypes, which are classified according to the categories proposed by Thomas and Howarth (2000), are described below the names of the enzymes. Note that most of the defects in chlorophyll catabolizing enzymes cause type C or type D stay-green phenotypes. Type A and type B stay-green phenotypes are observed with the mutants that have defects in other metabolic pathways or phytohormone signaling pathways
During senescence, chlorophyll b is first converted to chlorophyll a, and then all chlorophyll molecules seem to follow the same degradation pathway (Fig. 2). The conversion of chlorophyll b to chlorophyll a involves two steps. In the first step, the 7-formyl group of chlorophyll b is reduced to a hydroxymethyl group by chlorophyll b reductase (Kusaba et al. 2007). This enzyme is an NADPH-dependent short-chain dehydrogenase. All plants examined so far have two types of chlorophyll b reductase. NYC1 possesses putative membrane-spanning regions, while NOL does not. In rice, NYC1 and NOL seem to form a hetero dimer (Sato et al. 2009), while in Arabidopsis they appear to function independently (Horie et al. 2009). The second step of the chlorophyll b to chlorophyll a conversion is carried out by 7-hydroxymethyl chlorophyll a reductase (HCAR), which is an Fe–S containing flavoprotein. This enzyme reduces 7-hydroxymethyl-chlorophyll into chlorophyll a using ferredoxin as a reductant.
The first step of chlorophyll a breakdown involves the removal of Mg2+ to form pheophytin a. The enzyme responsible for this reaction has not yet been identified. Pheophytinase (PPH), a hydrolase belonging to the serine protease family, then catalyzes the removal of the phytol tail of pheophytin a to form pheophorbide a (Morita et al. 2009; Schelbert et al. 2009). Subsequently, the macrocycle ring of pheophorbide a is opened by pheophorbide a oxygenase (PaO), a Rieske-type 2Fe–2S oxygenase, to produce red chlorophyll catabolite (RCC) (Pruzinska et al. 2003; Tanaka et al. 2003). The C20/C1 double bond of RCC is then reduced to form primary fluorescent chlorophyll catabolite (pFCC) by RCC reductase (Pruzinska et al. 2005). This enzyme belongs to the ferredoxin-dependent bilin reductase family (Sugishima et al. 2009). All the described reactions, including the formation of pFCC, take place in the plastid while the remaining steps take place outside the plastid. In the latter steps of chlorophyll breakdown, pFCC is exported from the chloroplast and converted to fluorescent chlorophyll catabolites (FCCs) and non-fluorescent chlorophyll catabolites (NCCs) (see review, Hörtensteiner and Kräutler 2011; Christ et al. 2012).
Pigment color changes as the catabolic pathway proceeds. Chlorophyll a is just green and chlorophyll b appears slightly lighter in color when the pigments are dissolved in organic solvents. The color of these pigments are somewhat modified when they are bound to proteins within the cells. Pheophytin a and pheophorbide a appear dull green and RCC is red. pFCC and the downstream catabolites are all colorless in the visible light spectrum. Therefore, mutant or transgenic plants that accumulate pheophytin a or pheophorbide a still appear to have green leaves even though partial degradation of chlorophyll a and b has occurred.
The breakdown of chlorophyll requires an extra protein termed stay green (SGR) in addition to the catabolic chlorophyll enzymes already described. Overexpression of the SGR gene induces chlorophyll breakdown, thus this protein is likely a “master” regulator of chlorophyll degradation (Armstead et al. 2007; Jiang et al. 2007; Park et al. 2007; Ren et al. 2007; Aubry et al. 2008; Sakuraba et al. 2012b). SGR is reported to interact with LHC and chlorophyll-degrading enzymes, including NYC1, NOL, HCAR, PPH, PaO, and RCC reductase (Park et al. 2007; Sakuraba et al. 2012b). It is hypothesized that SGR induces chlorophyll breakdown by forming a large complex consisting of chlorophyll-degrading enzymes (Park et al. 2007; Sakuraba et al. 2012b).
Alterations in the chlorophyll degradation pathway often result in type C or type D stay-green phenotypes. In rice, absence of either NYC1 or NOL leads to the type C stay-green phenotype during natural senescence and under dark-induced senescence (Kusaba et al. 2007; Sato et al. 2009) indicating that both NYC1 and NOL proteins are necessary for chlorophyll b degradation. In contrast, in Arabidopsis only the absence of NYC1 results in the type C stay-green phenotype while the absence of NOL does not affect chlorophyll b degradation (Horie et al. 2009). This indicates that NOL is not essential in chlorophyll b degradation in Arabidopsis. Degradation of LHC proteins and thylakoid membranes is suppressed in the rice nyc1 and nol mutants and the Arabidopsis nyc1 mutant. As a result, thylakoid membranes with enlarged grana often remain in these mutants during senescence indicating that chlorophyll b to chlorophyll a conversion is essential for the degradation of LHC and thylakoid membranes during leaf senescence (Kusaba et al. 2007).
The hcar mutant also results in a stay-green phenotype different from the nyc1 and nol mutants (Meguro et al. 2011). In the hcar mutant, a small amount of 7-hydroxymethyl-chlorophyll and a large amount of pheophorbide a accumulated when senescence was induced by keeping the hcar mutant in darkness for more than 3 days. It is not clear why the hcar mutant accumulated pheophorbide a as well as 7-hydroxymethyl-chlorophyll. It can be explained if we assume that 7-hydroxymethyl-chlorophyll inhibits PaO, however, this hypothesis should be further examined. Substantial areas of the hcar leaves are already dead when plants are removed from the darkness as a consequence of pheophorbide a accumulation while plants are in the dark (unpublished results from A. Tanaka Lab). Therefore, the hcar mutant is classified as a type D stay-green. In addition, the whole leaves become bleached when senescing hcar mutant plants are returned to the light (Meguro et al. 2011). This is most likely because the accumulated pheophorbide a results in the generation of a high level of reactive oxygen species. Sakuraba et al. (2012b) offered another hypothesis to explain the stay-green phenotype of the hcar mutant. They suggested that HCAR forms a large protein complex comprised of chlorophyll-degrading enzymes together with SGR. Thus, a lack of HCAR would slow down the whole chlorophyll breakdown pathway as a result of lower levels of this essential protein complex being present. This hypothesis, however, does not explain why the hcar mutant accumulates a large amount of pheophorbide a. Further studies will be required to more completely understand the mechanism by which the hcar mutant results in a stay-green phenotype. The hmc1 mutant of Arabidopsis, which has a defect in the SufB subunit of a Fe–S cluster synthesizing complex, also accumulates 7-hydroxymethyl-chlorophyll and pheophorbide a and has a phenotype similar to the hcar mutant (Nagane et al. 2010). The hmc1 mutant is likely impaired in the synthesis of HCAR, as HCAR is one of the Fe–S containing enzymes, which would explain the basis of the observed stay-green phenotype.
Arabidopsis and rice mutants that lack PPH exhibit a stay-green phenotype (Morita et al. 2009; Schelbert et al. 2009). In these mutants, pheophytin a accumulates to a slightly greater amount than in wild-type plants and the rate of chlorophyll breakdown in the pph mutant appeared to slow down indicating that excess pheophytin a accumulation suppresses chlorophyll breakdown activity in a feedback manner (Morita et al. 2009; Schelbert et al. 2009). Photosynthetic activity during senescence decreases as fast in the pph mutant as in the wild type, indicating that the accumulation of pheophytin a only affects chlorophyll breakdown, but it does not affect the degradation of the photosynthetic apparatus and the Calvin-cycle enzymes. Thus, the pph mutant can be classified as a type C stay-green.
Rice lls1 and Arabidopsis acd1 mutants, as well as Arabidopsis transgenic plants with reduced PaO activity, accumulate a high level of pheophorbide a during dark-induced senescence (Pruzinska et al. 2003; Tanaka et al. 2003). Typically, approximately 30 % of chlorophyll is catabolized to pheophorbide a after 5 days of dark-induced senescence (Tanaka et al. 2003). Therefore, it is unlikely that lack of PaO feedback regulates chlorophyll breakdown. As in the hcar mutant, a substantial number of leaf cells are dead in the Arabidopsis transgenic plants that have reduced PaO activity during dark-induced senescence (Hirashima et al. 2009). It is likely that the plants only look green due to the color of pheophorbide a. Thus, the pheophorbide a-accumulating plants could be classified as type D stay-green plants. Pruzinska et al. (2003), however, reported that the rate of chlorophyll breakdown is slower in the lls1 mutant than in wild type, although the initial chlorophyll level is lower in the lls1 mutant. Sakuraba et al. (2012b) hypothesized that PaO is also part of a large protein complex, as previously described, involved in chlorophyll breakdown. Therefore, it is possible that PaO-deficient plants represent a mixed C and D type of stay-green.
Pruzinska et al. (2007) reported that the Arabidopsis acd2-2 mutant, lacking RCC reductase, did not show a stay-green phenotype. These data indicate that the lack of RCC reductase does not affect overall chlorophyll breakdown. Whether or not mutants with alterations further down stream in the chlorophyll degradation pathway from RCC exhibit stay-green phenotypes has not been determined.
Stay-green phenotypes are clearly exhibited in sgr mutants (Armstead et al. 2007; Jiang et al. 2007; Park et al. 2007; Ren et al. 2007; Sato et al. 2007; Aubry et al. 2008; Sakuraba et al. 2012b). In fact, Mendel’s famous stay-green peas were found to be sgr mutants (Armstead et al. 2007; Sato et al. 2007). Although degradation of the components of the photosynthetic apparatus is delayed in sgr mutants during senescence, other proteins in the chloroplast are degraded as fast as in wild-type plants (Park et al. 2007; Sato et al. 2007). Thus, sgr mutants are classified as type C stay-green plants.
Most of the mutants described thus far, with defects in chlorophyll breakdown, can be classified as type C or D stay-green plants. Although these mutants contribute to our understanding of chlorophyll catabolism, their ability to contribute to improved crop yields is questionable. Nevertheless, longer retention of (apparent) greenness might be useful when applied to some commercial plants used for landscaping, decoration, or display.
Chloroplast proteins and functional stay-green plants
Mutations in several other genes encoding chloroplast proteins lead to both functional and cosmetic stay-green phenotypes. Reduction in the level of chloroplast ribosomal protein S17 in the Arabidopsis ore4-1 mutant affects plant growth and retards the initiation and progress of senescence (Woo et al. 2002). These results indicate that plastids influence the senescence program of the cell.
Transgenic Arabidopsis plants that accumulate excessive amounts of chlorophyll b provide another example of the influence of plastid activities on senescence. Overexpression of the Arabidopsis CAO gene encoding chlorophyllide a oxygenase in tobacco elongates the duration of the vegetative growth leading to increased foliage and height of the plants (Biswal et al. 2012). Likewise, overexpression of a modified Arabidopsis CAO gene encoding truncated chlorophyllide a oxygenase lacking its own regulatory domain results in excessive accumulation of chlorophyll b (Yamasato et al. 2005; Sakuraba et al. 2012a) (Fig. 1b). These plants grow as fast as the wild type but the initiation of senescence is delayed for 1–2 weeks (Sakuraba et al. 2012a). One might expect that these plants should be categorized as type E stay-greens (Table 1). In other words, it is plausible to consider that it takes longer time for the plants to degrade chlorophyll b, and it might lead to slow-pigment breakdown phenotypes. However, the phenotypes of these plants are not simply represented by slow pigment degradation. Surprisingly, expression of senescence-related genes including those encoding senescence-inducible transcription factors is also suppressed (Sakuraba et al. 2012a). Accordingly, we consider that the truncated CAO-overexpressing lines mentioned above are categorized as type A stay-greens. These results demonstrate that chloroplast metabolism is an important factor influencing leaf senescence.
The initiation and progression of leaf senescence is delayed in tobacco ΔndhF plants that exhibit a reduced level of thylakoid NADH dehydrogenase (Zapata et al. 2005). In an analogy to the animal system in which mitochondrial NDH emits a cell-death signal, Zapata et al. (2005) suggested that chloroplast NDH is involved in leaf senescence. The mechanism that links chloroplast NDH with senescence, however, has not been elucidated (Table 2).
CND41 is an aspartic protease involved in the degradation of Rubisco (Kato et al. 2004). Antisense-RNA mediated gene suppression of the CND41 gene in tobacco resulted in delayed leaf senescence in lower leaves (Kato et al. 2004, 2005). It was suggested in these studies that impaired degradation of chloroplast proteins suppresses the mobilization of nitrogen from lower leaves to upper leaves and thus retards leaf senescence in lower leaves (Kato et al. 2004, 2005).
Yamatani et al. (2013) recently reported that a mutation in the thylakoid formation1 (THF1) gene results in a stay-green phenotype in the rice nyc4 mutant. This gene was first identified as a mutation causing leaf variegation in Arabidopsis (Wang et al. 2004). The THF1 gene is conserved among oxygenic, photosynthetic organisms including cyanobacteria and higher plants (Wang et al. 2004; Keren et al. 2005) and the THF1 gene product has been proposed to function in thylakoid formation and chloroplast development (Wang et al. 2004; Keren et al. 2005; Zhang et al. 2009) but its exact function still needs to be resolved. Membrane integrity and gene expression profiles of senescence-associated genes are similar between the wild-type and nyc4 mutant plants during dark-induced senescence (Yamatani et al. 2013). Thus, the nyc4 mutant is classified as a type C stay-green plant. Interestingly, the decline in Fv/Fm, a chlorophyll fluorescence parameter which represents the activity of PS2, is slower in the nyc4 mutant than the wild type during induced senescence. In corroboration of the Fv/Fm data, degradation of the D1 and D2 core PS2 subunits is nearly completely suppressed in the nyc4 mutant. Thus, THF1/NYC4 plays an important role in chlorophyll degradation and PS2 stability in the leaf senescence process.
Phytohormones
Ethylene
Ethylene affects a wide range of phenomenon including germination, seedling morphology (triple response), and fruit ripening. Treatment of plants with ethylene also promotes leaf senescence very efficiently. Since a dominant ethylene-insensitive mutant of the ethylene receptor gene ETR1 (etr1-1) exhibits a stay-green phenotype, ethylene has been thought to play an important role in leaf senescence (Grbić and Bleecker 1995). Furthermore, a mutant of EIN2, a positive regulator of ethylene signaling, was isolated as a stay-green mutant (ore3) (Oh et al. 1997). An Arabidopsis mutant in which the expression level of the ACS genes encoding a rate-limiting enzyme of ethylene biosynthesis is substantially reduced also exhibited a stay-green phenotype (Tsuchisaka et al. 2009). Thus, the role of ethylene as a key factor in the regulation of leaf senescence is well established. Some ethylene response factors (ERFs) are direct targets of the EIN3 transcription factor, a positive regulator of ethylene signaling. Expression of the senescence-regulating NAC transcription factor gene, ORE1, is also under control of EIN2 (Kim et al. 2009b). A detailed understanding of the mechanism by which ethylene promotes leaf senescence still needs to be elucidated.
Abscisic acid
Abscisic acid (ABA) has multiple functions involving the repression of germination, drought response, and promotion of leaf senescence. A leucine-rich repeat-containing membrane receptor kinase (RPK1) mutant shows reduced sensitivity to ABA in relation to germination and stomatal closure, while plants overexpressing RPK1 exhibit hypersensitivity to ABA, suggesting that RPK1 is a positive regulator in ABA signaling (Osakabe et al. 2005, 2010). Lee et al. (2011) recently reported that a prk1 mutant exhibited a stay-green phenotype during natural senescence and was characterized by delayed senescence specifically in response to ABA among several senescence-inducing treatments.
The protein phosphatase 2C (PP2C) family is an important negative regulator of ABA signaling. SAG113 encodes a protein that is a member of the PP2C family. The mutant of SAG113 (sag113) shows reduced sensitivity to ABA while plants overexpressing SAG113 exhibit hypersensitivity to ABA in relation to stomatal closure but not in repression of germination (Zhang and Gan 2012). Since sag113 exhibited a stay-green phenotype in response to ABA-induced senescence, the authors suggested that SAG113 is involved in ABA-mediated leaf senescence via regulation of water loss. SAG113 has also been reported to be a direct target of the senescence-regulating NAC transcription factor AtNAP (Zhang and Gan 2012).
Jasmonic acid
Jasmonic acid (JA) is a phytohormone that, among other things, is involved in wound response, disease resistance, and senescence (He et al. 2002). JA promotes leaf senescence during a dark treatment. Consistent with this characterization, a loss-of-function mutant of COI1 encoding the co-receptor of JA and the antisense transgenic plant of 3-ketoacyl-CoA thiolase 2 (KAT2) involved in JA synthesis have a stay-green phenotype in response to a dark incubation (Castillo and Leon 2008). However, Schommer et al. (2008) reported that coi1 did not exhibit a stay-green phenotype during natural senescence and that the JA deficient mutants of allene oxide synthase (aos) and oxophytodienoate reductase3 (opr3) also did not have a stay-green phenotype in response to dark incubation. In contrast, the activation-tagged mutant of miR319 (jaw-D) which targets teosinte branched/cycloidea/pcf (TCP) transcription factor genes does exhibit a stay-green phenotype (Schommer et al. 2008). Although jaw-D accumulates less JA than the wild-type, the causal relationship between delayed senescence and the level of JA is not necessarily clear. Additional evidence for this relationship needs to be validated.
Cytokinins
It is well-known that cytokinins delay leaf senescence. Transgenic plants that harbor an isopentenyltransferase (IPT; the rate-limiting enzyme of cytokinin synthesis) gene whose expression is driven by a senescence-inducible promoter exhibits a stay-green phenotype (Gan and Amasino 1995). The mutant line ore12-1, which constitutively expresses the cytokinin receptor gene AHK3, also displays a stay-green phenotype (Kim et al. 2006). In ore12-1, the cytokinin signal is activated via a type B response regulator, ARR2, resulting in delayed senescence. Similarly, Kim et al. (2012) reported that overexpression of the proteasome-mediated proteolysis-insensitive version of ARR2 delays leaf senescence. These reports collectively suggest that, among three cytokinin receptors and 12 type B response regulators, the AHK3-ARR2 signaling pathway plays an important role in leaf senescence.
Brassinosteroids
Brassinosteroids (BR) are also implicated in leaf senescence. DET2 encodes a steroid 5α-reductase that catalyzes an early step of BR synthesis. Its mutant det2 shows a stay-green phenotype (Chory et al. 1994). Furthermore, a mutant of BZR2/BES1, a transcription factor positively regulating BR signaling, also shows a stay-green phenotype (Yin et al. 2002). These and other lines of evidence suggest that BR promotes leaf senescence, however, little is known about the mechanism.
Auxins
Auxins are a class of phytohormones that play a critical role in the development and growth of plants. Overexpression of a gene (YUCCA6) coding for the rate-limiting enzyme of auxin synthesis results in a stay-green phenotype (Kim et al. 2011). The observation that endogenous natural auxin IAA content was reduced during dark incubation and that exogenously applied synthetic auxin (NAA) delayed leaf senescence supports the premise that auxin inhibits and delays leaf senescence. On the other hand, a mutant of the auxin-inducible gene Small Auxin Up RNA 36(SAUR36) has been reported to show a stay-green phenotype and inducible overexpression of SAUR36 causes precocious leaf senescence under natural conditions. These contradictory results rather suggest that auxin promotes rather than delays leaf senescence.
A mutant of auxin response factor 2 (ARF2) is reported to have a stay-green phenotype (Ellis et al. 2005; Okushima et al. 2005; Lim et al. 2010). The ARF family is divided into two types: activator and repressor. It is well-established that activator-type ARFs induce expression of auxin-inducible genes in response to auxin via auxin response elements (AuxREs). In contrast, how repressor-type ARFs function in auxin-regulated gene expression remains unclear. The current model predicts that repressor-type ARFs inhibit the expression of auxin-inducible genes via competition with activator-type ARFs for their interaction with AuxRE (Guilfoyle and Hagen 2012). The fact that arf2 has a stay-green phenotype appears to indicate that auxin represses leaf senescence. However, at present, it is not clear if ARF2 is involved in auxin-responsive gene expression (Ellis et al. 2005; Okushima et al. 2005; Lim et al. 2010).
Strigolactones
Strigolactones (SL) are a class of phytohormones that facilitate the establishment of the symbiosis of plant roots with arbuscular mycorrhizal fungi and also inhibit shoot branching. Recently, the αβ-hydrolase DAD2 (the orthologous protein of D14) was reported to play an important role in SL perception in concert with the F-box protein MAX2 (Hamiaux et al. 2012). MAX2 was originally identified as the causal gene of the stay-green mutant ore9 (Woo et al. 2001). Therefore, SL have been thought to promote leaf senescence. Although MAX2 is involved in several signal transduction pathways in addition to the SL signaling pathway (Nelson et al. 2011; Shen et al. 2012), the petunia dad1 mutant which is defective in strigolactone production, exhibits a stay-green phenotype, confirming the premise that SL promotes leaf senescence (Snowden et al. 2005).
Transcription factors
NAC transcription factors and stress signaling
Among families of plant transcription factors, the NAC family contains a number of senescence-inducible genes. In Arabidopsis, ORE1 and ORS1(ORE1 SISTER1) are induced by senescence and their mutants have a stay-green phenotype, suggesting that they are positive regulators of leaf senescence (Kim et al. 2009a, b; Balazadeh et al. 2011). Interestingly, a steady state level of ORE1 mRNA is regulated by micro RNA. The micro RNA, miR164, is expressed abundantly in young leaves and degrades ORE1 mRNA to prevent premature senescence. Reportedly, one of the direct targets of ORE1 is the senescence-inducible gene bifunctional nuclease1 (Matallana-Ramirez et al. 2013). Furthermore, it was recently reported that ORE1 binds GARP nuclear transcription factors Golden2-like 1 (GLK1) and GLK2, which are involved in the regulation of chloroplast development. This interaction represses the GLK transcriptional activity and the chloroplast maintenance activity, and it promotes senescence (Rauf et al. 2013). AtNAP is another NAC transcription factor gene, the mutant of which exhibits a delayed senescence phenotype. AtNAP binds the promoter of SAG113 and directly regulates its expression (Zhang and Gan 2012). In addition, loss-of-function mutants of senescence-inducible NTL4 with a transmembrane domain exhibit a stay-green phenotype under drought conditions (Lee et al. 2012). These senescence-inducible NAC transcription factors are thought to be positive regulators of leaf senescence. On the other hand, VNI2 and JUB are another type of senescence-inducible NAC transcription factors. Loss-of-function mutants of VNI2 and JUB1 display an early senescence phenotype and their overexpression in plants results in a stay-green phenotype (Yang et al. 2011; Wu et al. 2012) indicating that they are negative regulators of leaf senescence. Senescence-inducible negative regulators are thought to work in concert with positive regulators to fine-tune leaf senescence.
Senescence-inducible NAC transcription factors are also induced by hydrogen peroxide, salt treatment, and ABA. AtNAP is thought to promote leaf senescence by regulating ABA sensitivity via SAG113 (PP2C). NTL4 is believed to promote leaf senescence by ROS generation through the induction of a subset of Atrboh encoding NADPH oxidases. Meanwhile, the negative regulator of leaf senescence VNI2 directly induces the stress-inducible genes RA29 and COR15.
Overexpression of RA29 or COR15 results in a stay-green phenotype suggesting that VNI2 represses leaf senescence by inducing these and other genes. One of the direct targets of JUB1 is DREB2A, which induces RA29 directly. Furthermore, overexpression of the master regulators of stress responses CBF2/DREB1C and CBF3/DREB1A confer a stay-green trait in addition to stress resistance (Sharabi-Schwager et al. 2010). These lines of information suggest a close relationship between leaf senescence and stress signaling. For example, tobacco plants with IPT driven by a senescence-inducible promoter show very strong drought stress resistance as well as a stay-green phenotype (Rivero et al. 2007).
WRKY transcription factors and salicylic acid
WRKY is another transcription factor family that includes a number of members that are senescence-inducible.
In relation to leaf senescence, WRKY53 has been the most investigated (reviewed by Zentgraf et al. 2010). WRKY53 is induced during leaf senescence, and its overexpression in plants produces an early senescence phenotype, suggesting that WRKY 53 is a positive regulator of leaf senescence (Hinderhofer and Zentgraf 2001; Miao et al. 2004). WRKY53 is also regulated at the protein level via proteasome-mediated proteolysis (Miao and Zentgraf 2010). Miao and Zentgraf (2010) reported that the Epithiospecifying senescence regulator (ERS) and WRKY53 function antagonistically in leaf senescence. While WRKY53 is induced by salicylic acid (SA) and repressed by JA, ERS is induced by JA and repressed by SA. ERS interacts with WRKY53 and prevents DNA binding of WRKY53. Accumulation of SA or enhanced SA signaling promotes leaf senescence (Yoshimoto et al. 2009; Xiao et al. 2010; Vogelmann et al. 2012), suggesting that WRKY53 promotes leaf senescence through the SA signaling pathway.
The loss-of-function mutant of WRKY22 exhibits a stay-green phenotype when placed in the dark and its overexpression in plants results in early senescence, indicating that WRKY22 is a positive regulator of leaf senescence (Zhou et al. 2011). Although WRKY54 and WRKY70 are induced by senescence and SA, wrky70 exhibits an early senescence phenotype and the phenotype is enhanced in the double mutant wrky70/wrky54, suggesting that WRKY54 and WRKY70 are both negative regulators of leaf senescence (Ulker et al. 2007; Besseau et al. 2012). In addition, AtWRKY6 and WRKY30 are also associated with leaf senescence (Robatzek and Somssich 2002; Besseau et al. 2012). These positive and negative regulators may fine tune the regulation of leaf senescence through their complex interaction, including mutual transcription activation or heterodimer formation (Miao et al. 2004; Besseau et al. 2012). Most of the WRKY transcription factors described above are induced by SA and are thought to regulate leaf senescence via the SA signaling pathway. It is well-established that SA plays an important role in both local and systemic acquired disease resistance (Vlot et al. 2009). The involvement of WRKY genes in SA signaling may indicate a degree of commonality between disease resistance and leaf senescence.
Fine tuning of leaf senescence via coordinated actions of phytohormones and transcription factors
As indicated in this review, most phytohormones, including SA, have some relevance to leaf senescence. In addition, a number of reports, not mentioned in this review, have referred to the involvement of other signaling pathways in leaf senescence. Leaf senescence is thought to be a common programmed response occurring when leaves dye slowly (rapid death does not accompany senescence) which functions as a mechanism to salvage nutrients. In this context, it is possible that the effect of some phytohormones/stimuli on leaf senescence is an indirect consequence, and some of the inconsistencies described for the action of phytohormones might be attributed to differences in growing conditions. Leaf senescence is carefully regulated, even though it is a process that results in death, most likely due to the fact that rapid and uncontrolled cell death is not favorable for salvaging nutrients. The presence of senescence-inducible negative regulation strongly supports this idea. Plants contain a complex and finely tuned system which functions to regulate and control the progression of leaf senescence (Fig. 3).
Regulation of leaf senescence by nuclear and chloroplast signals. “Positive stimuli” that promote leaf senescence induce the expression of senescence-associated transcription factors (TFs) in the nucleus. “Positive TFs” induce the expression of senescence-associated genes (SAGs) that promote leaf senescence. “Negative TFs” induce SAGs that repress leaf senescence, finely tuning leaf senescence. The fact that senescence-associated TFs affect chloroplast senescence indicates that signals from the nucleus regulate chloroplast senescence. On the other hand, retention of chloroplast activity appears to repress progression of expression of senescence-inducible genes, indicating that signals from the chloroplast can regulate nuclear derived programs of senescence. Thus, the nucleus and chloroplast act coordinately to regulate leaf senescence. Negative stimuli, such as cytokinins, may repress the expression of senescence-associated TFs
Conclusion
Research on stay-green plants has revealed that a disruption in chlorophyll breakdown and other metabolic pathways within the chloroplast can result in delayed leaf senescence. These findings demonstrate that chloroplast metabolism somehow regulates the senescence program of cells. The future challenge will be to identify the entity that transmits the status of the chloroplast to the nucleus where multiple signaling pathways converge to modulate the transcriptional program related to senescence. Nearly all of the known phytohormones have an effect on the timing or progression of leaf senescence. These signals are mediated or interpreted by a network of transcription factors, several of which are members of the NAC and WRKY family of transcription factors, and interact as positive and negative regulators to fine tune the initiation and progression of leaf senescence. As illustrated in the aforementioned stay-green studies, our understanding regarding the mechanisms of leaf senescence has been much improved. As an outcome of such progress, transgenic tobacco plants overexpressing the CAO gene have been reported to photosynthesize for a longer duration and retain a larger biomass (Biswal et al. 2012). Nevertheless, the mechanisms concerning how an alteration in the CAO gene expression affects the initiation or progress of leaf senescence is not currently understood. Moreover, overexpression of the CAO gene has been reported to have a deleterious effect by making the transgenic plants more sensitive to strong light (Sakuraba et al. 2010). It is possible that stay-green phenotypes are affected by growth conditions, and as a result, optimization of growth conditions may be critical for accessing their maximum potential benefit. In order to extend the period of time that crop plants remain photosynthetically active, which could ultimately increase crop yields, we need to increase our understanding of mechanisms of leaf senescence, and to experimentally evaluate a combination of several strategies to regulate the process.
References
Armstead I, Donnison I, Aubry S, Harper J, Hörtensteiner S, James C, Mani J, Moffet M, Ougham H, Roberts L et al (2007) Cross-species identification of mendel’s I locus. Science 315:73
Aubry S, Mani J, Hörtensteiner S (2008) Stay-green protein, defective in Mendel’s green cotyledon mutant, acts independent and upstream of pheophorbide a oxygenase in the chlorophyll catabolic pathway. Plant Mol Biol 67:243–256
Balazadeh S, Kwasniewski M, Caldana C, Mehrnia M, Zanor M-I, Xue G-P, Mueller-Roeber B (2011) ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol Plant 4:346–360
Besseau SS, Li JJ, Palva ETE (2012) WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J Exp Bot 63:2667–2679
Biswal AK, Pattanayak GK, Pandey SS, Leelavathi S, Reddy VS, Govindjee, Tripathy BC (2012) Light intensity-dependent modulation of chlorophyll b biosynthesis and photosynthesis by overexpression of chlorophyllide a oxygenase in tobacco. Plant Physiol 159:433–449
Buchanan-Wollaston V (1997) The molecular biology of leaf senescence. J Exp Bot 48:181–199
Buchanan-Wollaston V, Earl S, Harrison E, Mathas E, Navabpour S, Page T, Pink D (2003) The molecular analysis of leaf senescence—a genomics approach. Plant Biotechnol J 1:3–22
Castillo M, Leon J (2008) Expression of the beta-oxidation gene 3-ketoacyl-CoA thiolase 2 (KAT2) is required for the timely onset of natural and dark-induced leaf senescence in Arabidopsis. J Exp Bot 59:2171–2179
Chory J, Reinecke D, Sim S, Washburn T, Brenner M (1994) A role for cytokinins in de-etiolation in Arabidopsis (det mutants have an altered response to cytokinins). Annu Rev Plant Physiol 104:339–347
Christ B, Schelbert S, Aubry S, Sussenbacher I, Muller T, Kräutler B, Hörtensteiner S (2012) MES16, a member of the methylesterase protein family, specifically demethylates fluorescent chlorophyll catabolites during chlorophyll breakdown in Arabidopsis. Plant Physiol 158:628–641
Dohleman FG, Long SP (2009) More productive than maize in the midwest: how does Miscanthus do it? Plant Physiol 150:2104–2115
Ellis CM, Nagpal P, Young JC, Hagen G, Guilfoyle TJ, Reed JW (2005) Auxin response factor1 and auxin response factor2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132:4563–4574
Gan SS, Amasino RMR (1995) Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270:1986–1988
Grbić V, Bleecker AB (1995) Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J 8:595–602
Guilfoyle TJT, Hagen GG (2012) Getting a grasp on domain III/IV responsible for auxin response factor-IAA protein interactions. Plant Sci 190:82–88
Hamiaux C, Drummond RSM, Janssen BJ, Ledger SE, Cooney JM, Newcomb RD, Snowden KC (2012) DAD2 is an alpha/beta hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr Biol 22:2032–2036
He YY, Fukushige HH, Hildebrand DFD, Gan SS (2002) Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Annu Rev Plant Physiol 128:876–884
Hinderhofer K, Zentgraf U (2001) Identification of a transcription factor specifically expressed at the onset of leaf senescence. Planta 213:469–473
Hirashima M, Tanaka R, Tanaka A (2009) Light-independent cell death induced by accumulation of pheophorbide a in Arabidopsis thaliana. Plant Cell Physiol 50:719–729
Horie Y, Ito H, Kusaba M, Tanaka R, Tanaka A (2009) Participation of chlorophyll b reductase in the initial step of the degradation of light-harvesting chlorophyll a/b-protein complexes in Arabidopsis. J Biol Chem 284:17449–17456
Hörtensteiner S (2006) Chlorophyll degradation during senescence. Annu Rev Plant Biol 57:55–77
Hörtensteiner S (2009) Stay-green regulates chlorophyll and chlorophyll-binding protein degradation during senescence. Trends Plant Sci 14:155–162
Hörtensteiner S, Kräutler B (2011) Chlorophyll breakdown in higher plants. Biochim Biophys Acta Bioenergetics 1807:977–988
Huang W, Chen Q, Zhu Y, Hu F, Zhang L, Ma Z, He Z, Huang J (2013) Arabidopsis Thylakoid Formation 1 is a critical regulator for dynamics of PSII-LHCII complexes in leaf senescence and excess light. Mol Plant (in press)
Jiang H, Li M, Liang N, Yan H, Wei Y, Xu X, Liu J, Xu Z, Chen F, Wu G (2007) Molecular cloning and function analysis of the stay green gene in rice. Plant J 52:197–209
Kato Y, Murakami S, Yamamoto Y, Chatani H, Kondo Y, Nakano T, Yokota A, Sato F (2004) The DNA-binding protease, CND41, and the degradation of ribulose-1,5-bisphosphate carboxylase/oxygenase in senescent leaves of tobacco. Planta 220:97–104
Kato Y, Yamamoto Y, Murakami S, Sato F (2005) Post-translational regulation of CND41 protease activity in senescent tobacco leaves. Planta 222:643–651
Keren N, Ohkawa H, Welsh EA, Liberton M, Pakrasi HB (2005) Psb29, a conserved 22-kD protein, functions in the biogenesis of photosystem II complexes in Synechocystis and Arabidopsis. Plant Cell 17:2768–2781
Kim HJH, Ryu HH, Hong SHS, Woo HRH, Lim POP, Lee ICI, Sheen JJ, Nam HGH, Hwang II (2006) Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc Natl Acad Sci USA 103:814–819
Kim C, Lee KP, Baruah A, Nater M, Göbel C, Feussner I, Apel K (2009a) (1)O2-mediated retrograde signaling during late embryogenesis predetermines plastid differentiation in seedlings by recruiting abscisic acid. Proc Natl Acad Sci USA 106:9920–9924
Kim J-H, Woo HR, Kim J, Lim PO, Lee IC, Choi SH, Hwang D, Nam HG (2009b) Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323:1053–1057
Kim JI, Murphy AS, Baek D, Lee S-W, Yun D-J, Bressan RA, Narasimhan ML (2011) YUCCA6 over-expression demonstrates auxin function in delaying leaf senescence in Arabidopsis thaliana. J Exp Bot 62:3981–3992
Kim K, Ryu H, Cho Y-H, Scacchi E, Sabatini S, Hwang I (2012) Cytokinin-facilitated proteolysis of Arabidopsis response regulator 2 attenuates signaling output in two-component circuitry. Plant J 69:934–945
Kusaba M, Ito H, Morita R, Iida S, Sato Y, Fujimoto M, Kawasaki S, Tanaka R, Hirochika H, Nishimura M et al (2007) Rice non-yellow coloring1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 19:1362–1375
Lee IC, Hong SW, Whang SS, Lim PO, Nam HG, Koo JC (2011) Age-dependent action of an ABA-inducible receptor kinase, RPK1, as a positive regulator of senescence in Arabidopsis leaves. Plant Cell Physiol 52:651–62
Lee S, Seo PJ, Lee H-J, Park C-M (2012) A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J 70:831–844
Lim PO, Kim HJ, Nam HG (2007) Leaf senescence. Annu Rev Plant Biol 58:115–136
Lim PO, Lee IC, Kim J, Kim HJ, Ryu JS, Woo HR, Nam HG (2010) Auxin response factor 2 (ARF2) plays a major role in regulating auxin-mediated leaf longevity. J Exp Bot 61:1419–1430
Matallana-Ramirez LP, Rauf M, Farage-Barhom S, Dortay H, Xue G-P, Dröge-Laser W, Lers A, Balazadeh S, Mueller-Roeber B (2013) NAC transcription factor ORE1 and senescence-induced bifunctional nuclease1 (BFN1) constitute a regulatory cascade in Arabidopsis. Mol Plant. doi:10.1093/mp/sst012
Meguro M, Ito H, Takabayashi A, Tanaka R, Tanaka A (2011) Identification of the 7-hydroxymethyl chlorophyll a reductase of the chlorophyll cycle in Arabidopsis. Plant Cell 23:3442–3453
Mendel G (1866) Versuche über Pflanzen-Hybriden. Verh Naturforsch Ver Brünn 4:3–47
Miao Y, Zentgraf U (2010) A HECT E3 ubiquitin ligase negatively regulates Arabidopsis leaf senescence through degradation of the transcription factor WRKY53. Plant J 63:179–188
Miao Y, Laun T, Zimmermann P, Zentgraf U (2004) Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol 55:853–867
Monje OA, Bugbee B (1992) Inherent limitations of nondestructive chlorophyll meters: a comparison of two types of meters. HortScience 27:69–71
Morita R, Sato Y, Masuda Y, Nishimura M, Kusaba M (2009) Defect in non-yellow coloring 3, an alpha/beta hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J 59:940–952
Nagane T, Tanaka A, Tanaka R (2010) Involvement of AtNAP1 in the regulation of chlorophyll degradation in Arabidopsis thaliana. Planta 231:939–949
Nelson N, Yocum C (2006) Structure and function of photosystems I and II. Annu Rev Plant Biol 57:521–565
Nelson DC, Scaffidi A, Dun EA, Waters MT, Flematti GR, Dixon KW, Beveridge CA, Ghisalberti EL, Smith SM (2011) F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc Natl Acad Sci USA 108:8897–8902
Oh SA, Park JH, Lee GI, Paek KH, Park SK, Nam HG (1997) Identification of three genetic loci controlling leaf senescence in Arabidopsis thaliana. Plant J 12:527–535
Okushima Y, Mitina I, Quach HL, Theologis A (2005) Auxin response factor 2 (ARF2): a pleiotropic developmental regulator. Plant J 43:29–46
Osakabe Y, Maruyama K, Seki M, Satou M, Shinozaki K, Yamaguchi-Shinozaki K (2005) Leucine-rich repeat receptor-like kinase1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell 17:1105–1119
Osakabe Y, Mizuno S, Tanaka H, Maruyama K, Osakabe K, Todaka D, Fujita Y, Kobayashi M, Shinozaki K, Yamaguchi-Shinozaki K (2010) Overproduction of the membrane-bound receptor-like protein kinase 1, RPK1, enhances abiotic stress tolerance in Arabidopsis. J Biol Chem 285:9190–9201
Park S, Yu J, Park J, Li J, Yoo S, Lee N, Lee S, Jeong S, Seo H, Koh H et al (2007) The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 19:1649–1664
Porra R, Thompson W, Kriedemann P (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlrophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975:384–394
Pruzinska A, Tanner G, Anders I, Roca M, Hörtensteiner S (2003) Chlorophyll breakdown: pheophorbide a oxygenase is a Rieske-type iron-sulfur protein, encoded by the accelerated cell death 1 gene. Proc Natl Acad Sci USA 100:15259–15264
Pruzinska A, Tanner G, Aubry S, Anders I, Moser S, Müller T, Ongania K-H, Kräutler B, Youn J-Y, Liljegren SJ et al (2005) Chlorophyll breakdown in senescent Arabidopsis leaves. Characterization of chlorophyll catabolites and of chlorophyll catabolic enzymes involved in the degreening reaction. Plant Physiol 139:52–63
Pruzinska A, Anders I, Aubry S, Schenk N, Tapernoux-Luthi E, Muller T, Krautler B, Hörtensteiner S (2007) In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell 19:369–387
Rauf M, Arif M, Dortay H, Matallana-Ramirez LP, Waters MT, Nam HG, Lim PO, Mueller-Roeber B, Balazadeh S (2013) ORE1 balances leaf senescence against maintenance by antagonizing G2-like-mediated transcription. EMBO Rep 14:382–388
Ren G, An K, Liao Y, Zhou X, Cao Y, Zhao H, Ge X, Kuai B (2007) Identification of a novel chloroplast protein AtNYE1 regulating chlorophyll degradation during leaf senescence in Arabidopsis. Plant Physiol 144:1429–1441
Rivero RM, Kojima M, Gepstein A, Sakakibara H, Mittler R, Gepstein S, Blumwald E (2007) Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc Natl Acad Sci USA 104:19631–19636
Robatzek S, Somssich IE (2002) Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev 16:1139–1149
Sakuraba Y, Yokono M, Akimoto S, Tanaka R, Tanaka A (2010) Deregulated chlorophyll b synthesis reduces the energy transfer rate between photosynthetic pigments and induces photodamage in Arabidopsis thaliana. Plant Cell Physiol 51:1055–1065
Sakuraba Y, Balazadeh S, Tanaka R, Mueller-Roeber B, Tanaka A (2012a) Overproduction of chl B retards senescence through transcriptional reprogramming in Arabidopsis. Plant Cell Physiol 53:505–517
Sakuraba Y, Schelbert S, Park S-Y, Han S-H, Lee B-D, Andrès CB, Kessler F, Hörtensteiner S, Paek N-C (2012b) Stay-green and chlorophyll catabolic enzymes interact at light-harvesting complex II for chlorophyll detoxification during leaf senescence in Arabidopsis. Plant Cell 24:507–518
Sato Y, Morita R, Nishimura M, Yamaguchi H, Kusaba M (2007) Mendel’s green cotyledon gene encodes a positive regulator of the chlorophyll-degrading pathway. Proc Natl Acad Sci USA 104:14169–14174
Sato Y, Morita R, Katsuma S, Nishimura M, Tanaka A, Kusaba M (2009) Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J 57:120–131
Schelbert S, Aubry S, Burla B, Agne B, Kessler F, Krupinska K, Hörtensteiner S (2009) Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell 21:767–785
Schommer C, Palatnik JF, Aggarwal P, Chételat A, Cubas P, Farmer EE, Nath U, Weigel D (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6:e230
Sharabi-Schwager M, Lers A, Samach A, Guy CL, Porat R (2010) Overexpression of the CBF2 transcriptional activator in Arabidopsis delays leaf senescence and extends plant longevity. J Exp Bot 61:261–273
Shen H, Zhu L, Bu QY, Huq E (2012) MAX2 affects multiple hormones to promote photomorphogenesis. Mol Plant 5:750–762
Snowden KC, Simkin AJ, Janssen BJ, Templeton KR, Loucas HM, Simons JL, Karunairetnam S, Gleave AP, Clark DG, Klee HJ (2005) The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell 17:746–759
Sugishima M, Kitamori Y, Noguchi M, Kohchi T, Fukuyama K (2009) Crystal structure of red chlorophyll catabolite reductase: enlargement of the ferredoxin-dependent bilin reductase family. J Mol Biol 389:376–387
Tanaka R, Tanaka A (2011) Chlorophyll cycle regulates the construction and destruction of the light-harvesting complexes. Biochim Biophys Acta 1807:968–976
Tanaka R, Hirashima M, Satoh S, Tanaka A (2003) The Arabidopsis-accelerated cell death gene ACD1 is involved in oxygenation of pheophorbide a: inhibition of the pheophorbide a oxygenase activity does not lead to the “stay-green” phenotype in Arabidopsis. Plant Cell Physiol 44:1266–1274
Thomas H (2013) Senescence, ageing and death of the whole plant. New Phytol 197:696–711
Thomas H, Howarth CJ (2000) Five ways to stay green. J Exp Bot 51:329–337
Thomas H, Ougham H, Canter P, Donnison I (2002) What stay-green mutants tell us about nitrogen remobilization in leaf senescence. J Exp Bot 53:801–808
Tsuchisaka A, Yu G, Jin H, Alonso JM, Ecker JR, Zhang X, Gao S, Theologis A (2009) A combinatorial interplay among the 1-aminocyclopropane-1-carboxylate isoforms regulates ethylene biosynthesis in Arabidopsis thaliana. Genetics 183:979–1003
Ulker B, Shahid Mukhtar M, Somssich IE (2007) The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta 226:125–137
Vlot AC, Dempsey DA, Klessig DF (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47:177–206
Vogelmann K, Drechsel G, Bergler J, Subert C, Philippar K, Soll J, Engelmann JC, Engelsdorf T, Voll LM, Hoth S (2012) Early senescence and cell death in Arabidopsis saul1 mutants involves the PAD4-dependent salicylic acid pathway. Plant Physiol 159:1477–1487
Wang Q, Sullivan RW, Kight A, Henry RL, Huang J, Jones AM, Korth KL (2004) Deletion of the chloroplast-localized Thylakoid formation1 gene product in Arabidopsis leads to deficient thylakoid formation and variegated leaves. Plant Physiol 136:3594–3604
Wang F, Wang G, Li X, Huang J, Zheng J (2008) Heredity, physiology and mapping of a chlorophyll content gene of rice (Oryza sativa L.). J Plant Physiol 165:324–330
Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG (2001) ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13:1779–1790
Woo HR, Goh C-H, Park J-H, de la Teyssendier Serve B, Kim J-H, Park Y-I, Nam HG (2002) Extended leaf longevity in the ore4-1 mutant of Arabidopsis with a reduced expression of a plastid ribosomal protein gene. Plant J 31:331–340
Wu A, Allu AD, Garapati P, Siddiqui H, Dortay H, Zanor M-I, Asensi-Fabado MA, Munné-Bosch S, Antonio C, Tohge T et al (2012) JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 24:482–506
Xiao S, Gao W, Chen Q-F, Chan S-W, Zheng S-X, Ma J, Wang M, Welti R, Chye M-L (2010) Overexpression of Arabidopsis acyl-CoA binding protein ACBP3 promotes starvation-induced and age-dependent leaf senescence. Plant Cell 22:1463–1482
Yamatani H, Sato Y, Masuda Y, Kato Y, Morita R, Fukunaga K, Nagamura Y, Nishimura M, Sakamoto W, Tanaka A, Kusaba M (2013) NYC4, the rice ortholog of Arabidopsis THF1, is involved in the degradation of chlorophyll – protein complexes during leaf senescence. Plant J 74:652–662
Yamasato A, Nagata N, Tanaka R, Tanaka A (2005) The N-terminal domain of chlorophyllide a oxygenase confers protein instability in response to chlorophyll B accumulation in Arabidopsis. Plant Cell 17:1585–1597
Yang S-D, Seo PJ, Yoon H-K, Park C-M (2011) The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 23:2155–2168
Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109:181–191
Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, Ohsumi Y, Shirasu K (2009) Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21:2914–2927
Zapata JM, Guéra A, Esteban-Carrasco A, Martín M, Sabater B (2005) Chloroplasts regulate leaf senescence: delayed senescence in transgenic ndhF-defective tobacco. Cell Death Differ 12:1277–1284
Zapata M, Rodríguez F, Garrido J (2000) Separation of chlorophylls and carotenoids from marine phytoplankton: a new HPLC method using a reversed phase C8 column and pyridine-containing mobile phases. Mar Ecol Prog Ser 195:29–45
Zentgraf U, Laun T, Miao Y (2010) The complex regulation of WRKY53 during leaf senescence of Arabidopsis thaliana. Eur J Cell Biol 89:133–137
Zhang K, Gan SS (2012) An abscisic acid-AtNAP transcription factor-SAG113 protein phosphatase 2C regulatory chain for controlling dehydration in senescing Arabidopsis leaves. Plant Physiol 158:961–969
Zhang L, Wei Q, Wu W, Cheng Y, Hu G, Hu F, Sun Y, Zhu Y, Sakamoto W, Huang J (2009) Activation of the heterotrimeric G protein alpha-subunit GPA1 suppresses the ftsh-mediated inhibition of chloroplast development in Arabidopsis. Plant J 58:1041–1053
Zhou X, Jiang Y, Yu D (2011) WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol Cells 31:303–313
Zhu X-G, Long SP, Ort DR (2010) Improving photosynthetic efficiency for greater yield. Annu Rev Plant Biol 61:235–261
Acknowledgments
We thank Yumi Nagashima and Hiroshi Yamatani (Hiroshima University) for the photographs in Fig. 1a and helpful discussion. We also thank Junko Kishimoto (Hokkaido University) for the photograph in Fig. 1b and the illustration that was used in Fig. 2. We would like to note that, while we were writing this manuscript, we have been inspired by the excellent review articles by Thomas and his coworkers (Thomas and Howarth 2000; Thomas et al. 2002; Thomas 2013).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kusaba, M., Tanaka, A. & Tanaka, R. Stay-green plants: what do they tell us about the molecular mechanism of leaf senescence. Photosynth Res 117, 221–234 (2013). https://doi.org/10.1007/s11120-013-9862-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11120-013-9862-x