Abstract
The discovery of the chlorophyll d-containing cyanobacterium Acaryochloris marina in 1996 precipitated a shift in our understanding of oxygenic photosynthesis. The presence of the red-shifted chlorophyll d in the reaction centre of the photosystems of Acaryochloris has opened up new avenues of research on photosystem energetics and challenged the unique status of chlorophyll a in oxygenic photosynthesis. In this review, we detail the chemistry and role of chlorophyll d in photosynthesis and summarise the unique adaptations that have allowed the proliferation of Acaryochloris in diverse ecological niches around the world.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Chemistry of chlorophyll
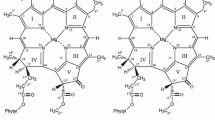
Chlorophylls (Chls) are a group of tetrapyrrolic pigments essential for oxygenic photosynthesis, participating in photon capture, transduction and conversion into chemical energy. Currently five Chls have been identified in nature—Chl a, b, c, d and the recently discovered f (Chen et al. 2010) (Fig. 1) isolated from the filamentous cyanobacterium Halomicronema hongdechloris (Chen et al. 2012). Chlorophylls all share common structural features: a tetrapyrrole ring which coordinates a Magnesium (Mg) atom, with an isocyclic fifth ring adjacent to the third ring (Fig. 1). Attached to this tetrapyrrolic ring is a long hydrophobic phytol tail (which is almost always absent in the Chl c family (Zapata et al. 2006); Fig. 1b). The degree of saturation of the tetrapyrrolic ring differs amongst chlorophylls; Chlorophyll c-type pigments have a completely unsaturated phytoporphyrin (double bond at C17–C18), whereas the other Chls are phytochlorins (reduced C17–C18 double bond; Fig. 1). Anoxygenic photosynthetic bacteria utilise bacteriochlorophylls, most of which are bacteriochlorins which are like phytochlorins but with a reduced C7–C8 double bond (Scheer 2006). These differences in saturation of the chlorophyll macrocycle have profound consequences on their electronic absorption spectra. For example the phytochlorins Chl a, Chl d, and Chl f have approximately equal absorption intensities around 400–450 nm (blue, Soret bands) and 650–700 nm (red, QY bands), with an absorbance ‘trough’ in the green spectral region (Fig. 2a). The more electronically symmetrical phytoporphyrins (Chl c-type) on the other hand, absorb only weakly in the red spectrum, and much more intensely around 450 nm (Zapata et al. 2006). The absorbance spectrum of the phytochlorin Chl b is intermediate between other phytochlorins and the phytoporphyrins, with a Soret:QY ratio of 2.8 in methanol (Fig. 2a). Different macrocycle peripheral groups can also modulate the electronic configuration, resulting in significant, biologically relevant changes in their absorbance spectra (Hoober et al. 2007) (Fig. 2a). The Mg atom coordinated by the macrocycle is important in mediating interactions with proteins as well as maximising the excited state lifetime of the molecule. Exceptions to this ubiquitous Mg atom can be found in two unrelated bacteria Acidiphilium rubrum and Candidatus Chloracidobacterium thermophilum, both of which contain bacteriochlorophyll a with a chelated zinc (Zn) atom rather than Mg (Wakao et al. 1996; Tsukatani et al. 2012). Less is known about the role of the phytyl tail of chlorophylls. Mutants in phytyl synthesis are sensitive to photo-oxidative stress and have reduced photosytsem stabilities (Bollivar et al. 1994; Shpilyov et al. 2013), suggesting a role for the flexible phytyl tail in maintaining photosystem integrity and correctly positioning the macrocycles within the antenna and reaction centre.
Molecular structures of chlorophylls. a Structures of phytochlorin chlorophylls with variable side chains (R1–R4) listed below. b Structures of the three most common phytoporphyrin (Chl c-type) chlorophylls with the variable side chains (R5–R6) are shown below. Carbon atom and ring numbering are as for the phytochlorin. Note the C17–C18 double bond (bold) and the absence of the phytyl tail in the Chl c structure. Phy phytyl tail (C20H39)
Online spectra and HPLC chromatography of chlorophylls. Total pigment was extracted from ground Nicotiana benthamiana leaves, Acaryochloris or H. hongdechloris cells with 100 % methanol and directly subjected to RP-HPLC analysis. a Online spectra were collected for each chlorophyll peak detected and plotted with data normalised to the Soret maxima. The Qy maximum for each chlorophyll is indicated. b Maximum absorbance between 620 and 720 nm of total pigment from the photosynthetic organisms was plotted against time. The identity of each chlorophyll peak is indicated
Detection of chlorophylls
Chlorophylls can readily be distinguished from one another based on their spectral properties (Fig. 2a). The detection of pigments such as chlorophylls relies on the absorption and/or fluorescence properties of the molecule, whether in situ as pigment-protein complexes, or extracted in a monomeric state in a solvent. Combined with a suitable separation technology such as HPLC, the spectral properties of a chlorophyll offers power tools for identification and classification as well as provide information regarding their functions. The extinction coefficient of a chlorophyll is an important measure, allowing fast and accurate quantitation based on its absorbance. The extinction coefficients of Chl a and Chl b were initially reported by Mackinney (1941), followed by a report for simultaneous Chl a and Chl b determination by Arnon (1949). These extinction coefficients have since been revised by Smith and Benitez (1955), the accuracy of which has been confirmed by atomic mass spectrometry of the magnesium atom (Porra et al. 1989). The extinction coefficient for Chl d has recently been revised, as well as the first report detailing the extinction coefficient for Chl f (Li et al. 2012).
Chlorophyll d—the major photopigment of Acaryochloris marina
Chl d was discovered 70 years ago (Manning and Strain 1943) and was initially believed to be a minor chlorophyll in red algae. Although its structure was proposed over 50 years ago (Holt and Morley 1959; Holt 1961), it was only in 1996, with the discovery of the predominantly Chl d-containing cyanobacterium Acaryochloris marina (named as Acaryochloris through the text) that the significance of Chl d became apparent, with Chl d constituting up to 90–99 % of total chlorophylls(Miyashita et al. 1996). The structure of Chl d is identical to Chl a, apart from a substitution of the C3 vinyl group in ring I of Chl a for a formyl group (Fig. 1a), causing a red shift in the Qy maximum absorbance peak, from 665 nm for Chl a to 697 nm for Chl d and a characteristic ‘double’ soret band at 400 and 455 nm in methanol (Fig. 2a). These spectral shifts are the result of an electronic redistribution along the y-axis of the tetrapyrrole ring, caused by the electronegative C3 formyl group (Hoober et al. 2007).
Function of chlorophyll d
Chlorophylls can broadly be divided into two groups based on their function in either the antenna system only or in both the antenna and reaction centre of the photosystems. Chl b, Chl c and most likely Chl f (Akutsu et al. 2011; Chen et al. 2012) are accessory pigments found in the antenna system of the photosynthetic organism, harvesting light and transferring energy; however, they are not involved in charge separation or electron transfer. The primary role of accessory pigments is to extend the photosynthetically active range of an organism. For example, Chl b is found in photosynthetic eukaryotes and prochlorophytes and its absorbance spectrum (Fig. 2a) allows these organisms to absorb light towards the middle (green) part of the visible spectrum. On the other hand Chl f, which has been found in the marine filamentous cyanobacterium H. hongdechloris (Chen et al. 2012) and a freshwater unicellular cyanobacterium from Lake Biwa, Japan (Akutsu et al. 2011) extends the photosynthetically available light for these organisms further into the far-red region of the spectrum (Fig. 2a).
Until relatively recently it was believed that the only redox active chlorophyll in the reaction centre was Chl a (and its derivatives). Following the ‘rediscovery’ of Chl d, however, there has been a shift in this paradigm and Chl d has been shown to act as a primary electron donor in the reaction centre of the Acaryochloris photosystem I (PS I) (Hu et al. 1998; Tomo et al. 2008) and probably photosystem II (PS II) (Nieuwenburg et al. 2003; Chen et al. 2005d; Tomo et al. 2007; Itoh et al. 2007). Analogous to the Chl a special pair in plant and other cyanobacterial PS I, the Chl d special pair is a Chl d/Chl d′ (a Chl d 132-C epimer) heterodimer (Tomo et al. 2008). Despite the fact that 90–99 % of the chlorophyll in Acaryochloris is Chl d (Miyashita et al. 1996), Chl a still plays a critical role in Acaryochloris, with small amounts of pheophytin a (Phe a; demetalated Chl a) being detected in PS II, and Chl a being detected in both PS I and PS II (Tomo et al. 2007, 2008). There is still some debate regarding the exact stoichiometry and role of Chl a in the Acaryochloris photosystems (Tomo et al. 2007, 2008, 2011; Tsuchiya et al. 2012a); however, a clearly defined role for Phe a acting as the primary electron acceptor in Acaryochloris PS II has being confirmed (Razeghifard et al. 2005). Furthermore, there have been no reports of Phe d being detected in Acaryochloris to date. In vitro redox potential measurements have demonstrated the higher oxidative potential of Phe d compared with Phe a which may explain the presence of Phe a and not Phe d in Acaryochloris (Kobayashi et al. 2007).
Synthesis of chlorophylls
The total synthesis of Chl a was described in exquisite detail in 1990 by Woodward and colleagues (Woodward et al. 1990). In brief, the first step in the process involves synthesising each of the four rings. ‘Left hand’ (rings I and IV) and ‘right hand’ (rings II and II) components are then synthesised and combined to generate a (porphyrin) tetrapyrrole ring. The remaining steps in the synthesis focus on reduction of the porphyrin to a chlorin, various side chain substitutions, and the formation of the isocyclic ring V. This chemical synthesis of Chl a requires 46 steps in total, with numerous acids, reducing agents, catalysts and solvents necessary for completion. From Chl a to Chl d the relatively simple C3 vinyl group substitution for formyl can be obtained using either a strong oxidising agent (Holt and Morley 1959) or a thiol under acidic conditions (Fukusumi et al. 2012). Fukusumi et al. (2012) achieved a Chl d yield of 31 % using reaction conditions, where Chl a was dissolved in tetrahydrofuran to which thiophenol and acetic acid were added. These methods in synthesising Chl d from Chl a demonstrate the general susceptibility of the C3 side chain to nucleophilic attack and oxidative cleavage, and offer hints as to how Chl d may be synthesised in vivo.
Biosynthesis of chlorophylls
Biosynthesis of Chl a
The synthesis of biological tetrapyrroles, including chlorophylls, is a metabolically expensive process. The first committed precursor in tetrapyrrole synthesis is 5-aminolevulinic acid (5-ALA), derived from either glutamate or glycine and succinyl-coenzyme A (Beale 2006; Jahn et al. 2006). Eight 5-ALA molecules are condensed through a series of reactions to form protoporphyrin IX, a tetrapyrrolic ring molecule from where heme and phycobilin synthesis branches from chlorophyll synthesis through the chelation of either Fe or Mg, respectively (Yaronskaya and Grimm 2006). For Chl a synthesis, the final few reactions catalyse the oxidative formation of the fifth isocyclic ring, reduce the C17–C18 and C81–C82 double bonds, and phytylate the molecule (Rudiger 2006). The genes encoding all the enzymes in this biosynthetic pathway have been elucidated in cyanobacteria (Bollivar 2006) and more recently higher plants, with the characterisation of a 3,8-divinyl protochlorophyllide a 8-vinyl reductase (DVR) from Arabidopsis thaliana (Nagata et al. 2005). Using an elegant bioinformatics approach, Ito and colleagues were also able to identify an unrelated gene in Synechocystis, a cyanobacterium which does not have an orthologue of DVR, that was essential for the reduction of the 8-vinyl group (Ito et al. 2008). It is of interest to note that the marine cyanobacterium Prochlorococcus lacks this reductase and uniquely uses 8-divinyl chlorophyll a and b for photosynthesis (Chisholm et al. 1992; Nagata et al. 2005) (Fig. 1a). There have been no reports to date describing 8-divinyl variants of Chl d or Chl f.
Biosynthesis of Chl b and Chl d
Chl a is found in almost all oxygenic photosynthetic organisms identified to date and is thought to be the progenitor chlorophyll, uniquely suited for its roles in photosynthesis(Björn et al. 2009). The other chlorophyll molecules are also most likely synthesised along the same biosynthetic pathway as Chl a, diverging at the last few steps of biosynthesis or alternatively being synthesised from Chl a itself. Chl b, Chl d and Chl f are identical to Chl a apart from a formyl substitution at the C7, C3 or C2 position in the macrocycle respectively (Fig. 1a). These substitutions affect the polarity of the molecule similarly (Fig. 2b) but distinctly perturb the electronic configuration in the macrocycle, as evidenced by their different absorption spectra (Fig. 2a). The enzyme responsible for the synthesis of Chl b, chlorophyll(lide) a-oxygenase (CAO) has been identified and characterised (Tanaka et al. 1998; Espineda et al. 1999; Oster et al. 2000). CAO is specific for Chlide a in vitro (Oster et al. 2000) however in vivo it may be that both Chl a and Chlide a act as a substrate and there is some evidence that protochlorophyllide a can also act as a substrate under unusual physiological conditions (Xu et al. 2002). Surprisingly, heterologous expression of a CAO gene in Acaryochloris led to the synthesis of a novel chlorophyll, termed P672 (or [7-formyl]-Chl d) based on its Qy maximum (Tsuchiya et al. 2012b). This new chlorophyll has formyl groups at the C3 and C7 positions, similar to Chl d and Chl b respectively, suggesting that both the as yet uncharacterised ‘Chl d synthase’ and the CAO work in concert to form P672. [7-formyl]-Chl d is incorporated into the antenna-PS II of CAO-expressing Acaryochloris however it does not function in primary electron transfer (Tsuchiya et al. 2012a). The amount of Chl d per PS II is reduced in these cells, whereas Chl a and Phe a levels remain unchanged (Tsuchiya et al. 2012a), providing strong evidence for the essential role of Chl a in Acaryochloris photosystems (see “PS I” section).
Chl a has five oxygen atoms in its structure, four of which are derived from H2O and the fifth from molecular oxygen (Walker et al. 1989; Porra and Scheer 2000). Based on the incorporation of two 18O-labelled oxygens from molecular oxygen into the Chl d molecule, Schliep et al. (2010) proposed that the oxygen in the C3 formyl group of Chl d is added via an oxygenase-type reaction. Furthermore, the timing of incorporation of the labelled oxygen into Chl a and Chl d indicates that Chl d is synthesised directly from Chl a and probably not from a Chl a precursor. Perhaps more evidence to support the synthesis of Chl d directly from Chl a may be found from the CAO-expressing Acaryochloris described by Tsuchiya et al. (2012a, b). They detected no decrease in the Chl a in these cells despite the newly synthesised [7-formyl]-Chl d constituting up to 10 % of the total chlorophyll. There was, however, a reduction in Chl d levels, suggesting that the substrate for the CAO enzyme is downstream of Chl a biosynthesis (but not Chl d biosynthesis). This interpretation would mean that Chl d is synthesised from Chl a, and that the CAO enzyme acts on Chl d which is somewhat controversial as the only confirmed substrate for CAO is Chlide a (Oster et al. 2000). Alternatively, CAO may be converting Chlide a to Chlide b, reducing the available Chlide a for Chl a biosynthesis. The ‘Chl d synthase’ enzyme(s) has yet to be characterised, however it has been suggested a cytochrome P450, which commonly catalyse mono-oxygenase type reactions, may be involved (Chen and Blankenship 2011). It is unknown whether Chl f is synthesised from Chl(ide) a or some other intermediate in the Chl a biosynthetic pathway however it seems logical that the reaction may be similar to Chl b synthesis, with the C2 methyl group being substituted for a formyl, possibly with a hydroxymethyl intermediate (Kräutler 2011).
Biology of Acaryochloris marina
Ecological distribution
Acaryochloris was first isolated from a squeezed extract of didemnid ascidians, Lissoclinum patella, collected from Palau Island (Miyashita et al. 1996). The phylogenetic study on small subunit rDNA sequences showed that Acaryochloris falls into the cyanobacteria radiation and forms a divergent clade, a new genus in cyanobacteria (Fig. 3) (Miyashita et al. 2003). Miyashita et al. were originally attempting to isolate and culture Prochloron, a symbiotic cyanobacterium containing Chl a and Chl b (Lewin and Withers 1975; Takaichi and Mochimaru 2007). Instead of Prochloron, they found small yellowish-green colonies formed by the Chl d-containing cyanobacterium Acaryochloris marina. For a long time after the discovery of Acaryochloris, it was presumed that it was a symbiont, similar to Prochloron, although it could be cultured and grown autophototrophically (Miller et al. 2005; Larkum and Kühl 2005). A microphotometric survey revealed that Acaryochloris grows on the underside of the didemnid ascidians where the visible light was strongly depleted by ascidian tissue and its symbiotic Prochloron. It is likely this far-red light enriched environment provides the selective pressure for the Chl d-containing organism (Kühl et al. 2005).
16S rRNA phylogenetic tree of Acaryochloris spp with other cyanobacteria and eukaryotic plastids. It was constracted with MEGA (molecular evolutionary genetics analysis) using Neighbour-joining method. The 16S rRNA sequences are aligned with multiple sequence aligment tools (ClustalW). The alignment was then manually edited based on the alignment of Mohr et al. (2010). The evolution distance was calculated with Jukes–Cantor model. The phylogenetic tree stability was evaluated by bootstrap replication at 10000 times. The sequence data of Acaryochloris sp. Awaji-1 and Acaryochloris sp. HICR111A are obtained from NCBI with accession number of AB112435 and EU873540, respectively. The sequence data of Acaryochloris sp. MPGRS1 is kindly provided by Yaqiong Li. The sequences of four plastids and the other cyanobacteria are downloaded form NCBI as described by Miyashita et al. (2003) and Mohr et al. (2010)
In 2004, a free living Acaryochloris strain was isolated as an epiphytic cyanobacterium from a range of red algae: Ahnfeltiopsis flabelliformis, Callophyllis japonica and Carpopeltis prolifera (Murakami et al. 2004). This discovery corrected the previous understanding that Chl d is a minor photopigment associated with red-algae (Manning and Strain 1943) and also raised the question of how widely Chl d and its containing organisms are distributed. We now know Acaryochloris spp, and therefore Chl d, are found widely through both aquatic and terrestrial ecological systems including: the intertidal zone of tropical reefs (Mohr et al. 2010), epibolic biofilms on colonial ascidians (Martínez-García et al. 2011), inside the tissues of didemnid ascidians (López-Legentil et al. 2011), eutrophic hypersaline lake (Miller et al. 2005), temperate fresh water lake and saline lakes (Kashiyama et al. 2008), high altitude lakes (Fleming and Prufert-Bebout 2010), terrestrial epilithic and endolithic biofilms (McNamara et al. 2006; De Los Ríos et al. 2007), endolithic biofilm under crustose coralline algae (Behrendt et al. 2011), sea sediments (Kashiyama et al. 2008) and stromatolites (Goh et al. 2008; Li et al. 2013). In addition, a recent environmental survey examining Acaryochloris associated with different marine invertebrates from the coast of the Republic of Palau identified numerous Acaryochloris phylotypes, even from the same invertebrate, and found no clear relationship between specific phylotypes associating with specific invertebrates, nor any clear geographical bias (Ohkubo and Miyashita 2012). Up to now, five strains of Acaryochloris have been successfully isolated and cultured: Acaryochloris MBIC11017 isolated from the squeezed extract of didemnid ascidian in Palau Island (Miyashita et al. 1996, 2003), Acaryochloris AWAJI-1 isolated as an epiphyte form red algae in Japan (Murakami et al. 2004), Acaryochloris sp.CCMEE 5410 isolated form eutrophic hypersaline lake, Salton Sea (Miller et al. 2005), Acaryochloris sp HICR111A isolated form intertidal zone surrounding dead coral in Heron Island, Australia (Mohr et al. 2010) and the recently isolated Acaryochloris sp MPGRS1, an epiphyte from red algae Gelidium caulacantheum collected from Georges River, Australia (Larkum et al. 2012). The 16s rDNA phylogenetic tree demonstrates the monophyletic nature of all five cultured Acaryochloris which are all clustered together, distinct from other cyanobacterial classes (Fig. 3).
The ecological significance of Acaryochloris and the contribution of Chl d-photosynthesis in the biosphere are still largely unknown. The diversity of ecological niches that are associated with Acaryochloris spp or Acaryochloris -like organisms causes difficulties in understanding the roles of Chl d-photosynthesis as global primary producers. All Acaryochloris spp are found to be associated with other organisms in their natural habitats although some of them are successfully cultured in the laboratory. More robust growth, at least in the case of Acaryochloris MBIC 11017, was observed when grown with some coexisting bacterial contaminations in the medium (Swingley et al. 2005).
Cell structure
Acaryochloris is a unicellular cyanobacterium, normally 1.8–2.1 μm × 1.5–1.7 μm in size (Fig. 4a, b). The cells are either spheroidal or ellipsoidal in shape and are yellow-greenish in colour due to the chlorophyll-binding protein complexes in thylakoid membranes (Fig. 4c). Amongst five cultured strains, Acaryochloris sp. HICR111A and Acaryochloris sp. MPGRS1 are relatively smaller in size, being 1–2 μm × 0.75–1 μm and 1.67 ± 0.23 μm × 1 ± 0.14 μm, respectively (Mohr et al. 2010; Larkum et al. 2012).
Confocal light and electron microscopic photography of A. marina cells. a Transmitted light and b corresponding confocal light microscopic images of Acaryochloris cells (λex = 488 nm, λem > 692 nm). c Transmission electron micrograph of a dividing A. marina cell. The majority of chlorophyll-derived fluorescence is observed around the periphery of the cells in b, corresponding to the thylakoid membrane (Tm) c. Cy cytoplasm, Ca carboxysome, Pg peptidoglycan (arrows), Om outer membrane (arrowhead)
Cyanobacteria are Gram-negative prokaryotes, having a layer of peptidoglycans as a part of the cell envelope. Most Gram-negative bacteria possess a peptidoglycan layer ~2–6 nm in thickness. Unicellular cyanobacteria on the other hand, have a thicker peptidoglycan layer of ~10 nm and filamentous cyanobacteria possess an even thicker peptidoglycan layer of ~15–35 nm (Hoiczyk and Hansel 2000). The peptidoglycan layer in Acaryochloris is about 10 nm, which is similar to other unicellular cyanobacteria, is sandwiched between the cytoplasmic membrane and the outer membrane (Marquardt et al. 2000).
There are 7–11 layers of thylakoid membranes peripherally surrounding the cytoplasm. These membranes are predominantly stacked and phycobiliprotein (PBP) arrays are located between the membranes in non-stacked area, although it is uncertain as whether they are attached to cytoplasm side of the membranes (Chen et al. 2002, 2009; Swingley et al. 2005). Instead of a usual phycobilisome in other cyanobacteria, an array structure of PBP was observered. This PBP array structure is present in Acaryochloris sp MBIC 11017, but absent in Acaryochloris CCMEE 5410 which lacks PBP proteins (Chen et al. 2009). The effect of iron availability on thylakoid membrane has been examined in Acaryochloris MBIC 11017. Under iron-enriched conditions, thylakoid membranes were evenly spaced from 20 to 35 nm apart by PBP arrays and the stacked structure was only observed around the end and ‘corners’ of the cell occasionally (Swingley et al. 2005). This dramatic change in thylakoid membrane and photosynthetic apparatus is in agreement with the previous study that Chl d-binding light-harvesting proteins are the major light-harvesting system for PS I and PS II under iron-stressed conditions (see ‘Light-harvesting system in Acaryocloris ’ section). Carboxysomes with a typical polyhedral shape ranging from 170 to 230 nm are observed in the central cytoplasmic part (Marquardt et al. 2000). Interestingly, the thylakoid membrane stacks in Acaryochloris are perforated by channel-like structures connecting central and peripheral cell portions. This ultrastructural feature has not been found in other organisms (Marquardt et al. 2000).
Pigment composition
Acaryochloris has an exceptional pigment complement. As well as containing Chl d as its major pigment, Acaryochloris also contains Chl a, zeaxanthin, α-carotene and phycobilins as minor pigments (Miyashita et al. 1997). MgDVP-like pigment and allophycocyanin are also detected as trace pigments under certain cultural conditions. Chl d′ and Phe a are present as minor components, but neither Chl a′ nor Phe d is found in Acaryochloris (Akiyama et al. 2001). Chl d makes up about 80 % of the total lipid-soluble pigment of the cell and more than 2 % of the cell dry weight (Miyashita et al. 1997). Chl a only presents as a minor pigment in Acaryochloris. Its content varies in different light conditions from 1 to 10 % of the total lipid-soluble pigment. The Chl a/Chl d ratio also varies, between 0.03 and 0.1, depending on the growth conditions (Mimuro et al. 2004), with cells grown under high light intensity and micro-aerobic condition showing a higher Chl a/Chl d ratio (Mimuro et al. 2004; Lin et al. 2013).
In all prokaryotes, α-carotene and its derivatives are only found in two genera of cyanobacteria: the 8-vinyl Chl a and 8-vinyl Chl b-containing Prochlorococcus and Acaryochloris. α-carotene in Acaryochloris replaces all the functions of ß-carotene in cyanobacteria. It has a unique C-6′(6′S)-chirality, which is the first evidence of the natural occurrence of (6′S)-α-carotene (Takaichi et al. 2012).
Genome diversity and regulation of gene expression
The genomes of Acaryochloris strain MBIC 11017 and CCMEE 5410 were sequenced in 2008 and 2011, respectively (Swingley et al. 2008; Miller et al. 2011). Both strains have enormous genomes compared to other unicellular cyanobacteria. The strain MBIC11017 has a genome of 8.36 Mb, with 8528 predicted ORFs, while the strain CCMEE 5410 genome is slightly smaller than MBIC11017, with a size of 7.88 Mb and 8383 predicted ORFs. They share 6112 putative orthologs, although there are more than 25 % predicted ORFs in each genome which was absent from the other (Miller et al. 2011). A preliminary genome study on Acaryochloris sp. HICR111A also showed a surprisingly expanded genome, estimated as ~8.37 Mb, however due to contamination the sequencing could not be completed (Mohr et al. 2010). In strain MBIC11017, along with the main circular chromosome DNA, there are 9 distinct plasmids which harbour >20 % of the total genome content. A comparison of Acaryochloris and other bacterial genome sizes and gene numbers are show in Table 1. The Acaryochloris genome is approximately four-fold larger than the Prochlorococcus genome, more than two times the size of Synechocystis PCC 6803, and of similar size to the genome of the filamentous cyanobacterium, Nostoc ATCC 29133.
The genomes of MBIC11017 and CCMEE 5410 are conserved and share a similar GC content of 47 %. About 89 % of ORFs on the strain MBIC11017 chromosome are homologous to strain CCMEE 5410. Differences, however, are concentrated on ORFs from plasmids with approximately 77 % of ORFs in MBIC11017 plasmids having no homologues in CCMEE 5410 genome (Swingley et al. 2008; Miller et al. 2011).
Comparative analysis of the strain MBIC 11017 and CCME 5410 genomes suggest both have an unusually high number of gene duplicates (Miller et al. 2011). They hypothesise that one reason for these gene duplicates is the presence of multiple copies of rec A, which encodes an enzyme necessary for homologous recombination. However, they also observe that these duplicates are rapidly removed from the genome (Swingley et al. 2008; Miller et al. 2011). Interestingly the rate of duplication and duplicate loss in Acaryochloris falls in the range of eukaryotes rather than bacteria and many of these duplicated genes may come from foreign origin through horizontal gene transfer (Miller et al. 2011). Irrespective of why, the expanded genome clearly allows a high level of gene expression plasticity and the large number of gene duplications provides the raw materials for neofunctionalization (Miller et al. 2011). The gene(s) encoding the ‘Chl d synthase’ may have evolved through such means. A large repertoire of ‘genetic options’ that comes with a large genome is probably important for the ability of Acaryochloris to exploit diverse ecological niches. It has also been proposed that organisms with a large genome are more likely to occupy a non competitive environment where resources (in case of Acaryochloris, the light source) are scarce but diverse in nature (Swingley et al. 2008; Konstantinidis and Tiedje 2004).
Important cellular metabolism
Nitrogen-fixation
The capacity of nitrogen-fixation has been reported in many cyanobacteria (Zehr 2011). Since the nitrogen-fixing enzyme complex, nitrogenase, is extremely oxygen sensitive, oxygen-evolving cyanobacteria have had to adopt strategies to achieve an anaerobic environment for this enzyme (Bergman et al. 1997).
For heterocystous cyanobacteria, oxygenic photosynthesis and nitrogen-fixation are separated spatially, with photosynthesis carried out in vegetative cells, whereas nitrogen-fixation takes place in specialised cells called heterocysts which have low oxygen permeability (Fay 1992). Other non-heterocystous solve this problem by separating photosynthesis and nitrogen- fixation by time. In these cyanobacteria, photosynthesis is performed during the light period (day time) and nitrogen-fixation happens in the dark periods (night) (Gallon 2005).
The best studied Acaryochloris strains, sp. MBIC11017 and sp. CCMEE 5410, do not have complete set of genes for the nitrogenase enzyme complex (Swingley et al. 2008; Miller et al. 2011) However, a complete set of genes encoding the nitrogenase complex was reported in the unfinished Acaryochloris sp HICR111A genome sequence (Mohr et al. 2010; Pfreundt et al. 2012). Phylogenetic comparison revealed that they are more closely related to the nitrogenase in Trichodesmium erythraeum, which suggest the genes may be recruited recently by horizontal gene transfer (Pfreundt et al. 2012). By measuring the gene expression and the nitrogenase activities, Pfreundt et al. (2012) confirmed nitrogenase activity in Acaryochloris sp HICR111A.
Although strains MBIC11017 and CCMEE 5410 do not have the essential set of nitrogenise genes, little is known regarding nitrogen metabolism in these strains. Recently, a new class of cyanobacterial PS II D1 protein was identified and designated as ‘rogue’ D1 proteins (rD1). Genes encoding the rD1 protein are almost exclusively found in dizaotroph or organisms that contain nitrogenase complexes, including Acaryochloris sp. HICR111A. Interestingly, Acaryochloris strain MBIC11017 which does not have nitrogenase genes is the only exception known by so far. The details on nitrogen metabolism are required in the future for understanding the relationship between photosynthesis and nitrogen metabolism, and the role of rD1 (Murray 2012).
Fatty acid content
Cyanobacteria are diverse in their fatty acid composition. Based on the pattern of desaturation of their fatty acids, cyanobacteria are divided into five groups: group 1 cyanobacteria- contain saturated and mono-unsaturated fatty acids; group 2 and group 3—cyanobacteria contain either α-linolenic acid (C18.3ω3) or γ-linolenic acid (C18.3ω6), respectively; group 4 cyanobacteria produce both α-linolenic acid (C18.3ω3) and γ-linolenic acid (C18.3ω6) as well as stearidonic acid (C18.4ω3) and group 5 cyanobacteria which contain polyunsaturated fatty acid with only two double bonds (Murata et al. 1992; Cohen et al. 1995). Gao et al. analysed the fatty acid composition of Acaryochloris MBIC 11017 and reported the most prominent single fatty acid was C16.0 with percentage of 52 % of total fatty acids extraction, although no C16 unsaturated fatty acid was detected. This strain also contains a mixture of C18 unsaturated fatty acids, predominantly stearidonic acid (C18.4ω3), which constitutes 19 % of the total fatty acids extracted. The minor C18 unsaturated fatty acids included C18.3ω3, C18.3ω6, C18.2ω6, C18.2, C18.1ω9, C18.1 and C18.0, with no C20 or higher fatty acids observed (Gao et al. 2010). Based on the presence of C18.4ω3, C18.3ω3, C18.3ω6, Acaryochloris MBIC 11017 belongs to group 4 cyanobacteria which including Synechocystis PCC 6803. However, these two specieses do not fall in the same group based on their 16s rRNA phylogenetic classification (Fig. 3).
Gao et al. also reported a fusion gene with an N-terminal catalase-related allene oxide synthase domain and a C-terminal lipoxygenase domain. Based on the origin of the oxygen in the 13S-hydroxyl group they concluded the intermediate in this catalase-related allene oxide synthase activity is ionic (Gao et al. 2009). Furthermore, they found the C-terminal lipoxygenase of this fusion gene has a unique catalytic activity that specifically utilizes the terminal pentadiene of omega-3 or omega-6 fatty acids. All omega -3 polyunsaturated fatty acids in Acaryochloris are oxygenated at the n-7 position which has never been reported before (Gao et al. 2010). Interestingly the α-carotene in Acaryochloris was recently found to have an C-6′(6′S)-chirality, which is also unique in nature (Takaichi et al. 2012). Considering the unique photopigments (Chl d and α-carotene) found in Acaryochloris, it may be that an unusual membrane environment to harbour them and their associated photosystems is necessary. It has been reported in cyanobacteria that the presence of unsaturated fatty acids accelerate the synthesis of D1 protein de novo, and thus protect the photosystem form high light damage (Gombos et al. 1997).
Hydrogenase and hydrogen production
Hydrogen is a potential energy source for the future. It is clean and one of its sources, water, is ubiquitous on earth. Photosynthetic hydrogen production utilizes the energy from sunshine and produce molecular hydrogen, which is both ‘ecofriendly’ and renewable. Oxygenic photosynthesis collects the solar energy via an antenna system and extracts electrons and protons from water at the oxygen evolution centre in PS II. The transmembrane proton gradient created during this process is utilized for ATP synthesis and the electrons are passed via an electron transfer pathway to ferredoxin, an iron-sulphur protein that is located at the reaction centre of PSI which further carries electrons to other pathways. Under normal aerobic conditions, these electrons reduce NADP+ to NADPH, an important source of reducing power for CO2 fixing in the Calvin-Benson cycle. However under anaerobic conditions, hydrogenase (or nitrogenase) can accept electrons from reduced ferredoxin and then convert protons to molecular hydrogen (or fixing N2).
There are two types of hydrogenase found in prokaryotes: the uptake hydrogenase which consumes the hydrogen generated during the nitrogen-fixation; and the bidirectional or reversible hydrogenase which can either consume or produce hydrogen (Allakhverdiev et al. 2010a).
Some cyanobacteria have both types of hydrogenase, some only have a single type of hydrogenase, either uptake or bidirectional, and others do not have any kind of hydrogenase. All hydrogenases in cyanobacteria have a NiFe reaction centre which is highly sensitive to oxygen only functional under anaerobic conditions. Despite this obstacle, hydrogenase activity has been detected in a wide range of genera in cyanobacteria under various culture conditions (Dutta et al. 2005).
Acaryochloris MBIC 11017 has a single bidirectional hydrogenase encoded by hoxEFUYH on the plasmid pREB4 (Swingley et al. 2008). The arrangement of these hox genes is similar to that of Synechocystis PCC 6803, where the bidirectional hydrogenase is encoded by the hoxEFUYH operon with the promoter upstream the hoxE gene (Kiss et al. 2009). However RT-PCR of the hox genes showed that their regulation pattern in Acaryochloris is actually closer to that of Synechocococcus elongatus PCC 7942 where hoxEF and hoxUYH fall into distinct operons with a stronger promoter for the latter one (Kiss et al. 2009, 2013). Anaerobic darkness, low light intensities and far red light illumination conditions all induce expression of the hox genes in Acaryochloris (Kiss 2012). Under anaerobic conditions, Boichenko et al. (2000) demonstrated the capability of H2 production in Acaryochloris for the first time, although the rate of H2 production was very low, approximately 80–250 μmol mol Chl−1. Interestingly, in cells grown under light that is depleted in the active spectral region below 650 nm (i.e. the active spectral region of Acaryochloris is 650–750 nm), the H2 production rate is increased three-fold.
A relatively lower availability of reducing equivalents for hydrogenase under dark hypoxic conditions has been reported (Kiss et al. 2013), although optimized culture conditions may be required for the activity of hydrogenase. The best condition for microorganisms to produce hydrogen was reported to be low irradiation, red light (Allakhverdiev et al. 2010a), which seem the similar cultural condition for Acaryochloris. The full set of gene encoding bidirectional hydrogenase located on one plasmid provide intriguing aspect for choosing Acaryochloris as a candidate for hydrogen fuel production. However, currently the level of hydrogen production is too low and culture conditions still need to be optimized.
Chl d-photosynthesis
Photosynthesis is the utilization of solar energy by plants, algae and certain bacteria for the synthesis of complex organic molecules. The overall reaction of the complicated process of photosynthesis can be expressed by the following equation:
Here H2A is the reductant, A is the electron acceptor and CH2O is the general formula of the carbohydrates, where the chemical energy is initially stored as organic carbon. H2A can be H2O, H2S, etc. where H2A = H2O, O2 is released, and this process is called oxygen-evolving photosynthesis, also named as oxygenic photosynthesis. Alternatively anoxygenic photosynthesis describes when a reductant other than H2O is used, i.e. no O2 is released. Oxygenic photosynthesis is one of the central events in the development of life on Earth because the accumulated free oxygen in the atmosphere is the essential element for the development of advanced eukaryotic life forms. All known oxygen-evolving photosynthetic organisms require two tandemly linked photosystems (PS), PS I and PS II. Each PS is comprised of an accessory antenna system, core-antenna system and a reaction centre. In PS II, light energy is used to drive the splitting of water, production of molecular oxygen and translocation of electrons across the membranes leading to an electrical potential (negative outside), which consequently establishes a proton gradient that in turn drives ATP synthesis. On the other hand, PS I uses energy from the absorption of a photon to reduce NADP+ to NADPH and also drive cyclic photophosphorylation. The products of this light reaction, ATP and NADPH, are required for CO2 fixation. The synthesis of stable carbon products (carbohydrates) is a light-independent process and hence known as the dark reactions. The photosynthetic products play an essential role in biomass production and carbon sequestration on Earth.
Light-harvesting system in Acaryochloris
Light-harvesting systems function at a very early step of photosynthesis and have developed individual systems to adapt to various light environments (Chen and Scheer 2012). There are two broad classes: peripheral membrane antenna complexes and integral-membrane antenna systems. Most cyanobacteria use the peripheral membrane phycobilin-binding proteins –phycobiliproteins (PBPs)- as their primary light-harvesting system, except prochlorophytes and Acaryochloris spp. Prochlorophytes are a unique group of cyanobacteria that carry out oxygenic photosynthesis using Chl a and Chl b. They lack PBPs and use the membrane-bound Chl a/b-binding protein as their major light-harvesting protein complexes (La Roche et al. 1996). The membrane-bound Chl a/b binding light-harvesting protein complex was designated as prochlorophyte chlorophyll- binding (Pcb) protein (La Roche et al. 1996) and was re-named as CBP to cover all accessory chlorophyll-binding proteins in cyanobacteria (Chen et al. 2008). The CBP proteins are only found in cyanobacteria, and are different from the light-harvesting complex (LHC) superfamily in plants and algae. This unique light-harvesting system has six tranmembrane helices and binds different types of chlorophylls, including Chl a, b, d and 8-vinyl Chl a and 8-vinyl Chl b.
Phycobiliproteins are made of an aggregated trimer of heterodimers that is composed of an α and a β-subunits and formed into hexamers by a tight face-to-face arrangement. The PBP large complexes are constructed by cylindrical cores with several radiating peripheral rods. There are four main classes of PBP: phycoerythrin (PE), phycoerythrocyanin (PEC), phycocyanin (PC) and allophycocyanin (APC). The cylindrical cores are composed of APC while the peripheral rods are composed of PC alone or with PE or PEC. It is known that the light energy absorbed by peripheral rods (PC, PE or PEC) is transferred to the special pair of Chl a in reaction centres through APC in the core, which is adjacent to the thylakoid membranes. There are two antenna systems coexisting in Acaryochloris, PBPs (Marquardt et al. 1997) and chlorophyll d-binding protein complexes (Chen et al. 2002). The two systems function complementarily to meet the changes of light environments (Miyashita et al. 1997; Duxbury et al. 2009) although PBPs are absent in two of the five isolated strains, Acaryochloris sp CCMEE5410 (Miller et al. 2005) and HICR111a (Mohr et al. 2010).
Acaryochloris has a unique PBP arrangement. It does not contain a typical phycobilisome (PBS). The first transmission electron microscopic observation revealed no detectable PBS in Acaryochloris, although there are detectable PC pigments (Miyashita et al. 1996). Hu et al. were the first group to isolate the rod structure of PBP and PBP-PS II complexes in Acaryochloris. Each aggregation consists of four discs and each disc is formed by three hexamers (α6β6). The spectral analysis suggested that PBPs are physically and functionally associated with PS II, not PS I (Hu et al. 1999). The PBP rod-arrays are located at stromal side of the thylakoids membranes where they are attached to PS II-antenna supercomplexes (Chen et al. 2009).
The energy transfer from simple PBPs to chlorophylls in the reaction centre is approximatly 70 ps, three times faster than the energy transfer from PBS to PS II in Chl a-containing cyanobacteria (Petrášek et al. 2005). No direct evidence support the connection between the PBPs and PS I, although the analogous CpcG2 protein was predicted in Acaryochloris based on sequence comparison (Chen and Cai 2007). Interestingly, excepting the putative genes encoded for ApcA and ApcB found in the Accaryochloris main chromosome, all PC-associated subunits (CpcA–CpcG) encoding genes are located on one plasmid, pREB3. It has been suggested that ApcA and ApcB form part of bottom disc with CpcA and CpcB together (Hu et al. 1999) and there are no other APC-associated subunits in Acaryochloris. It is still uncertain as to how PBPs associate with the thylakoid membranes because Acaryochloris lacks the core-membrane linker, ApcE. Notably, three copies of CpcG encoded genes in Acaryochloris genome belong to the same protein family, CpcG2, rather than CpcG1 group (Chen and Cai 2007). It is generally understood that CpcG1 plays an important role in attaching the rods to the PBS core when the core is attached to the membrane by core-membrane linker, ApcE. CpcG2 contains a hydrophobic C-terminus, which may play a functional role in attaching the rods to the membranes directly. CpcG2 is involved with the formation of simplified PBP structure that is associated with PS I instead of PS II (Kondo et al. 2007; Deng et al. 2012). The physiological function of CpcG2 in Acaryochloris is uncertain.
CBP-type light-harvesting protein complexes in Acaryochloris are reported to be associated with PS I and PS II (Chen et al. 2005c, d). PS I-antenna supercomplexes (trimer of PS I with a ring of CBP protein around) are not only detected under iron-stress conditions, as in other cyanobacteria (Bibby et al. 2001a, b), but can also be detected under normal culture conditions although the genes encoding CBP proteins are different from the iron-stress-induced CBP (Chen et al. 2005b, 2008).
PSII-antenna complexes were isolated from Acaryochloris. It contains a tetramer of PS II alongside two CBP subunits, the first tetrameric PS II structure reported (Chen et al. 2005a). The existing multiple copies of genes that encode the CBP light-harvesting-proteins suggest the capability to thrive under low light condition (Swingley et al. 2008; Chen et al. 2008). The fact that two isolates of Acaryochloris spp (CCMEE5410 and HICR111a) demonstrated the absence of PBPs suggests that PBPs are the result of an environmental adaptation and that the genes may have been recruited recently (Mohr et al. 2010; Miller et al. 2011). Further investigation is required for defining the relationship between the two antenna systems.
Photosystems in Acaryochloris
Acaryochloris spp are the only known organisms in which the essential function of Chl a in oxygenic photosynthesis has largely been replaced by Chl d. Chl d and Chl d′ function as the special pair in PS I instead of Chl a/Chl a′ (Akiyama et al. 2001; Hu et al. 1998) although the special pair of PSII is an ongoing controversy as to whether it is a Chl d/Chl d homodimer (Chen et al. 2005d; Tomo et al. 2007) or Chl a/Chl d heterodimer (Schlodder et al. 2007). Both PS II and PS I are multisubunit, pigment-protein complexes. Each PS II of cyanobacteria is composed of approximate 20 subunits and 35 chlorophylls (Umena et al. 2011). The first crystal structure of PS I from cyanobacteria showed that PS I exists as a trimer in vivo (but monomer in higher plants) and one monomer consists of at least 11 protein subunits and 96 chlorophylls (Jordan et al. 2001). The polypeptide composition of both PS I and PS II complexes in Acaryochloris are similar to that of well-known Chl a-containing cyanobacteria (Hu et al. 1998; Chen et al. 2005d; Tomo et al. 2008; Swingley et al. 2008). However, the pigment profiles are different due to the unique pigment composition in Acaryochloris.
PS II
The nature of PS II primary donors is an intriguing question as either way, the answer will challenge our current knowledge of energy balance in PS II. If there is Chl d in PS II centre, then whether there will be enough energy for water oxidation which is driven by PS II needs to be addressed. Alternatively, if Chl a play the primary donor in PS II, P680, as in other cyanobacteria, the uphill energy mechanism has to be considered because Chl d is the major photopigment in light-harvesting system. Mimuro et al. were the first team who studied the nature of the primary donor in PSII using Acaryochloris cell’s decay fluorescence measurement. The decay of the fluorescence component in the 685– 695 nm wavelength range with a life time of 15 ns lead to the favoured conclusion that Chl a is the primary donor in PS II and an uphill energy scheme must be applied in Acaryochloris when the energy absorbed by Chl d in light-harvesting complexes is transferred to the accessory Chl d in the reaction centre, then moderately uphill energy transferred to Chl a in the reaction centre (Mimuro et al. 2000).
The pigment composition provided additional support for the proposed P680 in PS II of Acaryochloris. The minimal ratio of Chl a to Phe a was about 1:1 under various culture conditions and the constant presence of Chl a strongly suggested that Chl a plays a critical role in Acaryochloris, maybe as P680 in PS II. However, these hypotheses on the identity of the primary donor of PSII in Acaryochloris were proposed on the basis of pigment analysis combined with whole cell picosecond fluorescence kinetics due to the lack of purified PS II complexes at that time (Mimuro et al. 1999, 2000, 2004; Akiyama et al. 2002). Even with more in depth pigment analysis on isolated Acaryochloris PS II there remains uncertainty regarding the ratio of Chl a to Phe a. An initial report on isolated PS II ‘crude cores’ determined a Chl a: Phe a ratio of 0.5, which is less than the two Chl a required if the special pair in PS II is composed by Chl a (Chen et al. 2005d), however more recent reports suggest this ratio may be closer to 1.5 or 1.0 (Tomo et al. 2007; Tsuchiya et al. 2012a). Fourier transform infrared spectroscopy (FTIR) analysis on purified PS II particle by Tomo et al. demonstrated that P713 of Chl d is the primary donor of PS II (Tomo et al. 2007). However, Itoh et al. suggested P725 of Chl d as the primary donor in PS II based on the laser-induced bleach at 725 nm with the recovery time of 25 μs and 1.2 ms using isolated thylakoid membrane (Itoh et al. 2007). The bleaching at ~713 nm was not observed by either Itoh et al. or by Schlodder et al. (2007) (Tomo et al. 2008). According to the theoretical calculation, Renger and Schlodder indicated the possibility that Chl a/Chl d heterodimer drives the water oxidation because Chl a is necessary for stabilization of the positive charge, and Chl d is the primary electron donor of PS II in Acaryochloris, i.e. in the reaction centre core, there are three Chl d, one Chl a and 2 Phe a (Renger and Schlodder 2008).
Electron paramagnetic resonance (EPR) spectral measurements using Acaryochloris cells revealed that electron transfer to the oxygen-evolving complex of the donor side of PS II is indistinguishable from Chl a-containing oxygenic photosynthetic organisms (higher plants, algae and cyanobacteria) (Razeghifard et al. 2005). The flash-induced oxygen evolution patterns in Acaryochloris cells suggested that the redox-potentials and kinetics within the oxygen-evolution centre of PS II and of redox-active tyrosine site (Yz) are the same as in higher plants (Shevela et al. 2006). The direct measurement on redox potential of Phe a and the energy gap between [PPSII·Phe a −·QA] and[PPSII·Phe a·Q −A ] indicated the overall energetic balance of PS II are similar between Chl a-driving PS II and Chl d-driving PS II (Allakhverdiev et al. 2010b, 2011). The shifted redox potential of Phe a and QA produced an energy gap of −325 mV, which is sufficient for water-splitting reaction driving by Chl d-PS II in Acaryochloris. On the basis of pigment analysis, the accessory Chl a and Chl az in Chl a-containing oxygenic photosynthetic organisms are all replaced with Chl d in Acaryochloris (Mimuro et al. 2004; Kobayashi et al. 2005), but the controversial results were reported based on spectral analysis (Tomo et al. 2007).
Since it is known that Phe a is the primary electron acceptor of PS II and no Phe d is detected in Acaryochloris, the function of Phe a plays the same function as in Chl a-containing cyanobacteria, a primary acceptor of PS II, although the nature of the primary donor of PS II of Acaryochloris is still under debate.
One type of photoprotection of PS II in cyanobacteria is the induction of additional copies of PsbA and PsbD genes, which encode a D1 and a D2 protein with the same/or highly similar amino acid sequence as the D1/D2 expressed under the normal conditions (Mulo et al. 2009). A large mRNA pool of psbA maintains the higher turnover rate of photodamaged D1 protein that is caused by the strongly oxidative chemistry of water splitting. The Acaryochloris genome encodes multiple copies of genes encoding PS II subunits, such as three psbA, three psbD and two psbE, four psbU and two psbV homologues (Swingley et al. 2008). The three copies of psbA genes produce two different D1 proteins, a normal one (D1:1) and the isoform (D1:2) (also see rD1 in “Nitrogen-fixation” section). Various stress conditions upregulate the isoform of D1 (Kiss et al. 2012; Murray 2012). Interestingly, Acaryochloris possesses a D2 protein as well, most cyanobacteria have two psbD copies for the same D2 protein. The isoform of D2 is induced in cells grown under a low visible light/high far-red light regime, indicating the important adaptational role for the D2 isoform in Acaryochloris.
PS I
In general, PS I has as special dimeric forms of Chl a, P700, and generates a strong reducing power to produce NADPH with electrons supplied from PS II. However, Acaryochloris uses Chl d as an energy sink in PS I, i.e. the special pair in PS I is Chl d, P740 (Hu et al. 1998). The polypeptide composition of PS I and the sequence of PsaA and PsaB in Acaryochloris show about 86 % homology which suggests the same architecture of PS I as other cyanobacteria. The image of isolated PS I supercomplexes confirmed that it has a trimer of PS I that form the mega-supercomplexes of antenna-PS I under certain culture conditions (Chen et al. 2005b). The PS I reaction centre complexes of Acaryochloris use Chl d both as the major antenna pigment and as the primary electron donor (Hu et al. 1998). The isolated PS I complexes contain ~180 Chl d and <1 Chl a per complex. The ratio of Chl d/P740 = 140 in the PS I complexes of Acaryochloris (Hu et al. 1998) is in a range similar to that of Chl a/P700 = 100–150 in higher plants, algae and cyanobacterial PS I (Akiyama et al. 2002). The FTIR difference spectral analysis strongly supports that P740 consist of two Chl d molecules (Sivakumar et al. 2003). Spectral analysis using EPR and Electron Spin Echo (ESE) technologies on isolated PS I complexes confirms the similarity between the Chl d-binding PS I in Acaryochloris and Chl a-binding PS I in other oxygenic photosynthetic organisms (Santabarbara et al. 2007). The role of Chl a as A0 in PS I was suggested according to the pigment composition found in isolated PS I, one Chl a per P740 (Hu et al. 1998). Recently, Tomo et al. (2008) isolated highly purified PS I complexes from Acaryochloris and indicated the details of pigment composition of PS I particle. There are 97 Chl d, 2 Chl a and 25 α-carotene, two phylloquinone per one Chl d′ (i.e. per monomer of PS I). Interestingly, the redox potential of P740 was estimated as 439 mV, similar to P700 of Chl a-PS I.
Electron carriers and carotenoids
Oxygenic photosynthesis requires PSI and PS II working in tandem and through a non-cyclic electron transfer chain (ET) to generate ATP. The same fluorescence transients as Chl a-containing species implies there is the same operation of intersystem electron transport chain in Acaryochloris (Schiller et al. 1997; Boichenko et al. 2000).
The PS I and PS II complexes in Acaryochloris are spatially separated and axenically clustered in thylakoid membrane. This phenomenon is also observed in green plants and prochlorophytes (van der Staay and Staehelin 1994). However, in those organisms the PSII is enriched in stacked thylakoid membranes, whereas in Acaryochloris the stacked area is devoid of PSII (Chen et al. 2009; Marquardt et al. 2000). This PS I–PS II spatial separation is also supported by detailed chlorophyll fluorescence measurements which indicate that there is no direct energy transfer from PS II to PS I (Itoh et al. 2007). Furthermore, with the aid of structure modelling Chen et al. (2009) revealed the clustered spatial separation of PS I and PS II in thylakoid membranes of Acaryochloris.
Based on the pigment composition of Chl d′ (representing PS I) and Phe a (representing PS II) in Acaryochloris, the ratio of PS I and PS II is estimated to be 1:1, which is higher than conventional cyanobacteria (Mimuro et al. 2004). Electron microscopic images also show that ~50 % of thylakoid membranes are covered by PBP arrays if they are associated with PS II. However, based on the spin densities on the YZ radical and the photo-oxidixed P740, the ratio of PS I to PS II is 1.8:1 (Itoh et al. 2007).
Interestingly, the same Phe a molecule in the different organisms demonstrated a different redox potential. The redox potential of Phe a in PS II of Synechocystis PCC 6803 is −536 ± 8 mV and in PS II of Acaryochloris is −478 ± 24 mV (Allakhverdiev et al. 2010b). Such different feature clearly indicates the conservation in the properties of oxygen evolution centre (Details see “PS II” section). The photoacoustic measurements indicate the energy storage efficiency of photosynthetic reactions driven by Chl d in Acaryochloris is comparable to or higher than the typical reactions driven by Chl a in the oxygenic photosynthetic organisms (Mielke et al. 2011).
There are two soluble electron carriers between the cyt b 6 f and PS I, plastocyanin (PC) and cyt c 6. The two electron carriers are usually found in a ratio of ~1:3 according to the amount of PS I (Hope 2000). There are two copies of petJ (encoding cyt c 6) and one petE (encoding PC) found in Acaryochloris genome. The expression of petE has been confirmed (Bailleul et al. 2008) although no purified PC is reported in Acaryochloris. On the other hand, purified cyt c 6 of Acaryochloris indicated that it is the major soluble electron carrier from cyt b 6 f to PS I (Bell et al. 2009). The function of isoform of cyt c 6 is unknown.
Conclusion
The discovery of Acaryochloris in 1996 precipitated a shift in our understanding of oxygenic photosynthesis. The presence of Chl d as its major photopigment, substituting many of the roles previously thought to be unique to Chl a, has for the first time allowed the comparative study of the energetics of oxygenic photosystems. The discoveries of red-shifted chlorophylls have highlighted the fundamental question of what is the energy threshold for oxygenic photosynthesis i.e. at what point is the energy of a photon insufficient to drive the process? The red-shifted absorption characteristic of Chl d has clear advantages in absorbing light filtered through Chl a-containing organisms, however challenges associated with the lower energy of these photons have had to be addressed by Acaryochloris. From its comparatively large genome, to its two complementary light- harvesting systems and unusual complement of accessory pigments, the distinctive adaptations of Acaryochloris are clearly advantageous for its proliferation in diverse environments across the planet. The discovery of Acaryochloris and more recent discovery of two Chl f-containing cyanobacteria raise the question of whether there are other as yet uncharacterised chlorophylls in oxygenic photosynthetic organisms, and whether the discovery of these too will challenge both and add to our understanding of this vital process.
Abbreviations
- Acaryochloris :
-
Acaryochloris marina
- APC:
-
Allophycocyanin
- Chl:
-
Chlorophyll
- EPR:
-
Electron paramagnetic resonance
- ESE:
-
Electron spin echo
- FTIR:
-
Fourier transform infrared spectroscopy
- ORF:
-
Open reading frame
- PBP:
-
Phycobiliprotein
- PC:
-
Phycocyanin
- PE:
-
Phycoerythrin
- PEC:
-
Phycoerythrocyanin
- Phe:
-
Pheophytin
- PS:
-
Photosystem
- QA :
-
The primary quinine electron acceptor of photosystem II
- YZ :
-
Redox-active tyrosine in photosystem II
References
Akiyama M, Miyashita H, Kise H, Watanabe T, Miyachi S, Kobayashi M (2001) Detection of chlorophyll d′ and pheophytin a in a chlorophyll d-dominating oxygenic photosynthetic prokaryote Acaryochloris marina. Anal Sci 17:205–208
Akiyama M, Miyashita H, Kise H, Watanabe T, Mimuro M, Miyachi S, Kobayashi M (2002) Quest for minor but key chlorophyll molecules in photosynthetic reaction centers–unusual pigment composition in the reaction centers of the chlorophyll d-dominated cyanobacterium Acaryochloris marina. Photosynth Res 74:97–107
Akutsu S, Fujinuma D, Furukawa H, Watanabe T, Ohnishi-Kameyama M, Ono H, Ohkubo S, Miyashita H, Kobayashi M (2011) Pigment analysis of a chlorophyll f-containing cyanobacterium strain KC1 isolated from Lake Biwa. Photochem Photobiol 33:35–40
Allakhverdiev SI, Thavasi V, Kreslavski VD, Zharmukhamedov SK, Klimov VV, Ramakrishna S, Los DA, Mimuro M, Nishihara H, Carpentier R (2010a) Photosynthetic hydrogen production. J Photochem Photobiol, C 11:101–113
Allakhverdiev SI, Tomo T, Shimada Y, Kindo H, Nagao R, Klimov VV, Mimuro M (2010b) Redox potential of pheophytin a in photosystem II of two cyanobacteria having the different special pair chlorophylls. Proc Natl Acad Sci USA 107:3924–3929
Allakhverdiev SI, Tsuchiya T, Watabe K, Kojima A, Los DA, Tomo T, Klimov VV, Mimuro M (2011) Redox potentials of primary electron acceptor quinone molecule (QA)—and conserved energetics of photosystem II in cyanobacteria with chlorophyll a and chlorophyll d. Proc Natl Acad Sci USA 108:8054–8058
Arnon DI (1949) Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol 24:1
Bailleul B, Johnson X, Finazzi G, Barber J, Rappaport F, Telfer A (2008) The thermodynamics and kinetics of electron transfer between cytochrome b6f and photosystem I in the chlorophyll d-dominated cyanobacterium, Acaryochloris marina. J Biol Chem 283:25218–25226
Beale SI (2006) Biosynthesis of 5-aminolevulinic acid. In: Grimm B, Porra RJ, Rudiger W, Scheer H (eds) Chlorophylls and bacteriochlorophylls: biochemistry, biophysics, functions and applications, vol 25. Springer, Dordrecht, pp 147–158
Behrendt L, Larkum AWD, Norman A, Qvortrup K, Chen M, Ralph P, Sorensen SJ, Trampe E, Kuhl M (2011) Endolithic chlorophyll d-containing phototrophs. ISME J 5:1072–1076
Bell PD, Xin Y, Blankenship RE (2009) Purification and characterization of cytochrome c6 from Acaryochloris marina. Photosynth Res 102:43–51
Bergman B, Gallon JR, Rai AN, Stal LJ (1997) N2 fixation by non-heterocystous cyanobacteria. FEMS Microbiol Rev 19:139–185
Bibby T, Nield J, Partensky F, Barber J (2001a) Oxyphotobacteria. Antenna ring around photosystem I. Nature 413(6856):590
Bibby TS, Nield J, Barber J (2001b) Iron deficiency induces the formation of an antenna ring around trimeric photosystem I in cyanobacteria. Nature 412:743–745
Björn L, Papageorgiou G, Blankenship R (2009) A viewpoint: why chlorophyll a. Photosynth Res 99:85–98
Boichenko V, Klimov V, Miyashita H, Miyachi S (2000) Functional characteristics of chlorophyll d-predominating photosynthetic apparatus in intact cells of Acaryochloris marina. Photosynth Res 65:269–277
Bollivar DW (2006) Recent advances in chlorophyll biosynthesis. Photosynth Res 90:173–194
Bollivar DW, Suzuki JY, Beatty JT, Dobrowolski JM, Bauer CE (1994) Directed muataional analysis of bacteriochlorophyll a biosynthesis in Rhodobacter capsulatus. J Mol Biol 237:622–640
Chen M, Blankenship RE (2011) Expanding the solar spectrum used by photosynthesis. Trends Plant Sci 16:427–431
Chen M, Cai Z-L (2007) Theoretical study on the thermodynamic properties of chlorophyll d-peptides coordinating ligand. Biochim Biophys Acta 1767:603–609
Chen M, Scheer H (2012) Extending the limits of natural photosynthesis and implications for technical light harvesting. J Porphyr Phthalocya 15:1–15
Chen M, Quinnell RG, Larkum AWD (2002) The major light-harvesting pigment protein of Acaryochloris marina. FEBS Lett 514:149–152
Chen M, Bibby TS, Nield J, Larkum AWD, Barber J (2005a) Iron deficiency induces a chlorophyll d-binding Pcb antenna system around photosystem I in Acaryochloris marina. Biochim Biophys Acta 1708:367–374
Chen M, Bibby TS, Nield J, Larkum AWD, Barber J (2005b) Structure of a large photosystem II supercomplex from Acaryochloris marina. FEBS Lett 579:1306–1310
Chen M, Hiller RG, Howe CJ, Larkum AWD (2005c) Unique origin and lateral transfer of prokaryotic chlorophyll-b and chlorophyll-d light-harvesting systems. Mol Biol Evol 22:21–28
Chen M, Telfer A, Lin S, Pascal A, Larkum AWD, Barber J, Blankenship RE (2005d) The nature of the photosystem II reaction centre in the chlorophyll d-containing prokaryote, Acaryochloris marina. Photochem Photobiol Sci 4:1060–1064
Chen M, Zhang Y, Blankenship RE (2008) Nomenclature for membrane-bound light-harvesting complexes of cyanobacteria. Photosynth Res 95:147–154
Chen M, Floetenmeyer M, Bibby TS (2009) Supramolecular organization of phycobiliproteins in the chlorophyll d-containing cyanobacterium Acaryochloris marina. FEBS Lett 583(15):2535–2539
Chen M, Schliep M, Willows RD, Cai ZL, Neilan BA, Scheer H (2010) A red-shifted chlorophyll. Science 329:1318–1319
Chen M, Li YQ, Birch D, Willows RD (2012) A cyanobacterium that contains chlorophyll f—a red-absorbing photopigment. FEBS Lett 586:3249–3254
Chisholm SW, Frankel SL, Goericke R, Olson RJ, Palenik B, Waterbury JB, West-Johnsrud L, Zettler ER (1992) Prochlorococcus marinus nov. gen. nov. sp.: an oxyphototrophic marine prokaryote containing divinyl chlorophyll a and b. Arch Microbiol 157:297–300
Cohen Z, Margheri MC, Tomaselli L (1995) Chemotaxonomy of cyanobacteria. Phytochemistry 40:1155–1158
De Los Ríos A, Grube M, Sancho LG, Ascaso C (2007) Ultrastructural and genetic characteristics of endolithic cyanobacterial biofilms colonizing Antarctic granite rocks. FEMS Microbiol Ecol 59:386–395
Deng G, Liu F, Liu X, Zhao J (2012) Significant energy transfer from CpcG2-phycobilisomes to photosystem I in the cyanobacterium Synechococcus sp. PCC 7002 in the absence of ApcD-dependent state transitions. FEBS Lett 586:2342–2345
Dutta D, De D, Chaudhuri S, Bhattacharya SK (2005) Hydrogen production by cyanobacteria. Microb Cell Fact 4:36
Duxbury Z, Schliep M, Ritchie R, Larkum AD, Chen M (2009) Chromatic photoacclimation extends utilisable photosynthetically active radiation in the chlorophyll d-containing cyanobacterium, Acaryochloris marina. Photosynth Res 101:69–75
Espineda CE, Linford AS, Devine D, Brusslan JA (1999) The AtCAO gene, encoding chlorophyll a oxygenase, is required for chlorophyll b synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 96:10507–10511
Fay P (1992) Oxygen relations of nitrogen fixation in cyanobacteria. Microbiol Rev 56:340
Fleming ED, Prufert-Bebout L (2010) Characterization of cyanobacterial communities from high-elevation lakes in the Bolivian Andes. J Geophys Res 115:G00d07
Fukusumi T, Matsuda K, Mizoguchi T, Miyatake T, Ito S, Ikeda T, Tamiaki H, Oba T (2012) Non-enzymatic conversion of chlorophyll-a into chlorophyll-d in vitro: a model oxidation pathway for chlorophyll-d biosynthesis. FEBS Lett 586:2338–2341
Gallon JR (2005) N2 fixation by non-heterocystous cyanobacteria. In: Klipp W, Masepohl B, Gallon J, Newton W (eds) Genetics and regulation of nitrogen fixation in free-living bacteria, vol 2., Nitrogen fixation: Origins, Applications, and Research progressSpringer, Houten, pp 111–139
Gao B, Boeglin WE, Zheng Y, Schneider C, Brash AR (2009) Evidence for an ionic intermediate in the transformation of fatty acid hydroperoxide by a catalase-related allene oxide synthase from the cyanobacterium Acaryochloris marina. J Biol Chem 284:22087–22098
Gao B, Boeglin WE, Brash AR (2010) Omega-3 fatty acids are oxygenated at the n-7 carbon by the lipoxygenase domain of a fusion protein in the cyanobacterium Acaryochloris marina. Biochim Biophys Acta 1801:58–63
Goh F, Allen MA, Leuko S, Kawaguchi T, Decho AW, Burns BP, Neilan BA (2008) Determining the specific microbial populations and their spatial distribution within the stromatolite ecosystem of Shark Bay. ISME J 3:383–396
Gombos Z, Kanervo E, Tsvetkova N, Sakamoto T, Aro EM, Murata N (1997) Genetic enhancement of the ability to tolerate photoinhibition by introduction of unsaturated bonds into membrane glycerolipids. Plant Physiol 115:551–559
Hoiczyk E, Hansel A (2000) Cyanobacterial cell walls: news from an unusual prokaryotic envelope. J Bacteriol 182:1191–1199
Holt AS (1961) Further evidence of the relation between 2-desvinyl-2-formyl-chlorophyll-a and chlorophyll-d. Can J Bot 39:327–331
Holt AS, Morley HV (1959) A proposed structure for chlorophyll d. Can J Chem 37:507–514
Hoober JK, Eggink LL, Chen M (2007) Chlorophylls, ligands and assembly of light-harvesting complexes in chloroplasts. Photosynth Res 94:387–400
Hope AB (2000) Electron transfers amongst cytochrome f, plastocyanin and photosystem I: kinetics and mechanisms. Biochim Biophys Acta 1456:5–26
Hu Q, Miyashita H, Iwasaki I, Kurano N, Miyachi S, Iwaki M, Itoh S (1998) A photosystem I reaction center driven by chlorophyll d in oxygenic photosynthesis. Proc Natl Acad Sci USA 95:13319–13323
Hu Q, Marquardt J, Iwasaki I, Miyashita H, Kurano N, Mörschel E, Miyachi S (1999) Molecular structure, localization and function of biliproteins in the chlorophyll a d containing oxygenic photosynthetic prokaryote Acaryochloris marina. Biochim Biophys Acta 1412:250–261
Ito H, Yokono M, Tanaka R, Tanaka A (2008) Identification of a novel vinyl reductase gene essential for the biosynthesis of monovinyl chlorophyll in Synechocystis sp. PCC6803. J Biol Chem 283:9002–9011
Itoh S, Mino H, Itoh K, Shigenaga T, Uzumaki T, Iwaki M (2007) Function of chlorophyll d in reaction centers of photosystems I and II of the oxygenic photosynthesis of Acaryochloris marina. Biochemistry 46:12473–12481
Jahn D, Moser J, Schubert W, Heinz DW (2006) Transfer RNA-dependent aminolevulinic acid formation: structure and function of glutamyl-trna synthetase, reductase and glutamate-1-semialdehyde-2,1-aminomutase. In: Grimm B, Porra RJ, Rudiger W, Scheer H (eds) Chlorophylls and bacteriochlorophylls: biochemistry, biophysics, functions and applications, vol 25. Springer, Dordrecht, pp 159–171
Jordan P, Fromme P, Witt HT, Klukas O, Saenger W, Krauß N (2001) Three-dimensional structure of cyanobacterial photosystem I at 2.5 Å resolution. Nature 411:909–917
Kashiyama Y, Miyashita H, Ohkubo S, Ogawa NO, Chikaraishi Y, Takano Y, Suga H, Toyofuku T, Nomaki H, Kitazato H, Nagata T, Ohkouchi N (2008) Evidence of global chlorophyll d. Science 321:658
Kiss É (2012) Expression of the genes encoding the subunits of the Photosystem II core and the Hox hydrogenase are aiding acclimation of the cyanobacterium Acaryochloris marina. University of Szeged (PhD Thesis). http://doktori.bibl.u-szeged.hu/1664/3/Kiss_%C3%89va_thesis.pdf
Kiss É, Kós PB, Vass I (2009) Transcriptional regulation of the bidirectional hydrogenase in the cyanobacterium Synechocystis 6803. J Biotechnol 142:31–37
Kiss É, Kós PB, Chen M, Vass I (2012) A unique regulation of the expression of the psbA, psbD, and psbE genes, encoding the D1, D2 and cytochrome b559 subunits of the Photosystem II complex in the chlorophyll d containing cyanobacterium Acaryochloris marina. Biochim Biophys Acta 1817:1083–1094
Kiss É, Kós PB, Chen M, Vass I (2013) Functioning of the bidirectional hydrogenase in different unicelluar cyanobacteria. In: Photosynthesis Research for Food, Fuel and the Future: Proceeding of 15th International Congress on Photosynthesis, Beijing, 2010. Springer (in press, ISBN 978-3-642-32033-0)
Kobayashi M, Watanabe S, Gotoh T, Koizumi H, Itoh Y, Akiyama M, Shiraiwa Y, Tsuchiya T, Miyashita H, Mimuro M (2005) Minor but key chlorophylls in photosystem II. Photosynth Res 84:201–207
Kobayashi M, Ohashi S, Iwamoto K, Shiraiwa Y, Kato Y, Watanabe T (2007) Redox potential of chlorophyll d in vitro. Biochim Biophys Acta 1767:596–602
Kondo K, Ochiai Y, Katayama M, Ikeuchi M (2007) The membrane-associated CpcG2-phycobilisome in Synechocystis: a new photosystem I antenna. Plant Physiol 144:1200–1210
Konstantinidis KT, Tiedje JM (2004) Trends between gene content and genome size in prokaryotic species with larger genomes. Proc Natl Acad Sci USA 101:3160–3165
Kräutler B (2011) A new factor in life’s quest for energy. Angew Chem Int Ed 50:2439–2441
Kühl M, Chen M, Ralph PJ, Schreiber U, Larkum AWD (2005) A niche for cyanobacteria containing chlorophyll d. Nature 433:820
La Roche J, Van der Staay G, Partensky F, Ducret A, Aebersold R, Li R, Golden S, Hiller R, Wrench P, Larkum A (1996) Independent evolution of the prochlorophyte and green plant chlorophyll a/b light-harvesting proteins. Proc Natl Acad Sci USA 93:15244–15248
Larkum AWD, Kühl M (2005) Chlorophyll d: the puzzle resolved. Trends Plant Sci 10:355–357
Larkum AWD, Chen M, Li Y, Schliep M, Trampe E, West J, Salih A, Kühl M (2012) A novel epiphytic chlorophyll d-containing cyanobacterium isolated from a mangrove-associated red alga. J Phycol 48:1320–1327
Lewin RA, Withers NW (1975) Extraordinary pigment composition of a prokaryotic alga. Nature 256:735–737
Li Y, Scales N, Blankenship RE, Willows RD, Chen M (2012) Extinction coefficient for red-shifted chlorophylls: chlorophyll d and chlorophyll f. Biochim Biophys Acta 1817:1292–1298
Li Y, Larkum A, Schliep M, kuhl M, Neilan B, Chen M (2013) Newly isolated Chl d-containing cyanobacteria. In: Photosynthesis research for food, fuel and the future: Proceeding of 15th International Congress on Photosynthesis, Beijing, 2010. Springer (in press, ISBN 978-3-642-32033-0)
Lin Y, Crossett B, Chen M (2013) Effects of anaerobic conditions on photosynthetic units of Acaryochloris marina. In: photosynthesis research for food, fuel and the future: Proceeding of 15th International Congress on Photosynthesis, Beijing, China, 2010. Springer (in press, ISBN 978-3-642-32033-0)
López-Legentil S, Song B, Bosch M, Pawlik JR, Turon X (2011) Cyanobacterial diversity and a new Acaryochloris-like symbiont from Bahamian sea-squirts. PLoS ONE 6:e23938
Mackinney G (1941) Absorption of light by chlorophyll solutions. J Biol Chem 140(2):315–322
Manning WM, Strain HH (1943) Chlorophyll d, a green pigment of red algae. J Biol Chem 151:1–19
Marquardt J, Senger H, Miyashita H, Miyachi S, Mörschel E (1997) Isolation and characterization of biliprotein aggregates from Acaryochloris marina, a Prochloron-like prokaryote containing mainly chlorophyll d. FEBS Lett 410:428–432
Marquardt J, Mörschel E, Rhiel E, Westermann M (2000) Ultrastructure of Acaryochloris marina, an oxyphotobacterium containing mainly chlorophyll d. Arch Microbiol 174:181–188
Martínez-García M, Koblížek M, López-Legentil S, Antón J (2011) Epibiosis of oxygenic phototrophs containing chlorophylls a, b, c, and d on the colonial ascidian Cystodytes dellechiajei. Microb Ecol 61:13–19
McNamara C, Perry TIV, Bearce K, Hernandez-Duque G, Mitchell R (2006) Epilithic and endolithic bacterial communities in limestone from a Maya Archaeological Site. Microb Ecol 51:51–64
Mielke S, Kiang N, Blankenship R, Gunner M, Mauzerall D (2011) Efficiency of photosynthesis in a Chl d-utilizing cyanobacterium is comparable to or higher than that in Chl a-utilizing oxygenic species. Biochim Biophys Acta 1807:1231–1236
Miller SR, Augustine S, Olson TL, Blankenship RE, Selker J, Wood AM (2005) Discovery of a free-living chlorophyll d-producing cyanobacterium with a hybrid proteobacterial/cyanobacterial small-subunit rRNA gene. Proc Natl Acad Sci USA 102:850–855
Miller SR, Wood AM, Blankenship RE, Kim M, Ferriera S (2011) Dynamics of gene duplication in the genomes of chlorophyll d-producing cyanobacteria: implications for the ecological niche. Genome Biol Evol 3:601–613
Mimuro M, Akimoto S, Yamazaki I, Miyashita H, Miyachi S (1999) Fluorescence properties of chlorophyll d-dominating prokaryotic alga, Acaryochloris marina: studies using time-resolved fluorescence spectroscopy on intact cells. Biochim Biophys Acta 1412:37–46
Mimuro M, Hirayama K, Uezono K, Miyashita H, Miyachi S (2000) Uphill energy transfer in a chlorophyll d-dominating oxygenic photosynthetic prokaryote, Acaryochloris marina. Biochim Biophys Acta 1456:27–34
Mimuro M, Akimoto S, Gotoh T, Yokono M, Akiyama M, Tsuchiya T, Miyashita H, Kobayashi M, Yamazaki I (2004) Identification of the primary electron donor in PS II of the Chl d-dominated cyanobacterium Acaryochloris marina. FEBS Lett 556:95–98
Miyashita H, Ikemoto H, Kurano N, Adachi K, Chihara M, Miyachi S (1996) Chlorophyll d as a major pigment. Nature 383:402
Miyashita H, Adachi K, Kurano N, Ikemot H, Chihara M, Miyach S (1997) Pigment composition of a novel oxygenic photosynthetic prokaryote containing chlorophyll d as the major chlorophyll. Plant Cell Physiol 38:274–281
Miyashita H, Ikemoto H, Kurano N, Miyachi S, Chihara M (2003) Acaryochloris marina gen. et sp. nov. (cyanobacteria), an oxygenic photosynthetic prokaryote containing Chl d as a major pigment. J Phycol 39:1247–1253
Mohr R, Vosz B, Schliep M, Kurz T, Maldener I, Adams DG, Larkum ADW, Chen M, Hess WR (2010) A new chlorophyll d-containing cyanobacterium: evidence for niche adaptation in the genus Acaryochloris. ISME J 4:1456–1469
Mulo P, Sicora C, Aro E-M (2009) Cyanobacterial psbA gene family: optimization of oxygenic photosynthesis. Cell Mol Life Sci 66:3697–3710
Murakami A, Miyashita H, Iseki M, Adachi K, Mimuro M (2004) Chlorophyll d in an epiphytic cyanobacterium of red algae. Science 303:1633
Murata N, Wada H, Gombos Z (1992) Modes of fatty-acid desaturation in cyanobacteria. Plant Cell Physiol 33:933–941
Murray J (2012) Sequence variation at the oxygen-evolving centre of photosystem II: a new class of ‘rogue’ cyanobacterial D1 proteins. Photosynth Res 110:177–184
Nagata N, Tanaka R, Satoh S, Tanaka A (2005) Identification of a vinyl reductase gene for chlorophyll synthesis in Arabidopsis thaliana and implications for the evolution of Prochlorococcus species. Plant Cell 17:233–240
Nieuwenburg P, Clarke RJ, Cai ZL, Chen M, Larkum AWD, Cabral NM, Ghiggino KP, Reimers JR (2003) Examination of the photophysical processes of chlorophyll d leading to a clarification of proposed uphill energy transfer processes in cells of Acaryochloris marina. Photochem Photobiol 77:628–637
Ohkubo S, Miyashita H (2012) Selective detection and phylogenetic diversity of Acaryochloris spp. That exist in association with didemnid ascidians and sponge. Microbes Environ 27:217–225
Oster U, Tanaka R, Tanaka A, Rudiger W (2000) Cloning and functional expression of the gene encoding the key enzyme for chlorophyll b biosynthesis (CAO) from Arabidopsis thaliana. Plant J 21:305–310
Petrášek Z, Schmitt FJ, Theiss C, Huyer J, Chen M, Larkum A, Eichler HJ, Kemnitz K, Eckert HJ (2005) Excitation energy transfer from phycobiliprotein to chlorophyll d in intact cells of Acaryochloris marina studied by time-and wavelength-resolved fluorescence spectroscopy. Photochem Photobiol Sci 4:1016–1022
Pfreundt U, Stal LJ, Vos B, Hess WR (2012) Dinitrogen fixation in a unicellular chlorophyll d-containing cyanobacterium. ISME J 6:1367–1377
Porra RJ, Scheer H (2000) O-18 and mass spectrometry in chlorophyll research: derivation and loss of oxygen atoms at the periphery of the chlorophyll macrocycle during biosynthesis, degradation and adaptation. Photosynth Res 66:159–175
Porra R, Thompson W, Kriedemann P (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975:384–394
Razeghifard MR, Chen M, Hughes JL, Freeman J, Krausz E, Wydrzynski T (2005) Spectroscopic studies of photosystem II in chlorophyll d-containing Acaryochloris marina. Biochemistry 44:11178–11187
Renger T, Schlodder E (2008) The primary electron donor of photosystem II of the cyanobacterium Acaryochloris marina is a chlorophyll d and the water oxidation is driven by a chlorophyll a/chlorophyll d heterodimer. J Phys Chem B 112:7351–7354
Rudiger W (2006) Biosynthesis of chlorophylls a and b: the last steps. In: Grimm B, Porra RJ, Rudiger W, Scheer H (eds) Chlorophylls and bacteriochlorophylls: biochemistry, biophysics, functions and applications, vol 25. Springer, Dordrecht, pp 189–200
Santabarbara S, Chen M, Larkum AWD, Evans MCW (2007) An electron paramagnetic resonance investigation of the electron transfer reactions in the chlorophyll d containing photosystem I of Acaryochloris marina. FEBS Lett 581:1567–1571
Scheer H (2006) An overview of chlorophylls and bacteriochlorophylls: biochemistry, biophysics, functions and applications. In: Grimm B, Porra RJ, Rudiger W, Scheer H (eds) Chlorophylls and bacteriochlorophylls: biochemistry, biophysics, functions and applications, vol 25. Springer, Dordrecht, pp 1–26
Schiller H, Senger H, Miyashita H, Miyachi S, Dau H (1997) Light-harvesting in Acaryochloris marina–spectroscopic characterization of a chlorophyll d-dominated photosynthetic antenna system. FEBS Lett 410:433–436
Schliep M, Crossett B, Willows RD, Chen M (2010) 18O labeling of chlorophyll d in Acaryochloris marina reveals that chlorophyll a and molecular oxygen are precursors. J Biol Chem 285:28450–28456
Schlodder E, Çetin M, Eckert HJ, Schmitt FJ, Barber J, Telfer A (2007) Both chlorophylls a and d are essential for the photochemistry in photosystem II of the cyanobacteria, Acaryochloris marina. Biochim Biophys Acta 1767:589–595
Shevela D, Nöring B, Eckert HJ, Messinger J, Renger G (2006) Characterization of the water oxidizing complex of photosystem II of the Chl d-containing cyanobacterium Acaryochloris marina via its reactivity towards endogenous electron donors and acceptors. Phys Chem Chem Phys 8:3460–3466
Shpilyov AV, Zinchenko VV, Grimm B, Lokstein H (2013) Chlorophyll a phytylation is required for the stability of photosystems I and II in the cyanobacterium Synechocystis sp. PCC 6803. Plant J 73:336–346
Sivakumar V, Wang RL, Hastings G (2003) Photo-oxidation of P740, the primary electron donor in photosystem I from Acaryochloris marina. Biophys J 85:3162–3172
Smith JAC, Benitez M (1955) Chlorophylls: analysis in plant materials. In: Paech K, Tracey MV (eds) Modern methods of plant analysis, vol 4. Springer, Berlin, pp 142–196
Swingley WD, Hohmann-Marriott MF, Le Olson T, Blankenship RE (2005) Effect of iron on growth and ultrastructure of Acaryochloris marina. Appl Environ Microbiol 71:8606–8610
Swingley WD, Chen M, Cheung PC, Conrad AL, Dejesa LC, Hao J, Honchak BM, Karbach LE, Kurdoglu A, Lahiri S, Mastrian SD, Miyashita H, Page L, Ramakrishna P, Satoh S, Sattley WM, Shimada Y, Taylor HL, Tomo T, Tsuchiya T, Wang ZT, Raymond J, Mimuro M, Blankenship RE, Touchman JW (2008) Niche adaptation and genome expansion in the chlorophyll d-producing cyanobacterium Acaryochloris marina. Proc Natl Acad Sci USA 105:2005–2010
Takaichi S, Mochimaru M (2007) Carotenoids and carotenogenesis in cyanobacteria: unique ketocarotenoids and carotenoid glycosides. Cell Mol Life Sci 64:2607–2619
Takaichi S, Mochimaru M, Uchida H, Murakami A, Hirose E, Maoka T, Tsuchiya T, Mimuro M (2012) Opposite chilarity of α-carotene in unusual cyanobacteria with unique chlorophylls, Acaryochloris and Prochlorococcus. Plant Cell Physiol 53:1881–1888
Tanaka A, Ito H, Tanaka R, Tanaka NK, Yoshida K, Okada K (1998) Chlorophyll a oxygenase (CAO) is involved in chlorophyll b formation from chlorophyll a. Proc Natl Acad Sci USA 95:12719–12723
Tomo T, Okubo T, Akimoto S, Yokono M, Miyashita H, Tsuchiya T, Noguchi T, Mimuro M (2007) Identification of the special pair of photosystem II in a chlorophyll d-dominated cyanobacterium. Proc Natl Acad Sci USA 104:7283–7288
Tomo T, Kato Y, Suzuki T, Akimoto S, Okubo T, Noguchi T, Hasegawa K, Tsuchiya T, Tanaka K, Fukuya M, Dohmae N, Watanabe T, Mimuro M (2008) Characterization of highly purified photosystem I complexes from the chlorophyll d-dominated cyanobacterium Acaryochloris marina MBIC 11017. J Biol Chem 283:18198–18209
Tomo T, Allakhverdiev SI, Mimuro M (2011) Constitution and energetics of photosystem I and photosystem II in the chlorophyll d-dominated cyanobacterium Acaryochloris marina. J Photochem Photobiol, B 104:333–340
Tsuchiya T, Akimoto S, Mizoguchi T, Watabe K, Kindo H, Tomo T, Tamiaki H, Mimuro M (2012a) Artificially produced 7-formyl -chlorophyll d functions as an antenna pigment in the photosystem II isolated from the chlorophyllide a oxygenase-expressing Acaryochloris marina. Biochim Biophys Acta 1817:1285–1291
Tsuchiya T, Mizoguchi T, Akimoto S, Tomo T, Tamiaki H, Mimuro M (2012b) Metabolic engineering of the Chl d-dominated cyanobacterium Acaryochloris marina: production of a novel Chl species by the introduction of the chlorophyllide a oxygenase Gene. Plant Cell Physiol 53:518–527
Tsukatani Y, Romberger SP, Golbeck JH, Bryant DA (2012) Isolation and characterization of homodimeric type-I reaction center complex from Candidatus Chloracidobacterium thermophilum, an aerobic chlorophototroph. J Biol Chem 287:5720–5732
Umena Y, Kawakami K, Shen JR, Kamiya N (2011) Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9Å. Nature 473:55–60
van der Staay GW, Staehelin LA (1994) Biochemical characterization of protein composition and protein phosphorylation patterns in stacked and unstacked thylakoid membranes of the prochlorophyte Prochlorothrix hollandica. J Biol Chem 269:24834–24844
Wakao N, Yokoi N, Isoyama N, Hiraishi A, Shimada K, Kobayashi M, Kise H, Iwaki M, Itoh S, Takaichi S, Sakurai Y (1996) Discovery of natural photosynthesis using Zn-containing bacteriochlorophyll in an aerobic bacterium Acidiphilium rubrum. Plant Cell Physiol 37:889–893
Walker CJ, Mansfield KE, Smith KM, Castelfranco PA (1989) Incorporation of atmospheric oxygen into the carbonyl functionality of the protochlorophyllide isocyclic ring. Biochem J 257:599–602
Woodward RB, Ayer WA, Beaton JM, Bickelhaupt F, Bonnett R, Buchschacher P, Closs GL, Dutler H, Hannah J, Hauck FP (1990) The total synthesis of chlorophyll a. Tetrahedron 46:7599–7659
Xu H, Vavilin D, Funk C, Vermaas W (2002) Small Cab-like proteins regulating tetrapyrrole biosynthesis in the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol Biol 49:149–160
Yaronskaya E, Grimm B (2006) The pathway from 5-aminolevulinic acid to protochlorophyllide and protoheme. In: Grimm B, Porra RJ, Rudiger W, Scheer H (eds) Chlorophylls and bacteriochlorophylls: biochemistry, biophysics, functions and applications, vol 25. Springer, Dordrecht, pp 173–188
Zapata M, Garrido JL, Jeffery SW (2006) Chlorophyll c pigments: current status. In: Grimm B, Porra RJ, Rudiger W, Scheer H (eds) Chlorophylls and bacteriochlorophylls: biochemistry, biophysics, functions and applications, vol 25. Springer, Dordrecht, pp 39–53
Zehr JP (2011) Nitrogen fixation by marine cyanobacteria. Trends Microbiol 19:162–173
Acknowledgments
MC holds Australia Research Council Future Fellowship. YL thanks Ms Yaqiong Li for her helps for 16 s rDNA sequence alignment and phylogentic analysis.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Loughlin, P., Lin, Y. & Chen, M. Chlorophyll d and Acaryochloris marina: current status. Photosynth Res 116, 277–293 (2013). https://doi.org/10.1007/s11120-013-9829-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11120-013-9829-y