Abstract
A short overview is given on the discovery of the chlorophyll d-dominated cyanobacterium Acaryochloris marina and the minor pigments that function as key components therein. In photosystem I, chlorophyll d′, chlorophyll a, and phylloquinone function as the primary electron donor, the primary electron acceptor and the secondary electron acceptor, respectively. In photosystem II, pheophytin a serves as the primary electron acceptor. The oxidation potential of chlorophyll d was higher than that of chlorophyll a in vitro, while the oxidation potential of P740 was almost the same as that of P700. These results help us to broaden our view on the questions about the unique photosystems in Acaryochloris marina.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A chlorophyll d-dominated cyanobacterium, Acaryochloris marina, was discovered by Miyashita, one of the authors of this review. In August 1993, while visiting Palau for two days, he came across one colony of ascidians, Lissoclinum patella, as a gift from Drs. M. Dionisio-sese and T. Maruyama. L. patella is a well-known host of Prochloron, a symbiotic cyanobacterium containing chlorophyll (Chl) a and Chl b (Fig. 1) without phycobiliproteins (PBP), but no one had ever succeeded in culturing Prochloron. The Prochloron cells were squeezed out from the ascidians, inoculated in a seawater-based IMK medium in a 24-well tissue culture plate, and then brought to Japan.
Though the Prochloron cells divided one or two times, they died within a few weeks, and the samples were left as it was. More than one month later, small yellowish-green colonies like green algae were unexpectedly found at the bottom of the wells. The microalga was ellipsoidal with 1–2 μm length; smaller than Prochloron in ascidians (spherical with 10–30 μm diameter). Miyashita assumed the yellowish-green microalga to be a free-living stage of Prochloron, and tried to incubate it in a fresh seawater-based medium, where the microalga grew well.
In December 1993, pigment analysis of the alga was performed. The dominant pigment exhibited apparently the same retention time as that of Chl b on the reversed-phase HPLC elution profile. Trace amounts of Chl a, zeaxanthin, and magnesium 3,8-divinylphaeoporphyrin a 5 monoethyl ester were also detected. At that point, there was no doubt that the microalga should be a free-living Prochloron, but a big surprise awaited Miyashita.
The absorption spectrum of the “Chl b-peak” was of an unknown feature, and completely different from that of Chl b (Fig. 2). A few days later, the same spectrum was found in a book “Photosynthetic Pigments of Algae” (K. S. Rowan); the pigment was Chl d! NMR analysis confirmed the judgment (Miyashita et al. 1997). Here, a new genus, Acaryochloris, being unicellular cyanobacterium containing Chl d (Fig. 1) as a major pigment, was established (Miyashita et al. 1996) and has opened a new window for the photosynthesis science.
Later, in a red seaweed, Ahnfeltiopsis flabelliformis, Murakami found small patches on the thalli surfaces, a cyanobacterium-like prokaryotic epiphyte containing Chl d, while Chl d was absent inside the thalli (Murakami et al. 2004). Miyashita cut the thalli into small pieces, and then put them in a tissue culture plate with a seawater-based medium. Familiar yellowish-green colonies appeared at the bottom of the well. The molecular phylogenetic analysis showed the cells were of an Acaryochloris sp. Epiphytic Acaryochloris sp. was found in several red algae around Awaji Island, Japan (Ohkubo et al. 2006). This finding showed that Chl d detected in red algae was due to epiphytic Acaryochloris sp., and the genus Acaryochloris was the only organism that synthesizes Chl d.
Minor but key components in Acaryochloris marina
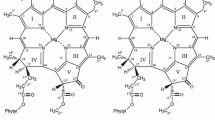
Though A. marina has Chl d as the major pigment, a few minor chlorophyllous pigments, Chl d′, Phe a, and Chl a (Fig. 1), are also present and function as key components in the reaction centers (RCs) of photosystem (PS) I and PS II (Akiyama et al. 2001). The content of Chl a varies according to light conditions (Boichenko et al. 2000; Akiyama et al. 2004; Mimuro et al. 2004), but at least one molecule of Chl a seems to be present in each RC (Akiyama et al. 2004; Kobayashi et al. 2005, 2007).
Photosystem I of A. marina
Chl d′, Chl a and phylloquinone in PS I
Chl d′, the 132-epimer of Chl d (Fig. 1), was always detected in A. marina as a minor component, while Chl a′ was absent (compare Fig. 3A with B) (Akiyama et al. 2001). P740, the primary electron donor of PS I in A. marina, was initially proposed to be a homodimer of Chl d (Hu et al. 1998), then a homodimer of Chl d′ (Akiyama et al. 2001), and finally a Chl d/d′ heterodimer (Fig. 4A) (Akiyama et al. 2002, 2004; Kobayashi et al. 2005, 2007), just like the Chl a/a′ for P700 in other cyanobacteria and higher plants (Fig. 4C) (Kobayashi et al. 1988; Jordan et al. 2001): a dimer model for P740 was supported by FTIR spectroscopy (Sivakumar et al. 2003). The homology of PsaA and PsaB between A. marina and other cyanobacteria is low (Swingley et al. 2008), which may reflect the replacement of Chl a by Chl d, also Chl a′ by Chl d′, in the PS I RC of A. marina (Fig. 4A), although the physiological significance of Chl d′ in P740, as well as Chl a′ in P700, is not yet clear. PsaA and PsaB in Acaryochloris marina as well as Prochlorococcus sp. have lower homology to those in common cyanobacteria. The phylogenetic tree for PsaA/B (Fig. 5) (Miyashita et al. unpublished data) shows that the branch length for the Acaryochloris (▼) as well as Prochlorococcus (▽) were longer than the others. The long branch means that the evolution rates of PsaA and PsaB in those cyanobacteria are faster than those in common cyanobacteria. The change of evolutionary rate in the protein with the same function is usually explained by the change of evolutional constraint of the protein. The reason of the low homology is inferred as the replacement of Chl a by Chl d and divinyl-chlorophyll a, respectively.
(A, B) Normal-phase and (C, D) reversed-phase HPLC profiles for (A, C) acetone/methanol extracts of A. marina; (B) a mixture of authentic Phe a, Phe d, Chl a′, Chl a, Chl d′, and Chl d; and (D) a mixture of authentic MQ-4, PhQ and MQ-7. Eluents of normal-phase and reversed-phase HPLC are n-hexane/2-propanol/methanol (100/0.7/0.2, v/v/v) and ethanol/methanol/water (86/14/3, v/v/v), respectively. Detection wavelength is (A, B) 650 nm and (C, D) 248 nm
It is interesting to note that the primary electron acceptor, A0, in PS I of A. marina is not Chl d but appears to be Chl a (Fig. 4A), which was first shown by laser photolysis experiment (Kumazaki et al. 2002), and then supported by flash-induced spectral analysis (Itoh et al. 2007). The results support our hypothesis that Chl a or its derivative is a general feature of A0 in the PS I-type RCs (see Fig. 2 in Akiyama et al. 2002), though the reason why Chl a functions as A0 in the PS I-type RCs is still unclear, and the role and localization of Chl a in PS I of A. marina remain controversial (Fig. 4B) (Tomo et al. 2008). Further investigation is required to settle these questions.
The secondary electron acceptor, A1, in A. marina was identified as phylloquinone (PhQ) (Fig. 4A) by HPLC, absorption, and mass analyses of cells (Ohashi et al. 2007, 2008a), and then supported by HPLC and absorption analyses of PS I (Tomo et al. 2008). The quinone from A. marina cells showed the same retention time as the authentic PhQ (Fig. 3C, D), and the same result was recently reported in PS I of A. marina. The molar ratio of quinone/Chl d′ was 2/1 both in cells (Fig. 3C) (Ohashi et al. 2007, 2008a) and in PS I (Tomo et al. 2008), indicating that two molecules of the quinone are present per PS I (Fig. 4A, B). The quinone exhibited the same absorption spectrum as the PhQ standard (Ohashi et al. 2007, 2008a; Tomo et al. 2008), indicating the presence of naphtoquinone framework in A1. The typical APCI-mass spectrum of the quinone is illustrated in Fig. 6A; the quinone showed a molecular ion peak [M + H]+ at m/z 451.3, which was the same value as the authentic PhQ (C31H46O2; mol wt. = 450.7, Fig. 6B). We thus concluded that A1 of A. marina is PhQ.
Redox potentials of Chl d and P740
The midpoint potential E m for P740 in PS I of A. marina was initially reported as +335 mV (Hu et al. 1998), being significantly lower than that of P700 (ca. +430 mV) in most Chl a-type PS Is (Ke 2001; Nakamura et al. 2005; Kato et al. 2008) (Fig. 7). Based on this, Chl d was supposed to have an oxidation potential E ox lower than that of Chl a. The longer wavelength of the Chl d QY-band (Fig. 2A) also appeared to support the view that Chl d is oxidized more easily than Chl a (see Fig. 1 in Ohashi et al. 2008b).
However, the E ox value of Chl d in acetonitrile was found to be higher than that of Chl a (Kobayashi et al. 2007). The E ox value order, Chl b (+0.93 V vs. SHE) > Chl d (+0.88 V) > Chl a (+0.81 V), is accounted for by invoking the inductive effect of substituents on the conjugated π-electron system on the macrocycle.
The pheophytins showed the same E ox order; Phe b (+1.25 V) > Phe d (+1.21 V) > Phe a (+1.14 V) (Kobayashi et al. 2007). The significantly higher potentials than those of the corresponding Chls are in line with electron density decrease on the π-system by replacement of magnesium with more electronegative hydrogen (Fig. 1). As a result, the E ox of Chl a is the lowest among the Chls and Phes (see Table 1 in Kobayashi et al. 2007).
Recently, the E m of P740 was re-examined and found to be +430 mV (Benjamin et al. 2007; Telfer et al. 2007) and +439 mV (Tomo et al. 2008), being much higher than the initial report (+335 mV) and almost equal to the Chl a-type P700 values of other cyanobacteria (Fig. 7), which is supported by the ENDOR analyses showing the location of spin density between P700+ and P740+ was almost same (Mino et al. 2005; Itoh et al. 2008). Significant differences in a few amino acids around P740 and P700 might also yield similar E m values. The difference in the oxidation potential shifts between Chl a/Chl a + → P700/P700+ and Chl d/d + → P740/P740+ in Fig. 7, suggests that the interaction between the special pair chlorophylls (Chl d and d′) of P740 might be stronger than that between Chl a and a′ of P700.
The stronger interaction between Chl d and d′ of P740 than Chl a and a′ of P700 is partially supported by absorption spectral properties. In diethyl ether, the wavelengths of QY maximum of Chl a and Chl d are 661 nm and 686 nm, respectively (Fig. 2A) (Kobayashi et al. 2006), and hence the excitation energy shifts caused by dimerization (Chl a → P700 and Chl d → P740) are roughly calculated to be 0.11 eV and 0.13 eV, respectively.
In purple bacterial special pairs, the energy shift of 0.13 eV is also calculated for BChl a (771 nm) → P840, and a much larger shift of 0.28 eV for BChl b (796 nm) → P970, suggesting that similarly stronger interaction is present in both P740 and P840, and much stronger interaction is present in P970. In this context, the interaction between two BChl g′ molecules in P798 of heliobacteria (Kobayashi et al. 1991) is presumed to be very weak, since the energy shift for BChl g′ (767 nm) → P798 is significantly small, ca. 0.07 eV.
It is well known that the special pair consists of two (bacterio)chlorophyll molecules that are axially coordinated by two histidine residues from the protein subunits (Fig. 8). In the case of the purple bacterial special pair, both BChl a and BChl b molecules have acetyl group at C3 of ring I, which are in direct contact with the Mg atoms of the other BChl in the special pair (see Fig. 8C) (Deisenhofer et al. 1984); the average distance between rings I of (BChl a)2 is 3.5 Å (Allen et al. 1987) and (BChl b)2 is closer to 3 Å (Deisenhofer et al. 1984). Chl d and d′ in P740 have formyl group at C3 of ring I (Fig. 1), and hence coordination of formyl group to Mg is also expected (Fig. 8B). In contrast, the coordination of vinyl group at C3 of ring I of Chl a and a′ to Mg was not observed in P700, where the vinyl group was configurated as avoiding steric hindrance against the macrocycle of the other Chl (Fig. 8A) (Jordan et al. 2001). The weak interaction expected between two BChl g′ molecules in P798 may be due to the presence of vinyl group at the same position, like Chl a and a′ in P700. Further support comes from the interplanar distance of 3.6 Å in P700 (Jordan et al. 2001), being longer than 3 - 3.5 Å in P840 and P970. A slightly stronger interaction is hence expected to exist in the special pair Chls d/d′ in P740 than the Chls a/a′ in P700, and other experimental results are expected to confirm the hypothesis.
The similarity of oxidation potentials, being around +430 mV for both P700 and P740, may be required for the use of a common electron donor, plastocyanin, for P700+ and P740+. The redox potential of plastocyanin is known to be affected by the amino acids around the active site, which are also responsible for binding to PS I. These amino acids are highly conserved among A. marina, other cyanobacteria, and higher plants (NCBI data base), and hence the redox potential of plastocyanin in A. marina seems to be similar in other cyanobacteria (Benjamin et al. 2007; Tomo et al. 2008). If this is the case, the lower reducing power of P740* may affect the energetics of electron flow on the acceptor side (Benjamin et al. 2007), and thus the redox potential of A0, Chl a (Fig. 4A) or Chl d (Fig. 4B), is expected to be lower than that in other cyanobacteria (see Fig. 7). Direct redox potential measurements for the electron transport components in vivo are strongly needed.
Photosystem II of A. marina
Phe a and Chl a in PS II
In the PS II RC of A. marina, the primary electron acceptor is not Phe d but Phe a (see Fig. 3A, C and 4A, B) (Akiyama et al. 2001, 2002, 2004; Mimuro et al. 2004; Kobayashi et al. 2005; Razeghifard et al. 2005; Chen et al. 2005; Tomo et al. 2007). It has not been clarified yet why A. marina uses Phe a instead of Phe d. One of the reasons might be the use of a common electron acceptor, plastoquinone, for pheophytin, because the reduction potential of Phe d (−0.63 V) was found to be significantly more positive than that of Phe a (−0.75 V) in acetonitrile (Kobayashi et al. 2007).
Identity of the special pair in PS II
The nature of the special pair in the PS II RC has been controversial, and three models have been proposed; (1) a Chl a homodimer, (2) a Chl a/d heterodimer (Fig. 4A), and a Chl d homodimer (Fig. 4B). The Chl a dimer model was first proposed on the basis of the origin of delayed fluorescence by Mimuro et al. (1999), and was supported by the same method (Mimuro et al. 2000, 2004; Akimoto et al. 2006), the estimated value of the redox potential (Boichenko et al. 2000), and the pigment stoichiometry analysis (Akiyama et al. 2001, 2002, 2004). The Chl a/d heterodimer model (Fig. 4A) was first proposed on the pigment stoichiometry analysis by Kobayashi et al. (2005), and was supported by the redox potential measurement of Chls a and d (Kobayashi et al. 2007), the flash-induced spectral experiment (Schlodder et al. 2007), and its theoretical analysis (Renger and Schlodder 2008). The Chl d homodimer model (Fig. 4B) was first proposed on the spectral analysis by Itoh et al. (2001), and was supported by the similar methods (Itoh et al. 2008; Nieuwenburg et al. 2003; Tomo et al. 2007) and pigment stoichiometry (Chen et al. 2005).
The Chl a homodimer model is most unlikely, because the minimum ratio of Chl a/Phe a in A. marina whole cells grown under low-light condition was 2/2 (Mimuro et al. 2004), indicating only one Chl a molecule is located in the PS II RC (Kobayashi et al. 2005) on the basis of Phe a/PS II = 2, PS I/PS II = 1 (Akiyama et al. 2001), and the presence of one Chl a molecule as an active A0 in PS I (Kumazaki et al. 2002). The model was also excluded by a quantitative explanation of the experimental data (Renger and Schlodder 2008).
It should be interesting to note that Itoh et al. (2007) and Schlodder et al. (2007) observed very similar difference spectra in the QY region with main bleaching around 727 nm, but their interpretations are completely different; the former assumed that the main bleaching arose from both a homodimeric Chl d special pair (Fig. 4B) designated by them as P727 and an electrochromic shift due to the accessory Chl d, while the latter inferred that the bleaching was solely caused by the electrochromic shift of the accessory Chl d and that the special pair is either a Chl a homo- or a Chl a/Chl d heterodimer (Fig. 4A). Tomo et al. (2007), however, reported a significantly different spectrum with main bleaching around 713 nm, and they assumed that the bleaching was derived from a homodimeric Chl d special pair (Fig. 4B), and named it P713. The difference between 727 nm and 713 nm may originate in different excitation conditions; Itoh’s and Schlodder’s groups used single turnover flashes, but Tomo’s group measured the difference between the absorbance during illumination for 1 s and in the dark. The bleaching around 713 nm might come from the accumulation of some denatured Chls, probably because their long illumination may lead to some denaturation of Chls. Hastings and Wang (2008) pointed out that the FTIR bands of Chl d identified by Tomo’s group in the P+-P spectrum might not belong to P+ but could be due to electrochromically shifted vibrational bands of pigments in the neighborhood of P+.
Schlodder et al. (2007) also observed the bleaching in the Soret region at 435 nm and the spectral changes in the near infrared region upon photo-oxidation identical with flash-induced absorbance changes measured for normal Chl a type PS II, implying that one of the special pair Chls is Chl a. A quantitative explanation of the experimental data requires one Chl a molecule as a special pair in the PS II RC of A. marina (Renger and Schlodder 2008), suggesting that the special pair in the PS II RC of A. marina is a Chl a/d heterodimer and the accessory is Chl d (Fig. 4A). To confirm the pigment arrangement in the PS II RC of A. marina, X-ray structural studies are strongly awaited.
Energy and electron transfer within PS II
It was shown that the primary charge separation in PS II is initiated by excitation of the accessory Chl a, AccChl a, of the D1-branch in Chl a-type organisms: P-Acc*-Phe → P-Acc+-Phe− → P+-Acc-Phe− (Diner and Rappaport 2002; Raszewski et al. 2005; Groot et al. 2005; Holzwarth et al. 2006) (Fig. 4C). Here, the replacement of AccChl a with AccChl d may be essential in PS II of A. marina (Fig. 4A) (Kobayashi et al. 2005, 2007, 2008), because if Acc was Chl a (see Fig. 4B), energy transfer from antenna Chl d to AccChl a would be difficult because of the extremely uphill process. Our model depicted in Fig. 4A was recently supported by the flash-induced absorbance difference spectrum measurement (Schlodder et al. 2007) and a theoretical analysis (Renger and Schlodder 2008). Further, the higher oxidation potential of Chl d than that of Chl a is favorable for the electron transfer from Chl a in P to AccChl d + (Kobayashi et al. 2007; Renger and Schlodder 2008) (Fig. 4A), since the lowest site energy was found for accessory chlorophyll in A. marina (Renger and Schlodder 2008). On the contrary, if the accessory chlorophyll was Chl a (Fig. 4B) (Tomo et al. 2007), electron transfer from Chl d in P to AccChl a + was unlikely, due to the lower oxidation potential of Chl a. The primary charge separation initiated by AccChl d is therefore most likely also in the PS II RC of A. marina, after energy transfer from antenna Chl d to AccChl d (Fig. 4A), although the model of Fig. 4B (Acc is Chl a) cannot be excluded at present.
The substitution frequencies in the PS I-RC proteins, PsaA and PsaB, as well as in the PS II core antenna proteins, CP47 (PsbB) and CP43 (PsbC), of A. marina are higher than those in other cyanobacteria (Swingley et al. 2008), indicating a rapid evolution due to Chl d incorporation into these proteins.
In contrast, the evolutionary rates in D1(PsbA) and D2(PsbD) proteins are comparable with those in other cyanobacteria, suggesting that amino acid substitution in D1/D2 proteins in A. marina has the same constraint as in other cyanobacteria, and that Phe a (not Phe d) may be used for the primary electron acceptor. Figure 9 shows the alignment of amino acid sequences of D2(PsbD) in cyanobacteria and representative plastids, where leucine(L) and isoleucine(I) interacting with Phe a are conserved in all D2. However, the alignment of amino acid sequences of D2 showed that the tryptophan (W) residue at position 191 in Synechocystis D2, which is known to interact with P680 and to be conserved in all D2, was substituted by leucine(L) in A. marina D2, while such conversion was not seen in D1. The evolutionary constraint on the tryptophan residue was changed in D2 of Acaryochloris, indicating a substitution of the D2-side special pair Chl a (P680) with Chl d in the PS II RC of A. marina (Fig. 4A), although this idea has not been widely accepted yet.
Abbreviations
- BChl:
-
Bacteriochlorophyll
- Chl:
-
Chlorophyll
- HPLC:
-
High performance liquid chromatography
- PBP:
-
Phycobiliproteins
- P680:
-
The primary electron donor of photosystem II
- P700:
-
The primary electron donor of Chl a-type photosystem I
- P740:
-
The primary electron donor of photosystem I in A. marina
- Phe:
-
Pheophytin
- PS:
-
Photosystem
- RC:
-
Reaction center
References
Akimoto S, Murakami A, Yokono M, Koyama K, Tsuchiya T, Miyashita H, Yamazaki I, Mimuro M (2006) Fluorescence properties of the chlorophyll d-dominated cyanobacterium Acaryochloris sp. strain Awaji. J Photochem Photobiol A 178:122–129. doi:10.1016/j.jphotochem.2005.09.031
Akiyama M, Miyashita H, Watanabe T, Kise K, Miyachi S, Kobayashi M (2001) Detection of chlorophyll d′ and pheophytin a in a chlorophyll d-dominating oxygenic photosynthetic prokaryote Acaryochloris marina. Anal Sci 17:205–208. doi:10.2116/analsci.17.205
Akiyama M, Miyashita H, Kise H, Watanabe T, Mimuro M, Miyachi S, Kobayashi M (2002) Quest for minor but key chlorophyll molecules in photosynthetic reaction centers—unusual pigment composition in the reaction centers of a chlorophyll d-dominated cyanobacterium Acaryochloris marina. Photosynth Res 74:97–107. doi:10.1023/A:1020915506409
Akiyama M, Gotoh T, Kise H, Miyashita H, Mimuro M, Kobayashi M (2004) Stoichiometries of chlorophyll d′/PSI and chlorophyll a/PSII in a chlorophyll d-dominated cyanobacterium Acaryochloris marina. Jpn J Phycol (Supplement for the Proceedings of Algae 2002) 52:67–72
Allen JP, Feher G, Yeates TO, Komiya H, Rees DC (1987) Structure of the reaction center from Rhodobacter sphaeroides R-26: the cofactors. Proc Natl Acad Sci USA 84:5730–5734. doi:10.1073/pnas.84.16.5730
Benjamin B, Finazzi G, Benson S, Barber J, Rappaport F, Telfer A (2007) Study of intersystem electron transfer in the chlorophyll d containing cyanobacterium Acaryochloris marina and a reappraisal of the redox properties of P740. Photosynth Res 91:155
Boichenko VA, Klimov VV, Miyashita H, Miyachi S (2000) Functional characteristics of chlorophyll d-predominating photosynthetic apparatus in intact cells of Acaryochloris marina. Photosynth Res 65:269–277. doi:10.1023/A:1010637631417
Chen M, Telfer A, Lin S, Pascal A, Larkum AWD, Barber J, Blankenship RE (2005) The nature of the photosystem II reaction centre in the chlorophyll d-containing prokaryote, Acaryochloris marina. Photochem Photobiol Sci 4:1060–1064. doi:10.1039/b507057k
Deisenhofer J, Epp O, Miki K, Huber R, Michel H (1984) X-ray structure analysis of a membrane protein complex. Electron density map at 3 Å resolution and a model of the chromophores of the photosynthetic reaction center from Rhodopseudomonas viridis. J Mol Biol 180:385–398. doi:10.1016/S0022-2836(84)80011-X
Diner BA, Rappaport F (2002) Structure, dynamics, and energetics of the primary photochemistry of photosystem II of oxygenic photosynthesis. Annu Rev Plant Biol 53:551–580. doi:10.1146/annurev.arplant.53.100301.135238
Groot ML, Pawlowicz NP, van Wilderen LJGW, Breton J, van Stokkum IHM, van Grondelle R (2005) Initial electron donor and acceptor in isolated photosystem II reaction centers identified with femtosecond mid-IR spectroscopy. Proc Natl Acad Sci USA 102:13087–13092. doi:10.1073/pnas.0503483102
Hastings G, Wang R (2008) Vibrational mode frequency calculations of chlorophyll-d for assessing (P740+-P740) FTIR difference spectra obtained using photosystem I particles from Acaryochloris marina. Photosynth Res 95:55–62. doi:10.1007/s11120-007-9228-3
Holzwarth AR, Müller MG, Reus M, Nowaczyk M, Sander J, Rögner M (2006) Kinetics and mechanism of electron transfer in intact photosystem II and in the isolated reaction center: pheophytin is the primary electron acceptor. Proc Natl Acad Sci USA 103:6895–6900. doi:10.1073/pnas.0505371103
Hu Q, Miyashita H, Iwasaki I, Kurano N, Miyachi S, Iwaki M, Itoh S (1998) A photosystem I reaction center driven by chlorophyll d in oxygenic photosynthesis. Proc Natl Acad Sci USA 95:13319–13323. doi:10.1073/pnas.95.22.13319
Itoh S, Iwaki M, Noguti T, Kawamori A, Mino H, Hu Q, Iwasaki I, Miyashita H, Kurano N, Miyachi S, Shen R (2001) Photosystem I and II reaction centers of a new oxygenic organism Acaryochloris marina that use chlorophyll d. In: PS2001 Proceedings of the 12th international congress on photosynthesis, CSIRO, Melbourne, Australia, pp S6–S028
Itoh S, Mino H, Itoh K, Shigenaga T, Uzumaki T, Iwaki M (2007) Function of chlorophyll d in reaction centers of photosystems I and II of the oxygenic photosynthesis of Acaryochloris marina. Biochemistry 46:12473–12481. doi:10.1021/bi7008085
Itoh S, Uzumaki T, Takaichi S, Iwaki M, Kumazaki S, Itoh K, Mino H (2008) Unidirectional electron transfer in chlorophyll d-containing photosystem I reaction center complex of Acaryochloris marina. In: Allen J, Gantt E, Golbeck J, Osmond B (eds) Energy from the sun. Springer, pp 93–96
Jordan P, Fromme P, Witt HT, Klukas O, Saenger W, Krauß N (2001) Three-dimensional structure of cyanobacterial photosystem I at 2.5 Å resolution. Nature 411:909–917. doi:10.1038/35082000
Kato Y, Nakamura A, Suzawa T, Watanabe T (2008) Unexpected difference in the P700 redox potential among oxygenic photosynthetic organisms revealed by spectroelectrochemistry. In: Allen J, Gantt E, Golbeck J, Osmond B (eds) Energy from the sun. Springer, pp 109–112
Ke B (2001) The primary electron donor of photosysten I-P700, Photosynthesis: photobiochemistry and photobiophysics. Kluwer Academic, Dordrecht, pp 463–478
Kobayashi M, Watanabe T, Nakazato M, Ikegami I, Hiyama T, Matsunaga T, Murata N (1988) Chlorophyll a′/P700 and pheophytin a/P680 stoichiometries in higher plants and cyanobacteria bacteria determined by HPLC analysis. Biochim Biophys Acta 936:81–89. doi:10.1016/0005-2728(88)90254-X
Kobayashi M, van de Meent EJ, Amesz J, Ikegami I, Watanabe T (1991) Bacteriochlorophyll g epimer as a possible reaction center component of heliobacteria. Biochim Biophys Acta 1057:89–96. doi:10.1016/S0005-2728(05)80087-8
Kobayashi M, Watanabe S, Gotoh T, Koizumi H, Itoh Y, Akiyama M, Shiraiwa Y, Tsuchiya T, Miyashita H, Mimuro M, Yamashita T, Watanabe T (2005) Minor but key chlorophylls in photosystem II. Photosynth Res 84:201–207. doi:10.1007/s11120-005-0474-y
Kobayashi M, Akiyama M, Kise H, Watanabe T (2006) Spectroscopy and structure determination. In: Grimm B, Porra RJ, Rüdiger W, Scheer H (eds) Chlorophylls and bacteriochlorophylls: biochemistry, biophysics, functions and applications. Springer, Dordrecht, pp 79–94
Kobayashi M, Ohashi S, Iwamoto K, Shiraiwa Y, Kato Y, Watanabe T (2007) Redox potential of chlorophyll d in vitro. Biochim Biophys Acta 1767:596–602. doi:10.1016/j.bbabio.2007.02.015
Kobayashi M, Ohashi S, Fukuyo S, Kasahara M, Watanabe T (2008) The oxidation potential of Chl a is the lowest—a new scheme for O2 evolution in PS II. In: Allen J, Gantt E, Golbeck J, Osmond B (eds) Energy from the sun. Springer, pp 113–116
Kumazaki S, Abiko K, Ikegami I, Iwaki M, Itoh S (2002) Energy equilibration and primary charge separation in chlorophyll d-based photosystem I reaction center isolated from Acaryochloris marina. FEBS Lett 530:153–157. doi:10.1016/S0014-5793(02)03446-4
Mimuro M, Akimoto S, Yamazaki I, Miyashita H, Miyachi S (1999) Fluorescence properties of chlorophyll d-dominating prokaryotic alga, Acaryochloris marina: studies using time-resolved fluorescence spectroscopy on intact cells. Biochim Biophys Acta 1412:37–46. doi:10.1016/S0005-2728(99)00048-1
Mimuro M, Hirayama K, Uezono K, Miyashita H, Miyachi S (2000) Uphill energy transfer in a chlorophyll d-dominating oxygenic photosynthetic prokaryote, Acaryochloris marina. Biochim Biophys Acta 1456:27–34. doi:10.1016/S0005-2728(99)00095-X
Mimuro M, Akimoto S, Gotoh T, Yokono M, Akiyama M, Tsuchiya T, Miyashita H, Kobayashi M, Yamazaki I (2004) Identification of the primary electron donor in PS II of the Chl d-dominated cyanobacterium Acaryochloris marina. FEBS Lett 556:95–98. doi:10.1016/S0014-5793(03)01383-8
Mino H, Kawamori A, Aoyama D, Tomo T, Iwaki M, Itoh S (2005) Proton ENDOR study of the primary donor P740+, a special pair of chlorophyll d in photosystem I reaction center of Acaryochloris marina. Chem Phys Lett 411:262–266. doi:10.1016/j.cplett.2005.06.033
Miyashita H, Ikemoto H, Kurano N, Adachi K, Chihara M, Miyachi S (1996) Chlorophyll d as a major pigment. Nature 383:402. doi:10.1038/383402a0
Miyashita H, Adachi K, Kurano N, Ikemoto H, Chihara M, Miyachi S (1997) Pigment composition of a novel oxygenic photosynthetic prokaryote containing chlorophyll d as the major chlorophyll. Plant Cell Physiol 38:274–281
Murakami A, Miyashita H, Iseki M, Adachi K, Mimuro M (2004) Chlorophyll d in an epiphytic cyanobacterium of red algae. Science 303:1633. doi:10.1126/science.1095459
Nakamura A, Suzawa T, Kato Y, Watanabe T (2005) Significant species-dependence of P700 redox potential as verified by spectroelectrochemistry: comparison of spinach and Theromosynechococcus elongatus. FEBS Lett 579:2273–2276. doi:10.1016/j.febslet.2005.02.076
Nieuwenburg P, Clarke RJ, Cai ZL, Chen M, Larkum AWD, Cabral NM, Ghiggino KP, Reimers JR (2003) Examination of the photophysical processes of chlorophyll d leading to a clarification of proposed uphill energy transfer processes in cells of Acaryochloris marina. Photochem Photobiol 77:628–637. doi :10.1562/0031-8655(2003)077<0628:EOTPPO>2.0.CO;2
Ohashi S, Tsuchiya T, Iwamoto K, Miyashita H, Watanabe T, Shiraiwa Y, Mimuro M, Kobayashi M (2007) Succession of co-factors in photosystem I. Photosynth Res 91:270
Ohashi S, Tsuchiya T, Iwamoto K, Miyashita H, Watanabe T, Shiraiwa Y, Mimuro M, Kobayashi M (2008a) Succession of co-factors in photosystem I. In: Allen J, Gantt E, Golbeck J, Osmond B (eds) Energy from the sun. Springer, pp 1177–1180
Ohashi S, Kasahara M, Fukuyo S, Nakazato M, Iwamoto K, Shiraiwa Y, Kato Y, Watanabe T, Kobayashi M (2008b) Redox potential of chlorophyll d. In: Allen J, Gantt E, Golbeck J, Osmond B (eds) Energy from the sun. Springer, pp 105–108
Ohkubo S, Miyashita H, Murakami A, Takeyama H, Tsuchiya T, Mimuro M (2006) Molecular detection of epiphytic Acaryochloris spp. on marine macroalgae. Appl Environ Microbiol 72:7912–7915. doi:10.1128/AEM.01148-06
Raszewski G, Saenger W, Renger T (2005) Theory of optical spectra of photosystem II reaction centers: location of the triplet state and the identity of the primary electron donor. Biophys J 88:986–998. doi:10.1529/biophysj.104.050294
Razeghifard MR, Chen M, Hughes JL, Freeman J, Krausz E, Wydrzynski T (2005) Spectroscopic studies of photosystem II in chlorophyll d-containing Acaryochloris marina. Biochemistry 44:11178–11187. doi:10.1021/bi048314c
Renger T, Schlodder E (2008) The primary electron donor of photosystem II of the cyanobacterium Acaryochloris marina is a chlorophyll d and the water oxidation is driven by a chlorophyll a/chlorophyll d heterodimer. J Phys Chem B 112:7351–7354. doi:10.1021/jp801900e
Schlodder E, Çetin M, Eckert HJ, Schmitt FJ, Barber J, Telfer A (2007) Both chlorophylls a and d are essential for the photochemistry in photosystem II of the cyanobacteria, Acaryochloris marina. Biochim Biophys Acta 1767:589–595. doi:10.1016/j.bbabio.2007.02.018
Sivakumar V, Wang R, Hastings G (2003) Photo-oxidation of P740, the primary electron donor in photosystem I from Acaryochloris marina. Biophys J 85:3162–3172
Swingley WD, Chen M, Cheung PC, Conrad AL, Dejesa LC, Hao J, Honchak BM, Karbach LE, Kurdoglu A, Lahiri S, Mastrian SD, Miyashita H, Page L, Ramakrishna P, Satoh S, Sattley WM, Shimada Y, Taylor HL, Tomo T, Tsuchiya T, Wang ZT, Raymond J, Mimuro M, Blankenship RE, Touchman JW (2008) Niche adaptation and genome expansion in the chlorophyll d-producing cyanobacterium Acaryochloris marina. Proc Natl Acad Sci USA 105:2005–2010. doi:10.1073/pnas.0709772105
Telfer A, Pascal A, Barber J, Schenderlein M, Schlodder E, Çetin M (2007) Electron transfer reactions in photosystem I and II of the chlorophyll d containing cyanobacterium, Acaryochloris marina. Photosynth Res 91:143
Tomo T, Okubo T, Akimoto S, Yokono M, Miyashita H, Tsuchiya T, Noguchi T, Mimuro M (2007) Identification of the special pair of photosystem II in a chlorophyll d-dominated cyanobacterium. Proc Natl Acad Sci USA 104:7283–7288. doi:10.1073/pnas.0701847104
Tomo T, Kato Y, Suzuki T, Akimoto S, Okubo T, Noguchi T, Hasegawa K, Tsuchiya T, Tanaka K, Fukuya M, Dohmae N, Watanabe T, Mimuro M (2008) Characterization of highly purified photosystem I complexes from the chlorophyll d-dominated cyanobacterium Acaryochloris marina MBIC 11017. J Biol Chem 283:18198–18209. doi:10.1074/jbc.M801805200
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ohashi, S., Miyashita, H., Okada, N. et al. Unique photosystems in Acaryochloris marina . Photosynth Res 98, 141–149 (2008). https://doi.org/10.1007/s11120-008-9383-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11120-008-9383-1