Abstract
Years of genetic, biochemical, and structural work have provided a number of insights into the oxygen evolving complex (OEC) of Photosystem II (PSII) for a variety of photosynthetic organisms. However, questions still remain about the functions and interactions among the various subunits that make up the OEC. After a brief introduction to the individual subunits Psb27, PsbP, PsbQ, PsbR, PsbU, and PsbV, a current picture of the OEC as a whole in cyanobacteria, red algae, green algae, and higher plants will be presented. Additionally, the role that these proteins play in the dynamic life cycle of PSII will be discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Photosystem II (PSII) is the multi-component enzyme of cyanobacteria, algae and plants that catalyzes the light-driven oxidation of water to molecular oxygen. This complex consists of more than 20 proteins including both integral membrane and extrinsically associated proteins. In addition to its protein components, PSII also has a large number of associated cofactors including chlorophylls, pheophytins, plastoquinones, manganese atoms, non-heme iron, calcium, chloride, and heme groups. Despite the large number of components, PSII can be divided into two functional domains (1) the electron transfer domain, comprised of the integral membrane helices and cofactors and (2) the oxygen evolving complex (OEC), located on the lumenal face of the complex including the extrinsic proteins, the loop regions of several membrane proteins and the catalytic inorganic manganese, calcium and chloride cluster.
The catalytic center of the OEC sequentially removes four electrons from two water molecules to form molecular oxygen. All of the crystal structures of cyanobacterial PSII show that the ligands to this catalytic center are provided by the intrinsic protein components (Kamiya and Shen 2003; Ferreira et al. 2004; Loll et al. 2005a). While numerous studies shown that the extrinsic proteins are not necessary for oxygen evolution activity in vitro, they are required to enhance oxygen evolution and serve important roles in vivo (reviewed in Seidler 1996).
The catalytic mechanism and architecture of PSII are remarkably conserved among a wide variety of organisms from unicellular cyanobacteria to multicellular plants, but the most heterogeneity among the photosynthetic organisms is found in the extrinsic proteins of the water oxidation machinery. As many as five proteins associate in different combinations depending on the type of organism. Only the PsbO protein (the 33 kDa or Manganese Stabilizing Protein) is ubiquitous to all oxygenic photosynthetic organisms. The other extrinsic proteins are PsbP (23, 24 kDa protein), PsbQ (16–18 kDa protein in plants, 20 kDa protein in red algae), PsbR (10 kDa protein), PsbU (12 kDa protein), PsbV (cytochrome c 550 ) and Psb27 (11 kDa protein). For clarity the nomenclature used in this manuscript refers to the proteins by their four letter name and not by their apparent molecular weight.
Table 1 shows the distribution of the different extrinsic proteins among the different types of photosynthetic organisms (cyanobacteria, red algae, green algae, and plants). The extrinsic proteins Psb28, which is not located in the lumen, and Sll1390 will not be discussed here because they have not been shown to be involved in the OEC. Recent reviews have addressed the evolutionary implications of sequence divergence and extrinsic protein distributions among the different organisms and so will not be discussed in detail here (Seidler 1996; De Las Rivas et al. 2004; De Las Rivas and Roman 2005; Enami et al. 2005; Thornton et al. 2005). However, these distributions highlight the way different types of organisms utilize the different combinations of extrinsic proteins to tune their PSII complexes for optimal activity. It also stimulates questions as to whether each protein performs the same function or has modified or specialized functions within the different types of organisms. However, the genomic analysis does not provide information as to whether all of the extrinsic proteins available to these organisms are simultaneously bound to their PSII complexes or whether only specific combinations of the proteins are present. These outstanding questions are all topics of intense research. Advances in structural biology, genetics, and biochemistry in a number of representative oxygenic photosynthetic model organisms will continue to further our knowledge of how the extrinsic proteins optimize PSII activity.
This review focuses on our current knowledge of the functions of the PSII extrinsic proteins and how they interact in the OECs of different oxygenic photosynthetic organisms to optimize the water oxidation reaction. Each protein will be discussed both individually and collectively in the context of the different types of PSII OECs—cyanobacterial, red algal, green algal and plant. Finally, the dynamic nature of PSII will be discussed as some extrinsic proteins may directly participate in the biogenesis and turnover of PSII.
Individual extrinsic lumenal subunits
PsbO
As shown in Table 1, the PsbO protein (33 kDa, Manganese Stabilizing Protein) is a component of the PSII OEC in every type of oxygenic photosynthetic organism. Biochemical and genetic studies have shown that the PsbO protein is a key structural component of many different types of OECs and functions to stabilize the manganese cluster and modulate the Ca2+ and Cl− requirements for oxygen evolution (reviewed in Bricker and Burnap 2005). The absence of the PsbO protein has varying effects on PSII depending on the organism. For example, plants and the green alga Chlamydomonas reinhardtii (referred to hereafter as Chlamydomonas) are not capable of photoautotrophic growth without PsbO (Mayfield et al. 1987; Yi et al. 2005). In contrast, cyanobacterial PsbO mutants can grow photoautotrophically, although at lower levels relative to wild type (Philbrick et al. 1991). While other papers in this issue will discuss PsbO in more detail, here we mention key unresolved questions regarding the PsbO protein in the context of the OEC.
In the current structural models of PSII, PsbO does not provide any ligand to the manganese cluster (Kamiya and Shen 2003; Ferreira et al. 2004; Loll et al. 2005a), but there is debate as to whether PsbO directly binds Ca2+ (reviewed in Seidler 1996). While the absence of the PsbO protein does not affect characteristic electron paramagnetic resonance (EPR) signals attributed to the Ca2+ ion of the OEC (Seidler and Rutherford 1996), recent analysis suggests that there may be another low affinity Ca2+ binding site within plant PsbO (Heredia and De Las Rivas 2003). Such observations could not be repeated with purified or recombinant cyanobacterial PsbO (Loll et al. 2005b), but a closer look at the anomalous diffraction data from cyanobacterial PsbO within PSII indicate a possible Ca2+ binding site (Murray and Barber 2006). Higher resolution PSII structures are necessary to fully resolve this issue.
Another controversy associated with PsbO, involves its stoichiometry within the PSII complex. Structural analyses of cyanobacterial PSII indicate one PsbO per PSII monomer (Kamiya and Shen 2003; Ferreira et al. 2004; Loll et al. 2005a), but reconstitution studies in plants indicate the molar ratio of PsbO in PSII is 2 (Xu and Bricker 1992; Popelkova et al. 2002b). These data can be reconciled by the recent observation that plant PsbO contains two sequence motifs at its N-terminus which determine binding stoichiometry, while cyanobacterial PsbO has only one such sequence (Popelkova et al. 2002a). However, structural studies of plant PSII show there is not sufficient electron density to accommodate two copies of the PsbO protein (Nield et al. 2002; Nield and Barber 2006). It is possible that one copy of the PsbO protein is lost upon sample preparation for structural analysis.
PsbP
The PsbP protein (23 or 24 kDa protein) was first determined to be a component of PSII in plants (reviewed in Seidler 1996), but homologs have been identified in the genomes across the entire spectrum of photosynthetic organisms from cyanobacteria to plants (Thornton et al. 2004; Table 1). In fact, psbP can even be found in the primitive cyanobacterium Gloeobacter violaceus (referred to hereafter as Gloeobacter), which lacks thylakoids, suggesting an ancient role for this protein in PSII. Genome analysis of Arabidopsis thaliana (referred to hereafter as Arabidopsis) yielded 10 copies of the psbP gene, and remarkably, eight of these were found to be expressed proteins in the thylakoid lumen (Table 1; Peltier et al. 2002; Schubert et al. 2002). Thus, plants must require a variety of PsbP isoforms to fine tune photosynthetic activity. Although psbP genes can be identified in all of the different classes of oxyphototrophs, the majority of studies have been conducted in plants and less is known about PsbP function in other organisms.
PsbP was first identified during release-reconstitution experiments in higher plants (reviewed in Seidler 1996). The PsbP and PsbQ proteins are removed by 1 M NaCl treatment with a concomitant decrease in oxygen evolution (Akerlund et al. 1982; Kuwabara and Murata 1983). Addition of Ca2+and Cl− to the assay medium restores PSII activity without the addition of these proteins (Ghanotakis et al. 1984a; Miyao and Murata 1985). Specifically, PsbP was found to modulate the Ca2+ requirement for PSII activity and has been hypothesized to act as a Ca2+ concentrator and prevent release of Ca2+ during turnover of PSII (Miyao and Murata 1984). Another study has shown that the kinetics of Ca2+ binding are altered in addition to the affinity in the absence of the PsbP and PsbQ proteins (Adelroth et al. 1995). However, PsbP has been implicated along with PsbQ to also modulate the Cl− requirement. Clearly the PsbP protein functions to modulate the ionic requirements for oxygen evolution, but it also plays a structural role to protect the manganese cluster from attack by exogenous reductants (Ghanotakis et al. 1984b).
The first genetic studies on the in vivo role of PsbP, conducted in the green alga Chlamydomonas reaffirmed the correlations between PsbP, Cl−, and activity levels (de Vitry et al. 1989; Rova et al. 1994, 1996). While the FUD39 mutant, which lacks the PsbP protein, accumulates wild type levels of PSII centers, high concentrations of Cl− were necessary to promote oxygen evolution, and there were significant defects in the light-driven assembly of the manganese cluster (termed photoactivation). This inefficient photoactivation process and decreased Cl− affinity resulted in a substantial amount of competing donor side damage. Together, these data suggest a specific role for PsbP and the Cl− ion in the functional assembly of the manganese cluster and stability of PSII.
Advances in RNAi technology have greatly facilitated genetic analysis in plants, especially in cases where the gene of interest is present in multiple copies. Thus, RNAi allowed for the in vivo characterization of PsbP in Nicotiana tabacum (referred to hereafter as Nicotiana) which contains four psbP genes (Ifuku et al. 2005). The PsbP knock-down plants exhibited a reduced variable fluorescence yield, lower oxygen evolution and decreased amounts of PsbQ, which has been shown to require PsbP for binding to PSII in plants. While the stability of the manganese cluster was also affected, it was rapidly reassembled in the light in contrast to the results seen in the Chlamydomonas mutant. However, photoactivation efficiency under low light in the transgenic plants was not extensively characterized. Differential RNAi technology was used to dissect the importance of the each of the different PsbP isoforms in Nicotiana (Ishihara et al. 2005). This study showed that all of the isoforms are required for optimal activity, but generally PSII activity was correlated with the total amount of PsbP protein. Together these results indicate that the PsbP protein is essential for normal in vivo PSII activity in plants because it is critical for OEC stabilization. The precise roles of the various PsbP isoforms remain unclear and they likely confer subtle changes in PSII activity that may be difficult to measure.
Recently, the PsbP protein was identified in PSII preparations from the cyanobacterium Synechocystis (Thornton et al. 2004). Sequence comparison revealed a key difference between transport of the plant and cyanobacterial PsbP proteins. The plant PsbP protein is translocated to the thylakoid lumen via the twin arginine translocation (TAT) pathway (Mould and Robinson 1991; Robinson and Bolhuis 2004) while cyanobacterial PsbP is predicted to be cleaved by signal peptidase II to yield an N-terminal lipid-modified cysteine (predicted by SignalP, (Bendtsen et al. 2004) and LipoP, (Juncker et al. 2003)). Currently, there is disagreement about the abundance of the PsbP protein in the thylakoid membranes of cyanobacteria. Thornton et al. (2004) determined the amount of PsbP to be approximately 3% of that of CP47 in the thylakoid membranes using quantitative immunoblot comparisons of the proteins relative to antigen standards. Using a different antibody, Ishikawa et al. (2005) determined the amount of PsbP in the thylakoid membranes to be equal to that of PsbO, but it is difficult to compare protein amounts from these two different antibodies without antigen standards. However, Ishikawa et al. (2005) do clearly show a lower amount of PsbP in purified PSII complexes relative to PsbO indicating that the protein is lost during the isolation procedure. Due to its N-terminal lipid modification, it is likely that membrane solubilization conditions will greatly influence the amount of PsbP detected in PSII complexes.
Mutational studies in cyanobacteria have yielded somewhat conflicting results as to the function of PsbP. Ishikawa et al. (2005) confirmed cyanobacterial PsbP is indeed associated with PSII and that its translation is highly dependent on the presence of PSII intrinsic components, but they observed no detectable photosynthetic phenotype in a mutant lacking PsbP. In contrast, other groups did observe photoautotrophic growth defects and decreased oxygen evolution in medium lacking either Ca2+ or Cl− for ΔpsbP cells (Thornton et al. 2004; Summerfield et al. 2005a). The ΔpsbP phenotype was not as severe as the phenotype of other cyanobacterial extrinsic protein mutants and differences in sample preparation or assay conditions could explain this phenotypic discrepancy. Summerfield et al. (2005a) also examined double deletion mutants of psbP in combination with each of the other cyanobacterial extrinsic proteins. In most cases, additional inactivation of psbP did not result any exacerbated phenotype. However, an increase in doubling time was observed for the ΔpsbO:ΔpsbP mutant under Cl−-limiting conditions. These results highlight the need for further analysis of PsbP in cyanobacteria to determine whether its function is conserved from cyanobacteria to plants.
The PsbP protein is not present in the current crystallographic models of cyanobacterial PSII (Kamiya and Shen 2003; Ferreira et al. 2004; Loll et al. 2005a). Structural studies of plant PSII complexes do not have the resolution to provide detailed information on the structure of PsbP within the complex, but most likely the position of PsbP is not analogous to any of the other extrinsic subunits observed in the current cyanobacterial models (Nield and Barber 2006). Many studies indicate that the mode of association of PsbP with PSII varies among different organisms. As described above for cyanobacteria, PsbP is predicted to contain an N-terminal lipid anchor, which confers some hydrophobic characteristics (Thornton et al. 2004). In plants, binding of the PsbP protein requires PsbO and is hypothesized to be a largely electrostatic interaction (reviewed in Seidler 1996). In contrast, PsbP from green algae has been shown to bind independently of the other extrinsic proteins (Suzuki et al. 2003). These variations in the binding properties of PsbP highlight structural adaptations for associating with distinct combinations of proteins in the OECs of different organisms. Refer to the sections for the organism-specific OECs for further discussion.
Currently, a high resolution (1.6 Å) crystal structure of PsbP from Nicotiana is available (Ifuku et al. 2004). The core of PsbP is an anti-parallel β-sheet with α-helices on either side; however, electron densities of the N-terminal 15 residues and two loop regions were not resolved. This result may indicate possible stabilizing conformational changes in PsbP upon binding. Because the N-terminus of PsbP is critical for ion retention in PSII (Ifuku and Sato 2001, 2002), no mechanism for its function could be proposed. Further analysis of the protein as a part of the PSII complex will be necessary to elucidate its role in ion retention.
Surprisingly, the structure of PsbP hints at a more exotic role for PsbP in plant PSII. PsbP is very similar to that of Mog1p, a regulatory protein for a Ran GTPase suggesting it may be a possible GTP/GDP-sensitive regulator (Ifuku et al. 2004; De Las Rivas and Roman 2005). While biochemical studies have not previously demonstrated such a role for the PsbP protein, recent studies have shown that GTP/GDP metabolism in the chloroplast thylakoid lumen regulates the turnover of PSII components (Spetea et al. 1999, 2000, 2004). Another study has also implicated the PsbP protein in the assembly of PSII, suggesting it plays a direct role in the light-induced assembly of the manganese cluster (Bondarava et al. 2005). Refer to the section on “PSII Biogenesis and Turnover” for further discussion on possible roles for the extrinsic proteins in this process.
PsbP clearly plays a structural role in the plant OEC to sequester the Ca2+ and Cl− ions and protect the manganese cluster from exogenous reducants, but the exact position of PsbP within the complex and mechanism for this function remains unknown. The studies described above demonstrate that PsbP is essential for normal in vivo PSII activity. The presence of psbP genes in a number of photosynthetic organisms emphasizes the need for more functional analyses of this protein in various organisms to determine whether its function is conserved or whether it has specialized functions in different organisms.
PsbQ
Genes for psbQ have been identified in a number of different photosynthetic organisms (Thornton et al. 2004; Table 1). However, there are some notable exceptions- Gloeobacter as well as the Prochlorococci strains. Arabidopsis contains multiple copies of psbQ genes, four of which have been identified as expressed proteins in the thylakoid lumen (Peltier et al. 2002; Schubert et al. 2002). Because of the apparently random distribution of psbQ genes, it has been hypothesized that PsbQ is the protein most recently incorporated into the OEC (De Las Rivas et al. 2004).
The first analyses of PsbQ were release-reconstitution studies in spinach, which indicate that PsbQ plays a role in modulating the Ca2+ and Cl− requirements for optimal oxygen evolution (Akerlund et al. 1982; Kuwabara and Murata 1983; Ghanotakis et al. 1984a; Miyao and Murata 1985). While PsbP was shown to contribute mainly to the Ca2+ requirement, recent results implicate PsbQ in Ca2+ retention as well (Ifuku and Sato 2002; Barra et al. 2005). PsbQ in concert with PsbP functions to lower the Cl− requirement for optimal activity (Akabori et al. 1984; Ghanotakis et al. 1984a; Miyao and Murata 1985). Along with the PsbO and PsbP proteins, PsbQ plays a structural role within the plant OEC to protect the manganese cluster from inactivation by reductants in the thylakoid lumen (Ghanotakis et al. 1984a).
Recently, genetic studies in plants have investigated the role of the PsbQ protein in vivo. RNAi was used to knock-down PsbQ in Nicotiana (Ifuku et al. 2005). Although transgenic plants exhibited strong, stable gene silencing, they did not display any observable phenotype under the conditions assayed. Another study utilizing RNAi in Arabidopsis found that under normal growth conditions, PsbQ mutant plants were similar to wild type, akin to the results in Nicotiana, but detailed analysis of their photosynthetic machinery indicated the OEC was quite unstable (Yi et al. 2006). Under low light, the mutant plants died after 3–4 weeks, indicating that PsbQ is essential for growth under these conditions. It is possible that in the absence of PsbQ the manganese cluster more readily dissociates from PSII, but cannot be reassembled into the complex efficiently enough to maintain the PSII activity required for survival in low light.
The 20 kDa PSII extrinsic protein in red algae has recently been renamed PsbQ′ because it has low but significant homology to PsbQ from green algae (Ohta et al. 2003). This protein was originally identified in PSII preparations from Cyanidium caldarium and it is released from these complexes along with PsbO, PsbU and PsbV upon treatment with 1 M CaCl2 (Enami et al. 1995, 1998). While PsbQ′ can bind PSII independently, it alone does not enhance oxygen evolution. These results suggest that PsbQ′ is not directly involved in water oxidation in red algae. However, it is important for the association of the PsbV and PsbU proteins (Enami et al. 1998).
PsbQ in cyanobacteria was first identified during a proteomic analysis of PSII (Kashino et al. 2002). Unlike the other cyanobacterial extrinsic proteins, it was not removed by treatment with 1 M CaCl2 or 1 M Tris-HCl, pH 8.0 (Kashino et al. 2002). Sequence analysis of provided an explanation for the hydrophobic nature of cyanobacterial PsbQ; it is predicted to be cleaved by signal peptidase II to yield an N-terminal lipid-modified cysteine (predicted by SignalP, (Bendtsen et al. 2004) and LipoP, (Juncker et al. 2003)). Indeed, a recent study has confirmed the hydrophobic nature of PsbQ but also showed that it is lumenally exposed (Kashino et al. 2006). This is in contrast to the PsbQ homologs from plants and algae, which are easily removed by treatments with high salt.
Data suggests that PsbQ is a stoichiometric component of PSII in cyanobacteria (Thornton et al. 2004). Inactivation of the psbQ gene in cyanobacteria resulted in a mutant with photosynthetic defects under Ca2+- and Cl− -limiting conditions, which is consistent with the biochemical studies of plant PsbQ (Thornton et al. 2004; Summerfield et al. 2005b). However, the phenotype observed for the ΔpsbQ mutant was less severe relative to that of other cyanobacterial extrinsic protein mutants like ΔpsbV or ΔpsbO (Thornton et al. 2004; Summerfield et al. 2005b). Double mutants where psbQ was inactivated in combination with each of the other cyanobacterial extrinsic proteins showed exacerbated photosynthetic defects. In fact, the ΔpsbQ:ΔpsbV mutant could not grow photoautotrophically in nutrient replete medium and assembled low amounts of PSII centers (30% relative to wild type). Interestingly, photoautotrophic growth could be restored when cells were grown at pH 10.0 (vs. pH 7.5), but the mechanism behind this pH-sensitivity remains unknown (Summerfield et al. 2005b).
A more detailed analysis of the PSII complexes in the ΔpsbQ mutant showed a partial loss of the PsbV protein and destabilization of the manganese cluster (Kashino et al. 2006). This finding is consistent with previous results of the PsbQ′ protein from red algae described above. While much of the cyanobacterial ΔpsbQ phenotype could be explained by the loss of the PsbV protein, the results from the ΔpsbQ:ΔpsbV double mutant suggest a synergistic relationship between the cyanobacterial PsbQ and PsbV proteins or perhaps an additional role for PsbQ.
Currently, there is little structural information on the PsbQ protein within the PSII OEC. While it has been found in the genomes of the cyanobacterial strains used for PSII crystallization, it is not present in the current structural models and there is no unassigned electron density that could be attributed to PsbQ (Kamiya and Shen 2003; Ferreira et al. 2004; Loll et al. 2005a). Therefore, it is possible that the PsbQ protein is lost during the purification procedure for crystallography. Analysis of low resolution plant PSII structural reconstructions in comparison to the cyanobacterial models indicate that the position of the PsbQ protein does not correspond to that of any currently resolved cyanobacterial extrinsic proteins (Nield and Barber 2006). Evidence for an independent binding site for PsbQ supports the hypothesis that PSII complexes in some organisms contain PsbQ in addition to PsbO, PsbU, PsbV and even PsbP.
High resolution crystal structures of isolated spinach PsbQ are available and have shed new light on its association with PSII (Calderone et al. 2003; Balsera et al. 2005). PsbQ can be divided into two structural domains; a C-terminal four helix bundle with an asymmetric charge distribution and a flexible N-terminus. Structural features of the PsbQ N-terminal region include two short β-strands surrounding a large flexible loop region (residues 14–33) and a polyproline type II motif (Balsera et al. 2005), which may obtain a more rigid conformation upon binding PSII. Sequence divergence in the N-terminal region may explain the differences in binding characteristics of PsbQ among the different organisms (Balsera et al. 2005). In contrast, the C-terminus region is more conserved and includes a number of conserved positively charged residues which have been implicated as binding determinants for PsbQ in plants (Gao et al. 2005; Meades et al. 2005). Interestingly, the crystal structures of PsbQ show two bound Zn2+ atoms, where the coordinating residues of one atom are entirely conserved in the plant PsbQ proteins (Calderone et al. 2003; Balsera et al. 2005). While Zn2+ was specifically required for crystal growth, the biological significance of this result has yet to be determined.
The cumulative data on PsbQ indicate it is a key structural component of OECs from a number of different organisms, but many questions remain regarding its function and its mode of association with PSII. High resolution structures of PSII complexes containing PsbQ are necessary to provide insights into its role in the ionic requirement for oxygen evolution and its location relative to the other protein components of the OEC. Targeted mutagenesis of the N-terminal structural elements of PsbQ together with reconstitution analysis will unravel the differences in binding requirements among various photosynthetic organisms.
PsbR
PsbR, also known as the 10 kDa PSII polypeptide, is present in plants and green algae and still remains something of a mystery. Though it does contain a lumenal targeting sequence like the other OEC components, it is quite unusual in its primary structure. PsbR contains much shorter transit sequence relative to the other extrinsic proteins and lacks the N-terminal hydrophobic loop thought to be necessary for cleavage (Lautner et al. 1988; Webber et al. 1989). However, PsbR does have a similar motif at the C-terminus. It has been suggested that the shortened N-terminal sequence acts to target PsbR into the chloroplast where it is cleaved off in the stroma and that the C-terminal region acts as a non-cleavable signal for lumen import (Webber et al. 1989). The final protein product is predicted to contain a C-terminal transmembrane span and a lumenally exposed 70 amino acid N-terminus.
The function of PsbR remains unclear. Certainly PsbR is necessary for the optimization of electron transfer and water oxidation. PSII activity is impaired with the loss of PsbR function, as shown by multiple lines of evidence. Release-reconstitution experiments have shown that a concomitant decrease of oxygen evolution with the removal of PsbR, that can be partially restored by the addition of CaCl2 (Ljungberg et al. 1986). Analysis of PsbR mutants showed a similar decrease in oxygen evolution and the decrease in oxygen evolution became more dramatic when the plants were grown under low light (Stockhaus et al. 1990, Suorsa et al. 2006). Together, these studies suggest that PsbR, like many of the other extrinsic proteins, enhances oxygen evolution and may have a role in maintaining a proper ion environment for PSII function. Additionally loss of PsbR results in an increased PSII excitation pressure and a higher PSI:PSII ratio, indicating diminished electron transfer from the plastoquinone pool (Suorsa et al. 2006).
Interestingly, the lack of PsbR results in a specific post transcriptional reduction in PsbQ and PsbP, with nearly undetectable levels under low light conditions, perhaps suggesting the PsbR is important for PsbP docking or that the presence of PsbR is required for stable assembly of PsbP and PsbQ (Suorsa et al. 2006). Also, the absence of PsbR, PsbP, and PsbQ in ΔpsbJ mutant further suggests that these low molecular weight proteins are interdependent for the proper assembly of the PSII complex (Swiatek et al. 2003; Suorsa et al. 2006).
Less is known about PsbR in green algae. PsbR in Chlamydomonas contains a unique N-terminal region which is phosphorylated at Ser43 in State 2 or light induced thylakoids (Turkina et al. 2006). This is the first evidence that PsbR undergoes phosphorylation in any photosynthetic organism. Several proteins of the light harvesting system, as well as D2, were also be phosphorylated in these conditions in Chlamydomonas. Interestingly phosphorylation of the core proteins under light stress is important for PSII repair in higher plants. In PsbR mutant plants, there is a decrease in phosphorylation of LHCII, D1, D2, and CP43 (Turkina et al. 2006). This suggests a possible role for PsbR in PSII repair.
PsbR enhances oxygen evolution and might play a role in PSII repair. Additionally, PsbR, PsbJ, PsbQ, and PsbP may interact to form a stable PSII. However, the nature of the interaction between PsbR and PSII remains unknown. The stoichiometry is unclear in both plants and green algae. Additionally, because the current structures available for PSII lack the PsbR protein, they are not helpful in approximating its location. Based on the presence of the leader sequence and experimental Tris washes, PsbR is predicted to be in the lumen (Ljungberg et al. 1984a; Harper et al. 1998). Moreover, shown associations of PsbR with CP47, PsbO, and PsbP support this localization (Ljungberg et al. 1984b; Harper et al. 1998).
PsbU
PsbU is found in most cyanobacteria and red algae (Table 1) and was formerly referred to as the 12 kDa and 9 kDa polypeptide in these organisms, respectively (Stewart et al. 1985). Cyanobacterial PsbU has a single N-terminal loop that forms a cleavable transit sequence while algal PsbU has a two-part transit sequence to allow transport across the chloroplast envelope and into the lumen (Shen et al. 1997; Ohta et al. 1999).
The function of PsbU is generally assigned as enhancing the structural stability of PSII and shielding the manganese cluster. Indeed, the removal of PsbU affects the lumenal side, the core, and the stromal side of PSII. The donor side of PSII is impaired in the absence of PsbU as shown by decreases in photosynthetic efficiency (Inoue-Kashino et al. 2005). Release-reconstitution experiments originally identified PsbU by the reduction in oxygen evolution associated with its removal (Shen et al. 1997). This decrease in activity can be partially restored by the addition of Cl− and is unaltered by additional Ca2+ (Shen et al. 1997; Inoue-Kashino et al. 2005). This suggests that PsbU, along with PsbP and PsbQ, modulates the Cl− requirement. The higher light intensity needed for PSII efficiency in ΔpsbU and decreased maximal rate of photosynthesis support that there is a defect in forward electron flow in the absence of PsbU (Balint et al. 2006). Additionally, Q −A reoxidation and thermoluminescence studies show slowed kinetics and temperature shifts for ΔpsbU only in the presence of the herbicide DCMU (3-(3′,4′-dichlorphenyl)-1,1-dimethylurea) indicating that recombination between Q −A and the S2-state is also impaired (Shen et al. 1997; Inoue-Kashino et al. 2005; Balint et al. 2006).
Interestingly, even the stromal surface of the PSII complex is affected by the loss of PsbU, as evidenced by the increased uncoupling of the light harvesting machinery, the phycobilisome, and PSII (Veerman et al. 2005). This indicates that the binding of PsbU induces structural changes to PSII, perhaps promoting the binding of the phycobilisome.
PsbU enhances the overall stability of PSII in several ways. PsbU specifically stabilizes the ion environment for oxygen evolution as shown by decreased growth of cyanobacterial and red algal PsbU mutants in medium lacking Ca2+ and or Cl− (Shen et al. 1997; Ohta et al. 1999; Inoue-Kashino et al. 2005). Additionally, the loss of PsbU results in a PSII core more susceptible to photodamage, resulting in rapid degradation of D1 (Inoue-Kashino et al. 2005; Balint et al. 2006). Interestingly, the presence of PsbU also protects PSII from dark inactivation (Shen et al. 1998; Veerman et al. 2005). PsbU also contributes to the thermal stability of the OEC, as PsbU mutants in both thermophilic and mesophilic cyanobacteria do not have the ability to acclimate to higher temperatures and exogenously applied PsbU enhances thermostability (Nishiyama et al. 1997, 1999). Lastly, Balint et al. (2006) have also proposed that PsbU provides protection from reactive oxygen species, as PsbU mutants have enhanced mechanisms to detoxify exogenously applied H2O2.
PsbU mutants have altered electron flow in both the forward and reverse directions. Additionally, PsbU serves to stabilize PSII against heat, photodamage, dark inactivation, reactive oxygen species, and in low ionic solutions. However, the specific mechanisms for these actions of PsbU remain unknown. In light of the variety of effects PsbU has on cyanobacterial and red algal PSII, it is unclear why plant and green algal PSII no longer retain PsbU.
PsbV
PsbV, also referred to as cytochrome c 550 , is found in cyanobacteria and red algae (Table 1). Additionally, its homology to other c-type cytochromes makes it the only extrinsic protein with similarity to anoxygenic and non-photosynthetic bacterial proteins (Raymond and Blankenship 2004). Like the other lumenal proteins, it contains a localization signal that is similar to that of PsbO and PsbU in cyanobacteria (Shen et al. 1995). Genome sequence of the red algae Cyanidioschyzon merolae shows that psbV (as well as psbO) are both nuclear-encoded but have rather different targeting signals with only 15% identity (Matsuzaki et al. 2004).
The function of PsbV, like that of PsbU, is regarded as stabilizing PSII structure and electron transfer. In cyanobacteria, PsbV can be removed by treatment with high salt buffers, resulting in a decrease of oxygen evolution (Shen and Inoue 1993).
The cyanobacterial PsbV mutant also has a severe growth phenotype. Under normal conditions growth of the PsbV mutant is severely retarded and the absence of Ca2+ or Cl− in the growth media eliminates the capacity for growth, suggesting that PsbV aids in maintaining the proper ion environment within the OEC (Shen and Inoue 1993; Shen et al. 1995, 1998). Suppressor screens using ΔPsbV strains in the absence of Cl− have shown that disruption of the ion transporter encoded in Synechocystis by slr0753 reestablishes photoautotrophic growth (Kobayashi et al. 2006).
The PsbV mutant specifically has defects in the catalytic cycle of water oxidation (Shen et al. 1998; Kimura et al. 2002). Loss of PsbV results in a decrease in oxygen evolution rates (Kimura et al. 2002). Additionally, thermoluminescence studies show that the PsbV mutant has either a lower efficiency of s-state transition or fewer reaction centers capable of s-state transition (Shen et al. 1995). Also, the overall stability of PSII is reduced in the PsbV mutant, as the PsbV mutant has fewer assembled PSII centers (Shen et al. 1995). Dark treatment reduces photosynthetic capacity dramatically in the PsbV mutant, suggesting increased dark inactivation (Shen et al. 1998). Like PsbU, PsbV has also been shown to enhance thermostability (Nishiyama et al. 1994). Thus, PsbV functions in several capacities to protect and stabilize the OEC and the manganese cluster.
The discovery of a second expressed copy of PsbV (∼44% identity) in several cyanobacteria raises additional questions as to the function of PsbV. PsbV2, like PsbV, exhibits the spectral properties of a six-coordinate, low-spin c-type cytochrome (Kerfeld et al. 2003). PsbV2 can functionally rescue a ΔpsbV mutant in Synechocystis with the exception of reduced growth in the absence of Ca2+ or Cl− (Katoh et al. 2001). The role of this second PsbV, found only in a small subset of cyanobacteria, remains unclear.
The PsbV protein is also associated with PSII complexes in red algae, where it also plays a role in the ionic requirement for oxygen evolution (Enami et al. 1998). Less is known about the protein architecture of the red algal OEC, but studies suggest the binding properties of algal PsbV differ from that of cyanobacteria (Enami et al. 1998, 2003). Refer to the “Red Algal OEC” section for further details.
In addition to its typical roles as a protein component of the OEC, PsbV is a particularly interesting cytochrome. It is a water-soluble c-type monoheme cytochrome, with a very low reduction potential (−240 mV) (Pettigrew and Moore 1987; Krogmann and Smith 1990). The general structure of the heme environment does not account for this difference, as EPR and resonance Raman spectroscopy showed that the heme has a bishistidine ligation that is similar to other c-type cytochromes (Vrettos et al. 2001). Three factors may partially explain the low potential. One of these is solvent exposure of the heme, which generally reduces the potential of cytochromes (Tezcan et al. 1998). Indeed, Roncel et al. (2003) have shown that PsbV bound to PSII has a higher potential (−80 mV), in agreement with studies by Vrettos et al. (2001). Additionally, the presence of ionizable residues may affect the reduction potential. This ionizable group must be in the vicinity of the heme and has been proposed to be either a tyrosine or asparigine residue (Roncel et al. 2003; Ishikita and Knapp 2005). Lastly, the bishistidine heme ligation typically has a lower potential than the other two possible ligands, lysine or methionine. Alteration of PsbV the histidine ligands resulted in a small increase in potential without affecting PSII activity (Kirilovsky et al. 2004; Li et al. 2004; Andrews et al. 2005). Interestingly, the mutation of His-92 to methionine does cause some destabilization of the PSII complex, as evidenced by easier detachment of PsbV, a heightened dark inactivation response, and a decreased in functional PSII centers (Kirilovsky et al. 2004; Li et al. 2004). Taken together these factors begin to explain, but cannot fully account for the low reduction potential of PsbV. Furthermore, its exact function is unclear as mutations that increase the reduction potential of PsbV do not exhibit PSII defects (Kirilovsky et al. 2004; Andrews et al. 2005).
In light of the fact that the heme of PsbV does not play a role in PSII electron transfer and given that plant and green algal OECs do not contain any cytochrome equivalents, it has been hypothesized that PsbV plays another, as-yet uncharacterized role in cyanobacteria and red algae. It has been proposed that PsbV may be involved in anaerobic removal of electrons from carbohydrate reserves or fermentation for increased survival during long dark and anaerobic conditions (Krogmann and Smith 1990; Krogmann 1991). This is supported by experimental evidence from Shen and Inoue (1993) that PsbV, in the presence of dithionite, can accept electrons from ferredoxin II. It has also been proposed that PsbV may accept electrons from ferredoxin during NADPH oxidation and in cyclic phosphorylation.
PsbV plays an important role in the OEC and perhaps has an additional uncharacterized function. Loss of PsbV results in decreased oxygen evolution and in decreased stability of PSII. It has been theorized that PsbV has another role in cyanobacteria and red algae. The discovery of PsbV2, which only partially complements the loss of PsbV, has raised questions as to the necessity of more than one PsbV protein. Additionally, the unusually low potential of PsbV, especially in light of the fact that plants and green algae do not require an extrinsic cytochrome at all, raises questions as to the function of the cytochrome. It is possible that PsbV is also electron acceptor to remove excess electrons under conditions where photosynthesis is not maximal.
Psb27
The Psb27 subunit was first identified as part of a purified PSII preparation from Synechocystis 6803 (Ikeuchi et al. 1995). This 11 kDa protein was first named PsbZ; however, according to a new nomenclature was changed to Psb27 (Kashino et al. 2002). A distinctly different smaller protein is now referred to as PsbZ (Swiatek et al. 2001; Inoue-Kashino et al. 2005). Homologs of psb27 are present in all oxygenic photosynthetic organisms except Gloeobacter, which lacks a separate thylakoid membrane system (Table 1).
This small basic (pI = 9) protein is predicted by to be targeted to the thylakoid lumen in Synechocystis 6803 and cleaved by signal peptidase II to yield an N-terminal lipid modification [predicted by SignalP (Bendtsen et al. 2004) and LipoP (Juncker et al. 2003)]. Indeed, Psb27 was not removed by washes that typically deplete cyanobacterial PSII of its extrinsic subunits (Kashino et al. 2002). One of the two Arabidopsis Psb27 homologs (At1g03600) was found in a proteomic analysis of the thylakoid lumen (Peltier et al. 2002; Schubert et al. 2002). In those studies, Psb27 was predicted to be targeted to the thylakoid lumen via the TAT pathway suggesting a possible difference in its interaction with PSII in higher plants. Chen et al. (2006) reported that the other Arabidopsis homologue (At1g05835) is expressed at low levels. However, sequence analysis revealed this copy is predicted to be chloroplast-localized but it lacks the lumenal transit peptide for transport across the thylakoid membrane (Chen et al. 2006).
Psb27 is not present in the recent PSII crystal structures (Kamiya and Shen 2003; Ferreira et al. 2004; Loll et al. 2005a). The Psb27 protein is hypothesized to function in the PSII biogenesis because it was found to accumulate on mutant PSII complexes arrested early in the PSII assembly pathway (Roose and Pakrasi 2004). Indeed, a recent study of a psb27 loss of function mutant in Arabidopsis showed that while Psb27 is not essential for PSII activity, it is involved in PSII recovery after photoinhibition (Chen et al. 2006). Refer to the “PSII Biogenesis and Turnover” section for further discussion on Psb27.
OEC systems in different organisms
Cyanobacterial OEC
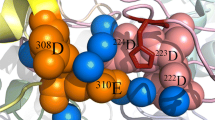
The cyanobacterial OEC contains 5 lumenal proteins: PsbO, PsbU, PsbV, PsbP, and PsbQ. In regards to PsbO, PsbV, and PsbU, it represents the best understood OEC, due to numerous release-reconstitution experiments and crystal structures (Zouni et al. 2001; Kamiya and Shen 2003; Ferreira et al. 2004; Loll et al. 2005a). PsbP and PsbQ remain more ambiguous as they are not present in the crystal structures and there are fewer reconstitution experiments due to their relatively recent discovery in cyanobacteria. Figure 1A represents a cartoon model of the cyanobacterial OEC, which incorporates the known components as well as PsbP and PsbQ.
Models for the OECs in different organisms. Based on the current structural, biochemical and genetic data discussed in the manuscript, the structures of the OECs in cyanobacteria (A), red algae (B), green algae (C), and plants (D) are modeled in cartoon form. The intrinsic components D1, D2, CP43 and CP47 are labeled and the large lumenal portions of CP43 and CP47 are shown as solid loops. The manganese cluster is also represented by small dots on the lumenal side of the D1 protein. The extrinsic proteins are labeled as O (PsbO), P (PsbP), Q (PsbQ), R (PsbR), U (PsbU), and V (PsbV). The cyanobacterial OEC (A) shows the presence of five extrinsic proteins, but PsbP may not be a stoichiometric component of these complexes. In the green algal (C) and plant (D) OEC models, the PsbR protein is positioned behind the PsbP and PsbQ proteins such that it is closer to the intrinsic PsbJ protein beyond the CP43 and D2 proteins (see text). These models are designed to aid the reader in conceptualizing the relative locations of the indicated subunits among the different organisms, but by no means replace the more detailed structural analyses discussed in the manuscript. Refer to the text for more detailed discussions about the individual proteins or organismal OECs
The binding order of PsbO, PsbU, and PsbV has been well established. It has long been known that PsbO can rebind to extrinsic protein-depleted PSII in the absence of any other proteins. PsbV can also bind independently, but is stabilized by the presence of PsbO and PsbU (Shen and Inoue 1993). PsbU cannot rebind at all in the absence of either PsbO or PsbV (Shen and Inoue 1993), but can bind with the addition of PsbV (Shen et al. 1997; Eaton-Rye et al. 2003). Thus, it is thought that PsbO binds first, followed by PsbV, and that PsbU binds third.
Recent crystal structures have provided specific information on the interactions among PsbO, PsbU and PsbV. PsbO is a β-barrel consisting of 8 anti-parallel β-strands with a loop between strands 5 and 6 that is involved with binding PsbO to PSII (De Las Rivas and Barber 2004; Ferreira et al. 2004). PsbO is positioned over the CP47 side of the reaction center (Zouni et al. 2001). This is in agreement with experiments showing that deletions in the E loops of CP47 combined with the deletion of PsbO abolished photoautotrophic growth (Morgan et al. 1998; Clarke and Eaton-Rye 1999), and site directed mutagenesis studies that show a binding domain for PsbO at CP47 Arg384 and Arg385 (Putnam-Evans and Bricker 1992; Putnam-Evans et al. 1996; Qian et al. 1997). Additionally, PsbO stabilizes the AB loop and C-terminus of D1 (the location of many of the manganese cluster ligands) and interacts with the large E loop of CP47 (Ferreira et al. 2004; Nield and Barber 2006).
PsbV is mainly alpha helical with a two-stranded beta sheet near the N-terminus (Kerfeld et al. 2003). PsbV is located over the CP43 side of the reaction center with extensive contacts with the E loop of CP43 (Zouni et al. 2001; Kamiya and Shen 2003; Ferreira et al. 2004; Loll et al. 2005a). Deletions in the E loop of CP43 result in a loss of photoautotrophic growth and PSII activity, suggesting that CP43 stabilizes the OEC (Kuhn and Vermaas 1993). The specific mutation R305S in CP43 prevents strong association of PsbV with PSII, although the amount of PsbV is unaffected on a cellular basis (Bricker et al. 2002).
PsbU is composed of five or more short α-helices, with no homology to other structures in the database (Kamiya and Shen 2003). The crystal structures place PsbU between PsbO and PsbV with a majority of its contacts to these two proteins (Kamiya and Shen 2003; Ferreira et al. 2004; Loll et al. 2005a). While there is a significant amount of distance between PsbU and the membrane, it interacts with PsbV, PsbO, CP47 and CP43 (Eaton-Rye 2005). Combinatorial mutants of the CP47 E loop and ΔpsbU affect PSII stability, not growth, whereas the combination of the CP47 E loop deletions and ΔpsbV abolish photoautotrophic growth. In light of crystal structure data, the latter OEC mutant must be severely destabilized due to reduced binding of PsbU and loss of PsbV (Morgan et al. 1998; Clarke and Eaton-Rye 1999).
PsbP and PsbQ proteins have recently been identified as cyanobacterial PSII components (Kashino et al. 2002; Thornton et al. 2004). Their binding properties are significantly different from that of PsbP and PsbQ in other systems, in that, they are not easily removed by salt-washing treatments (Kashino et al. 2002). Cyanobacterial PsbP and PsbQ are predicted to contain an N-terminal lipid-modified cysteine (see above discussion of PsbP and PsbQ). While the precise lipid moiety has not been identified, a recent study demonstrated that cyanobacterial PsbQ has hydrophobic characteristics, but is still highly exposed on the lumenal face of the thylakoid membrane (Kashino et al. 2002).
The PsbQ protein is hypothesized to be a stoichiometric component of PSII complexes in cyanobacteria, but there is some debate over the stoichiometry of PsbP (Thornton et al. 2004; Ishikawa et al. 2005). One recent study indicates that the absence of PsbQ destabilizes PsbV, suggesting a location near PsbV (Enami et al. 1998; Kashino et al. 2006). Less is known about the possible location of PsbP. With the prediction of the N-terminal lipid anchor and no other constraints from reconstitution or genetic studies, the cyanobacterial PsbP protein could bind anywhere on the lumenal side of the complex in cyanobacteria.
Despite the wealth of experimental information available for the cyanobacterial OEC, there are still many unanswered questions. It remains unclear what the role of the PsbV cytochrome is, especially since none if the extrinsic proteins in the plant OEC are cytochromes. Resolving the locations and stoichiometries of PsbP and PsbQ in cyanobacterial PSII will be a topic of intense future research. And although this portrayal of the cyanobacterial OEC was based on that of Synechocystis and Thermosynechococcus, there are other cyanobacterial species that contain variations on this theme. Most interesting are Gloeobacter and certain Prochlorococci strains such as MED4 and SS120. The genome sequences of these species do not contain psbU or psbV, raising interesting questions as to the structures and efficacy of these unusual PSIIs.
Red algal OEC
Red algae have an OEC comprised of PsbO, PsbU, PsbV, and PsbQ′. Currently there are only low resolution electron microscopy models and a handful of release-reconstitution studies to aid in defining the OEC structure in red algae. Figure 1B illustrates the model of red algal OEC.
Electron microscopy and single particle analysis of the Porphyridium cruentum revealed that the location of PsbO, PsbV and PsbU in red algae is similar to that observed in the structures of cyanobacteria (Kamiya and Shen 2003; Bumba et al. 2004; Ferreira et al. 2004; Loll et al. 2005a). PsbQ′ was not seen in this structure and its location relative to the other subunits could not be determined (Bumba et al. 2004). While the protein content of cyanobacterial and red algal OECs are similar, there are likely differences between these types OECs because of the differences in their extrinsic protein binding patterns. As in cyanobacteria, PsbO can bind to PSII independently (Enami et al. 1998). PsbQ′ can also partially bind independently, but with no restorative effect on oxygen evolution (Enami et al. 1998). In contrast to cyanobacteria, red algal PsbV and PsbU require the presence of all four of the extrinsic proteins for association (Enami et al. 1998). There must be extensive interactions among all four extrinsic proteins in red algae as all are needed for full binding and recovery of activity (Enami et al. 1998). Notably, cross-reconstitution experiments with red algal and cyanobacterial components indicated that the binding of the extrinsic proteins is determined by the intrinsic proteins (Enami et al. 2003).
Red algae are considered to represent a transitional state between cyanobacterial and photosynthetic eukaryotes. They have thylakoids more similar to those of cyanobacteria, rather than the stacked thylakoids of green algae and plants. Thus, it is intriguing that red algae lack PsbP, which is found in cyanobacteria, green algae, and plants. A higher resolution structure of the red algal OEC would aid in understanding the similarities of the red algal OEC to that of cyanobacteria, green algae and plants.
Green algal OEC
The protein content of the green algal OEC is similar to that of plants (PsbO, PsbP, PsbQ, and PsbR), but there are significant differences in the way these proteins interact with the PSII complex and each other. Currently, there is only one low resolution structural study of the OEC from green algae (Nield et al. 2000); however, many biochemical and genetic studies in Chlamydomonas have been useful in characterizing the interactions among the OEC proteins. Based on these studies, Fig. 1C shows a cartoon structure of the OEC in green algae.
Release-reconstitution studies of Chlamydomonas PSII complexes have shown all three algal proteins can bind to PSII independently of each other (Suzuki et al. 2003). Upon addition of PsbO, activity was only partially restored and activity was highly dependent on the addition of Ca2+ and Cl− ions. The addition of PsbP alone or in combination with PsbQ also restored activity in contrast to observations for plant PSII. While the PsbQ protein was able to bind independently, it alone does not restore any level of oxygen evolution.
These findings are consistent with previous genetic studies of a Chlamydomonas mutant lacking the PsbO protein, in which PsbP and PsbQ proteins still bind to PSII (de Vitry et al. 1989). However, PsbQ could not associate with PSII complexes in a mutant lacking the PsbP protein (de Vitry et al. 1989). Therefore, while in vitro conditions indicate that it is possible for PsbQ to associate independently of the other extrinsic proteins, this interaction does not occur in vivo.
Cross-reconstitution studies using plant and green algal components have provided new insights into the functional exchangeability of the extrinsic proteins (Suzuki et al. 2005). Algal PsbP and PsbQ proteins could not bind independently to spinach PSII, and spinach PsbP and PsbQ only bound to non-specific sites on algal PSII. Like the cross-reconstitution studies between cyanobacteria and red algae, these results suggest that the determinants for independent binding reside on the intrinsic PSII components. They also highlight that the majority of the PsbQ binding sites reside on the PsbO and PsbP proteins from the same organism.
The green algal OEC is quite similar to that of plants, but further structural studies will be useful in defining the details within the algal OEC that confer altered binding properties to its extrinsic proteins.
Plant OEC
The protein components of the plant OEC include PsbO, PsbP, PsbQ and PsbR. Currently only low resolution electron microscopy structural models are available for plant PSII (Nield et al. 2002; Bumba and Vacha 2003; Nield and Barber 2006). Yet, together with detailed reconstitution studies and high resolution crystal structures of some of the individual subunits, a clearer picture of the OEC is emerging. Figure 1D is a cartoon model of the plant OEC.
In contrast to many of the other systems, the plant extrinsic proteins appear to have a strict binding order. While the PsbO protein can bind to PSII independently, PsbP requires the presence of the PsbO protein and PsbQ requires the presence of both PsbO and PsbP (Miyao and Murata 1983a, 1983b). One recent study challenges this model, as treatment with HgCl2 could effectively remove the PsbO protein leaving the PsbP and PsbQ proteins intact (Yu et al. 2006). It is possible that these proteins have more extensive interactions with the intrinsic components than originally predicted. Alternatively, conformational changes in the intrinsic components upon PsbO binding could allow for the association of PsbP and PsbQ, and it is possible that treatment with HgCl2 removes PsbO without affecting the conformational state of PSII.
The PsbP and PsbQ proteins can be removed by washing with 1 M NaCl, suggesting a strong electrostatic component for their association with the complex. A number of studies have investigated the roles of certain types of residues using chemically modified proteins in reconstitution experiments. Negatively charged carboxylate groups on the PsbO have been shown to be necessary for the binding of PsbP (Bricker and Frankel 2003) and positively charged groups on PsbP were essential for its association with PsbO (Tohri et al. 2004). Positively charged groups on the PsbQ protein have also been shown to be critical for its association with PsbP (Gao et al. 2005; Meades et al. 2005). The structure of PsbP revealed an asymmetric surface charge distribution which can explain the chemical modification data (Ifuku et al. 2004). The positive face of the protein is predicted to interact with the negative PsbO surface, while the negative side of PsbP interacts with the PsbQ protein. Interestingly, the N-termini of each of these proteins are critical for binding (Kuwabara et al. 1986; Eaton-Rye and Murata 1989; Ifuku and Sato 2001, 2002).
Comparisons of the plant PSII structural data to that of cyanobacteria indicate that the location of the PsbO protein is similar between the two systems. On the other hand, the locations of the PsbU and PsbV proteins do not correspond to the density assigned to PsbP and PsbQ (Nield and Barber 2006). The individual X-ray structures for PsbP and PsbQ were fit into the electron density observed for the plant PSII complex along with one copy of PsbO per PSII monomer. According to this fit, the PsbP protein interacts with the PsbO subunit as well as the lumenal face of the CP43 protein, and PsbQ spans between the PsbO and PsbP proteins. While this type of analysis can stimulate new hypotheses regarding the structure of the OEC in plants, higher resolution structures with assignments for all of the extrinsic proteins are necessary to determine how each of these proteins contributes to the water oxidation reaction.
A number of studies have indicated significant conformational changes occur upon the binding and release of the extrinsic proteins. Investigations of the extrinsic proteins individually have demonstrated they have highly flexible domains in solution, which are likely to be stabilized upon binding to the complex (Calderone et al. 2003; Ifuku et al. 2004; Balsera et al. 2005). The intrinsic core components of PSII are also affected by the extrinsic proteins. Specifically, removal of PsbP and PsbQ affect the peripheral antenna proteins and further removal of the PsbO protein also destabilizes the dimeric structure of PSII. These linked conformational changes may be significant for the assembly and disassembly of the PSII complex.
Currently, little is known about the structure of the PsbR protein, as it is unaccounted for in the PSII complexes used for structural studies and there is no structure of the purified protein. The PsbR protein is considered an extrinsic subunit, but there is also evidence that the C-terminus of the protein is hydrophobic. Biochemical analysis has indicated the PsbR protein is in close proximity to the PsbO, PsbP and CP47 proteins. Genetic analyses also implicated a role for PsbR in the stable association of the PsbP and PsbQ proteins (Suorsa et al. 2006). Mutational studies have, in turn, shown that the intrinsic PsbJ subunit is necessary for the association of PsbR. Analysis of PSII assembly intermediates suggest that PsbR can bind independently of the other extrinsic subunits as it is found in CP43-less monomers which lack these proteins (Rokka et al. 2005). Given the current PSII structural models and the constraints outlined above, it is difficult to assign a location for PsbR. Clearly, the experimental evidence suggests a location in the vicinity of PsbO, PsbP and CP47, but the requirement for the PsbJ protein cannot be easily resolved given that it is closer to the CP43-side of the complex. Additional mutational and biochemical studies should further elucidate the position of PsbR relative to the other plant OEC components.
A number of different techniques have contributed to our understanding of the structure of the plant OEC, and advances in plant genetic analysis and structural methods will be key tools for resolving certain details. Obviously, the interactions among the different extrinsic subunits and the PSII core need to be more explicitly defined and higher resolution structures of plant PSII complexes, which retain the extrinsic subunits, are required. Also, plants have a number of different isoforms for each extrinsic protein, but it is not clear why so many isoforms are necessary or in what context they bind to PSII complexes. This issue will likely be a challenge for future structural studies as well as biochemical analyses.
Photosystem II biogenesis and turnover
PSII requires precise and regulated assembly to ensure the proper positioning of all the essential redox active cofactors. Studies have shown that the assembly pathway is clearly an ordered step-wise association of the PSII subunits. Furthermore, PSII assembly is a frequent process; as a consequence of normal PSII activity, the core D1 protein is irreversibly damaged and must be replaced with a newly synthesized copy (Baena-Gonzalez and Aro 2002; Aro et al. 2005). Defining the details of this cycle is the focus of intense research (Rokka et al. 2005). In this section, the roles of some of the extrinsic proteins in the dynamic PSII life cycle will be discussed.
In addition to its role as a structural component of the plant OEC, PsbP may play a more direct role in the light-driven assembly of the manganese cluster. Upon release of the three extrinsic proteins, PsbP sequesters manganese ions, and photoactivation assays showed that the manganese-containing PsbP protein specifically facilitated manganese cluster assembly and restoration of oxygen evolution. While these results agree with previous characterization of the PsbP-deficient mutant in Chlamydomonas, no defects in photoactivation were observed in the Nicotiana PsbP-RNAi plants (Ifuku et al. 2005). Additional studies are necessary to reconcile the in vitro and in vivo data.
Little is known about the function of the Psb27 protein, but data suggest it may play a role in PSII assembly. While it is a component of cyanobacterial PSII complexes isolated using a histidine-tagged CP47 protein, it was shown to be more abundant on PSII complexes in which the precursor D1 protein did not undergo the necessary C-terminal cleavage event to yield the mature functional D1 protein (Roose and Pakrasi 2004). This population of PSII complexes lacks the manganese cluster and the PsbO, PsbQ, PsbU, and PsbV extrinsic proteins. These findings suggest that Psb27 associates with PSII complexes at an early step in assembly and may exclude the binding of the other extrinsic proteins. It is not clear whether the Psb27 protein plays a role in the pD1-processing event or serves another function in the biogenesis pathway. A recent study of a psb27 loss of function mutant in Arabidopsis also supports the hypothesis that Psb27 has an important role in the repair of half the PSII life cycle (Chen et al. 2006). The mutant plants did not show any obvious phenotype under normal lab conditions, but they were significantly impaired in their recovery after light-induced damage. Together, these data indicate that Psb27 only transiently associates with PSII during assembly, but is not part of the functional complex. Additional experiments are necessary to determine its exact role in the repair pathway.
In addition to addressing the forward pathway of assembly, it is also necessary to consider the disassembly steps required for the removal of the damaged D1 protein. The current PSII structural models position the D1 protein in the center of the PSII core with numerous interactions with the manganese cluster and extrinsic proteins (Ferreira et al. 2004; Loll et al. 2005a). Thus, the OEC must be disassembled to remove the damaged D1 protein. Notably, the interaction of the extrinsic proteins with PSII is affected by the presence of the assembled manganese cluster (Kavelaki and Ghanotakis 1991). Furthermore, the release of the extrinsic proteins results in significant changes in the intrinsic components and destabilizes the dimeric form of PSII (Boekema et al. 2000). Consequently, release of the manganese cluster would ensure an efficient disassembly of the entire OEC with additional effects on the core components. Interestingly, under normal conditions free extrinsic proteins in the thylakoid lumen are not targeted for degradation, which could facilitate reassembly.
Experiments have identified a GTP requirement for the primary proteolytic cleavage of the damaged D1 protein. It has been reported that the primary cleavage event occurs in isolated PSII complexes, suggesting that perhaps a PSII subunit or co-purifying protein is responsible (Salter et al. 1992; De Las Rivas et al. 1993). Subsequent reports have shown that the PsbO protein can bind GTP indicating this ubiquitous extrinsic protein may be involved in regulating D1 degradation. The current models of PSII do not provide any additional evidence for this hypothesis. Interestingly, recent analysis of the PsbP protein indicates it is structurally similar to Mog1p, a regulator of Ran-GTPase in yeast (Ifuku et al. 2004). This result presents yet another possible role of the PsbP protein in the PSII life cycle. More experiments are necessary to determine whether the PSII extrinsic proteins play a more active role in the turnover of the D1 protein.
Conclusion
The unique reaction of water oxidation is similar among all photosynthetic organisms. Yet the protein complement of the OEC varies significantly. Future studies should focus on assigning functions to the individual extrinsic proteins in different organisms. Also, higher resolution structures of PSII complexes from a variety of sources are necessary to provide a complete picture of the various OECs as a whole. However, a static structure cannot tell the entire story of PSII, as it is a dynamic complex undergoing constant assembly and degradation. Future studies should also work toward increasing our understanding of PSII as a dynamic enzyme.
Abbreviations
- PSII:
-
Photosystem II
- OEC:
-
Oxygen evolving complex
- EPR:
-
Electron paramagnetic resonance
References
Adelroth P, Lindberg K, Andreasson LE (1995) Studies of Ca2+ binding in spinach photosystem II using 45Ca2+. Biochemistry 34:9021–9027
Akabori K, Imaoka A, Toyoshima Y (1984) The role of lipids and 17-kDa protein in enhancing the recovery of O2 evolution in cholate-treated thylakoid membranes. FEBS Lett 173:36–40
Akerlund HE, Jansson C, Andersson B (1982) Reconstitution of photosynthetic water splitting in inside-out thylakoid vesicles and identification of a participating polypeptide. Biochim Biophys Acta 681:1–10
Andrews H, Li Z, Altuve-Blanco A, Rivera M, Burnap RL (2005) Expression, mutagenesis, and characterization of recombinant low-potential cytochrome c550 of photosystem II. Biochemistry 44:6092–6100
Aro EM, Suorsa M, Rokka A, Allahverdiyeva Y, Paakkarinen V, Saleem A, Battchikova N, Rintamaki E (2005) Dynamics of photosystem II: a proteomic approach to thylakoid protein complexes. J Exp Bot 56:347–356
Baena-Gonzalez E, Aro EM (2002) Biogenesis, assembly and turnover of photosystem II units. Philos Trans R Soc Lond B Biol Sci 357:1451–1459; discussion 1459–1460
Balint I, Bhattacharya J, Perelman A, Schatz D, Moskovitz Y, Keren N, Schwarz R (2006) Inactivation of the extrinsic subunit of photosystem II, PsbU, in Synechococcus PCC 7942 results in elevated resistance to oxidative stress. FEBS Lett 580:2117–2122
Balsera M, Arellano JB, Revuelta JL, de las Rivas J, Hermoso JA (2005) The 1.49 A resolution crystal structure of PsbQ from photosystem II of Spinacia oleracea reveals a PPII structure in the N-terminal region. J Mol Biol 350:1051–1060
Barra M, Haumann M, Dau H (2005) Specific loss of the extrinsic 18 KDa protein from photosystem II upon heating to 47 degrees C causes inactivation of oxygen evolution likely due to Ca release from the Mn-complex. Photosynth Res 84:231–237
Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides:SignalP 3.0. J Mol Biol 340:783–795
Boekema EJ, van Breemen JF, van Roon H, Dekker JP (2000) Conformational changes in photosystem II supercomplexes upon removal of extrinsic subunits. Biochemistry 39:12907–12915
Bondarava N, Beyer P, Krieger-Liszkay A (2005) Function of the 23 kDa extrinsic protein of photosystem II as a manganese binding protein and its role in photoactivation. Biochim Biophys Acta 1708:63–70
Bricker TM, Frankel LK (2003) Carboxylate groups on the manganese-stabilizing protein are required for efficient binding of the 24 kDa extrinsic protein to photosystem II. Biochemistry 42:2056–2061
Bricker TM, Burnap RL (2005) The extrinsic proteins of photosystem II In: Wydrzynski T, Satoh K (eds). photosystem II: the light-driven water:plastoquinone oxidoreductase, Dordrecht: Springer, pp 95–120
Bricker TM, Young A, Frankel LK, Putnam-Evans C (2002) Introduction of the 305Arg–>305Ser mutation in the large extrinsic loop E of the CP43 protein of Synechocystis sp. PCC 6803 leads to the loss of cytochrome c(550) binding to photosystem II. Biochim Biophys Acta 1556:92–96
Bumba L, Vacha FE (2003) Electron microscopy in structural studies of photosystem II. Photosynth Res 77:1–19
Bumba L, Havelkova-Dousova H, Husak M, Vacha F (2004) Structural characterization of photosystem II complex from red alga Porphyridium cruentum retaining extrinsic subunits of the oxygen-evolving complex. Eur J Biochem 271:2967–2975
Calderone V, Trabucco M, Vujicic A, Battistutta R, Giacometti GM, Andreucci F, Barbato R, Zanotti G (2003) Crystal structure of the PsbQ protein of photosystem II from higher plants. EMBO Rep 4:900–905
Chen H, Zhang D, Guo J, Wu H, Jin M, Lu Q, Lu C, Zhang L (2006) A Psb27 homologue in Arabidopsis thaliana is required for efficient repair of photodamaged photosystem II. Plant Mol Biol 61:567–575
Clarke SM, Eaton-Rye JJ (1999) Mutation of Phe-363 in the photosystem II protein CP47 impairs photoautotrophic growth, alters the chloride requirement, and prevents photosynthesis in the absence of either PSII-O or PSII-V in Synechocystis sp. PCC 6803. Biochemistry 38:2707–2715
De Las Rivas J, Barber J (2004) Analysis of the Structure of the PsbO protein and its implications. Photosynth Res 81:329–343
De Las Rivas J, Roman A (2005) Structure and evolution of the extrinsic proteins that stabilize the oxygen-evolving engine. Photochem Photobiol Sci 4:1003–1010
De Las Rivas J, Balsera M, Barber J (2004) Evolution of oxygenic photosynthesis: genome-wide analysis of the OEC extrinsic proteins. Trends Plant Sci 9:18–25
De Las Rivas J, Shipton CA, Ponticos M, Barber J (1993) Acceptor side mechanism of photoinduced proteolysis of the D1 protein in photosystem II reaction centers. Biochemistry 32:6944–6950
de Vitry C, Olive J, Drapier D, Recouvreur M, Wollman F (1989) Posttranslational events leading to the assembly of photosystem II protein complex: a study using photosynthesis mutants from Chlamydomonas reinhardtii. J Cell Biol 109:991–1006
Eaton-Rye JJ (2005) Requirements for different combinations of the extrinsic proteins in specific cyanobacterial photosystem II mutants. Photosynth Res 84:275–281
Eaton-Rye JJ, Murata N (1989) Evidence that the amino-terminus of the 33 kDa extrinsic protein is required for binding to the Photosystem II complex. Biochim Biophys Acta 977:219–226
Eaton-Rye JJ, Shand JA, Nicoll WS (2003) pH-dependent photoautotrophic growth of specific photosystem II mutants lacking lumenal extrinsic polypeptides in Synechocystis PCC 6803. FEBS Lett 543:148–153
Enami I, Kikuchi S, Fukuda T, Ohta H, Shen JR (1998) Binding and functional properties of four extrinsic proteins of photosystem II from a red alga, Cyanidium caldarium, as studied by release-reconstitution experiments. Biochemistry 37:2787–2793
Enami I, Murayama H, Ohta H, Kamo M, Nakazato K, Shen JR (1995) Isolation and characterization of a photosystem II complex from the red alga Cyanidium caldarium: association of cytochrome c-550 and a 12 kDa protein with the complex. Biochim Biophys Acta 1232:208–216
Enami I, Iwai M, Akiyama A, Suzuki T, Okumura A, Katoh T, Tada O, Ohta H, Shen J-R (2003) Comparison of binding and functional properties of two extrinsic components, cyt c550 and a 12 kDa protein, in cyanobacterial PSII with those in red algal PSII. Plant Cell Physiol 44:820–827
Enami I, Suzuki T, Tada O, Nakada Y, Nakamura K, Tohri A, Ohta H, Inoue I, Shen JR (2005) Distribution of the extrinsic proteins as a potential marker for the evolution of photosynthetic oxygen-evolving photosystem II. FEBS J 272:5020–5030
Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S (2004) Architecture of the photosynthetic oxygen-evolving center. Science 303:1831–1838
Gao JP, Yong ZH, Zhang F, Ruan KC, Xu CH, Chen GY (2005) Positive charges on lysine residues of the extrinsic 18 kDa protein are important to its electrostatic interaction with spinach photosystem II membranes. Acta Biochim Biophys Sin (Shanghai) 37:737–742
Ghanotakis DF, Babcock GT, Yocum CF (1984a) Calcium reconstitutes high rates of oxygen evolution in polypeptide depleted photosystem II preparations. FEBS Lett 167:127–130
Ghanotakis DF, Topper JN, Yocum CF (1984b) Structural organization of the oxidizing side of photosystem II. Exogenous reductants reduce and destroy the Mn-complex in photosystems II membranes depleted of the 17 and 23 kDa polypeptides. Biochim. Biophys. Acta 767:524–531
Harper R, Bassi R, Testi MG, Schafer C (1998) Nearest-neighbor analysis of a photosystem II complex from Marchantia polymorpha L. (liverwort), which contains reaction center and antenna proteins. FEBS Lett 255:196–205
Heredia P, De Las Rivas J (2003) Calcium-dependent conformational change and thermal stability of the isolated PsbO protein detected by FTIR spectroscopy. Biochemistry 42:11831–11838
Ifuku K, Sato F (2001) Importance of the N-terminal sequence of the extrinsic 23 kDa polypeptide in photosystem II in ion retention in oxygen evolution. Biochim Biophys Acta 1546:196–204
Ifuku K, Sato F (2002) A truncated mutant of the extrinsic 23-kDa protein that absolutely requires the extrinsic 17-kDa protein for Ca2 + retention in photosystem II. Plant Cell Physiol 43:1244–1249
Ifuku K, Nakatsu T, Kato H, Sato F (2004) Crystal structure of the PsbP protein of photosystem II from Nicotiana tabacum. EMBO Rep 5:362–367
Ifuku K, Yamamoto Y, Ono TA, Ishihara S, Sato F (2005) PsbP protein, but not PsbQ protein, is essential for the regulation and stabilization of photosystem II in higher plants. Plant Physiol 139:1175–1184
Ikeuchi M, Inoue Y, Vermaas W (1995) Characterization of photosystem II subunits from the cyanobacterium Synechocystis sp. PCC 6803. In: Mathis P (ed). Photosynthesis: from light to biosphere, Dordrecht: Kluwer Academic Publishers, pp 297–300
Inoue-Kashino N, Kashino Y, Satoh K, Terashima I, Pakrasi HB (2005) PsbU provides a stable architecture for the oxygen-evolving system in cyanobacterial photosystem II. Biochemistry 44:12214–12228
Ishihara S, Yamamoto Y, Ifuku K, Sato F (2005) Functional analysis of four members of the PsbP family in photosystem II in Nicotiana tabacum using differential RNA interference. Plant Cell Physiol 46:1885–1893
Ishikawa Y, Schroder WP, Funk C (2005) Functional analysis of the PsbP-like protein (sll1418) in Synechocystis sp. PCC 6803. Photosynth Res 84:257–262
Ishikita H, Knapp EW (2005) Redox potential of cytochrome c550 in the cyanobacterium Thermosynechococcus elongates. FEBS Lett 579:3190–3194
Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A (2003) Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci 12:1652–1662
Kamiya N, Shen JR (2003) Crystal structure of oxygen-evolving photosystem II from Thermosynechococcus vulcanus at 3.7-A resolution. Proc Natl Acad Sci U S A 100:98–103
Kashino Y, Inoue-Kashino N, Roose JL, Pakrasi HB (2006) Absence of the PsbQ protein results in destabilization of the PsbV protein and decreased oxygen evolution activity in cyanobacterial photosystem II. J Biol Chem 281:20834–20841
Kashino Y, Lauber WM, Carroll JA, Wang Q, Whitmarsh J, Satoh K, Pakrasi HB (2002) Proteomic analysis of a highly active photosystem II preparation from the cyanobacterium Synechocystis sp. PCC 6803 reveals the presence of novel polypeptides. Biochemistry 41:8004–8012
Katoh H, Itoh S, Shen JR, Ikeuchi M (2001) Functional analysis of psbV and a novel c-type cytochrome gene psbV2 of the thermophilic cyanobacterium Thermosynechococcus elongatus strain BP-1. Plant Cell Physiol 42:599–607
Kavelaki K, Ghanotakis DF (1991) Effect of the manganese complex on the binding of the extrinsic proteins (17, 23 and 33 kDa) of photosystem II. Photosynth Res 29:149–155
Kerfeld CA, Sawaya MR, Bottin H, Tran KT, Sugiura M, Cascio D, Desbois A, Yeates TO, Kirilovsky D, Boussac A (2003) Structural and EPR characterization of the soluble form of cytochrome c-550 and of the psbV2 gene product from the cyanobacterium Thermosynechococcus elongatus. Plant Cell Physiol 44:697–706
Kimura A, Eaton-Rye JJ, Morita EH, Nishiyama Y, Hayashi H (2002) Protection of the oxygen-evolving machinery by the extrinsic proteins of photosystem II is essential for development of cellular thermotolerance in Synechocystis sp. PCC 6803. Plant Cell Physiol 43:932–938
Kirilovsky D, Roncel M, Boussac A, Wilson A, Zurita JL, Ducruet JM, Bottin H, Sugiura M, Ortega JM, Rutherford A (2004) Cytochrome c550 in the cyanobacterium Thermosynechococcus elongatus. J Biol Chem 279:52869–52880
Kobayashi M, Katoh H, Ikeuchi M. (2006) Mutations in a putative chloride efflux transporter gene suppress the chloride requirement of photosystem II in the cytochrome c550-deficient mutant. Plant Cell Physiol 47:799–804
Krogmann DW (1991) The low-potential cytochrome c of cyanobacteria and algae. Biochim Biophys Acta 1058:35–37
Krogmann DW, Smith S (1990) Current Research in Photosynthesis. In: Raltxheffsky M (ed) Kluwer Academic Publishers, Dordrecht, The Netherlands, 11:687–690
Kuhn MG, Vermaas WF (1993) Deletion mutations in a long hydrophilic loop in the photosystem II chlorophyll-binding protein CP43 in the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol Biol 23:123–133
Kuwabara T, Murata N (1983) Quantitative analysis of the inactivation of photosynthetic oxygen evolution and the release of polypeptides and manganese in the photosystem II particles of spinach chloroplasts. Plant Cell Physiol 24:741–747
Kuwabara T, Murata T, Miyao M, Murata N (1986) Partial degradation of the 18-kDa protein of the photosynthetic oxygen-evolving complex: a study of a binding site. Biochim Biophys Acta 850:146–155
Lautner A, Klein R, Ljungberg U, Reilander H, Bartling D, Andersson B, Reinke H, Beyreuther K, Herrmann RG (1988) Nucleotide sequence of cDNA clones encoding the complete precursor for the “10-kDa” polypeptide of photosystem II from spinach. J Biol Chem 263:10077–10081
Li Z, Andrews H, Eaton-Rye JJ, Burnap RL (2004) In situ effects of mutations of the extrinsic cytochrome c550 of photosystem II in Synechocystis sp. PCC6803. Biochem, 43:14161–14170
Ljungberg U, Akerlund HE, Andersson B (1986) Isolation and characterization of the 10-kDa and 22-kDa polypeptides of higher plant photosystem 2. Eur J Biochem 158:477–482
Ljungberg U, Akerlund HE, Larsson C, Andersson B (1984a) Identification of polypeptides associated with the 23 and 33 kDa proteins of photosynthetic oxygen evolution. Biochim Biophys Acta 767:145–152
Ljungberg U, Akerlund HE, Andersson B (1984b) The release of a 10-kDa polypeptide from everted photosystem II thylakoid membranes by alkaline Tris. FEBS Lett 175:255–258
Loll B, Kern J, Saenger W, Zouni A, Biesiadka J (2005a) Towards complete cofactor arrangement in the 3.0 A resolution structure of photosystem II. Nature 438:1040–1044
Loll B, Gerold G, Slowik D, Voelter W, Jung C, Saenger W, Irrgang KD (2005b) Thermostability and Ca2 + binding properties of wild type and heterologously expressed PsbO protein from cyanobacterial photosystem II. Biochemistry 44:4691–4698
Matsuzaki M, Misumi O, Shin IT, Maruyama S, Takahara M, Miyagishima SY, Mori T, Nishida K, Yagisawa F, Nishida K, Yoshida Y, Nishimura Y, Nakao S, Kobayashi T, Momoyama Y, Higashiyama T, Minoda A, Sano M, Nomoto H, Oishi K, Hayashi H, Ohta F, Nishizaka S, Haga S, Miura S, Morishita T, Kabeya Y, Terasawa K, Suzuki Y, Ishii Y, Asakawa S, Takano H, Ohta N Kuroiwa H, Tanaka K, Shimizu N, Sugano S, Sato N, Nozaki H, Ogasawara N, Kohara Y, Kuroiwa T (2004) Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428:653–657
Mayfield SP, Bennoun P, Rochaix JD (1987) Expression of the nuclear encoded OEE1 protein is required for oxygen evolution and stability of photosystem II particles in Chlamydomonas reinhardtii. EMBO J 6:313–318
Meades GD Jr, McLachlan A, Sallans L, Limbach PA, Frankel LK, Bricker TM (2005) Association of the 17-kDa extrinsic protein with photosystem II in higher plants. Biochemistry 44:15216–15221
Miyao M, Murata N (1983a) Partial disintegration and reconstitution of the photosynthetic oxygen evolution system. Binding of 24 kilodalton and 18 kilodalton polypeptide. Biochim Biophys Acta 725:87–93
Miyao M, Murata N (1983b) Partial reconstitution of the photosynthetic oxygen evolution system by rebinding of the 33-kDa polypeptide. FEBS Lett 164:375–378
Miyao M, Murata N (1984) Calcium ions can be substituted for the 24-kDa polypeptide in photosynthetic oxygen evolution. FEBS Lett 168:118–120
Miyao M, Murata N (1985) The Cl− effect on photosynthetic oxygen evolution: interaction of Cl− with 18-kDa, 24-kDa and 33-kDa proteins. FEBS Lett 180:303–308
Morgan TR, Shand JA, Clarke SM, Eaton Rye JJ (1998) Specific requirements for cytochrome c-550 and the manganese-stabilizing protein in photoautotrophic strains of Synechocystis sp. PCC 6803 with mutations in the domain Gly-351 to Thr-436 of the chlorophyll-binding protein CP47. Biochemistry 37:14437–14449
Mould R, Robinson C (1991) A proton gradient is required for the transport of two lumenal oxygen- evolving proteins across the thylakoid membrane. J Biol Chem 266:12189–12193
Murray JW, Barber J (2006) Identification of a calcium-binding site in the PsbO protein of photosystem II. Biochemistry 45:4128–4130
Nield J, Barber J (2006) Refinement of the structural model for the Photosystem II supercomplex of higher plants. Biochim Biophys Acta 1757:353–361
Nield J, Balsera M, De Las Rivas J, Barber J (2002) Three-dimensional electron cryo-microscopy study of the extrinsic domains of the oxygen-evolving complex of spinach: assignment of the PsbO protein. J Biol Chem 277:15006–15012
Nield J, Kruse O, Ruprecht J, da Fonseca P, Buchel C, Barber J (2000) Three-dimensional structure of Chlamydomonas reinhardtii and Synechococcus elongatus photosystem II complexes allows for comparison of their oxygen-evolving complex organization. J Biol Chem 275:27940–27946
Nishiyama Y, Los DA, Murata N (1999) PsbU, a protein associated with photosystem II, is required for the acquisition of cellular thermotolerance in Synechococcus species PCC 7002. Plant Physiol 120:301–308
Nishiyama Y, Hayashi H, Watanbe T, Murata N (1994) Photosynthetic oxygen evolution is stabilized by cytochrome c550 against heat inactivation in Synechococcus sp PCC 7002. Plant Physiol 105:1313–1319
Nishiyama Y, Los DA, Hayashi H, Murata N (1997) Thermal protection of the oxygen-evolving machinery by PsbU, an extrinsic protein of photosystem II, in Synechococcus species PCC 7002. Plant Physiol 115:1473–1480
Ohta H, Suzuki T, Ueno M, Okumura A, Yoshihara S, Shen JR, Enami I (2003) Extrinsic proteins of photosystem II: an intermediate member of PsbQ protein family in red algal PS II. Eur J Biochem 270:4156–4163
Ohta H, Okumura A, Okuyama S, Akiyama A, Iwai M, Yoshihara S, Shen JR, Kamo M, Enami I (1999) Cloning, expression of the psbU gene, and functional studies of the recombinant 12-kDa protein of photosystem II from a red alga Cyanidium caldarium. Biochem Biophys Res Commun 260:245–250
Peltier JB, Emanuelsson O, Kalume DE, Ytterberg J, Friso G, Rudella A, Liberles DA, Soderberg L, Roepstorff P, von Heijne G, van Wijk KJ (2002) Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 14:211–236
Pettigrew GW, Moore GR (1987) Cytochromes c: biological aspects. Berlin Heidelberg New York: Springer
Philbrick JB, Diner BA, Zilinskas BA (1991) Construction and characterization of cyanobacterial mutants lacking the manganese-stabilizing polypeptide of photosystem II. J Biol Chem 266:13370–13376
Popelkova H, Im MM, Yocum CF (2002a) N-terminal truncations of manganese stabilizing protein identify two amino acid sequences required for binding of the eukaryotic protein to photosystem II and reveal the absence of one binding-related sequence in cyanobacteria. Biochemistry 41:10038–10045
Popelkova H, Im MM, D’Auria J, Betts SD, Lydakis-Simantiris N, Yocum CF (2002b) N-terminus of the photosystem II manganese stabilizing protein: effects of sequence elongation and truncation. Biochemistry 41:2702–2711
Putnam-Evans C, Bricker TM (1992) Site-directed mutagenesis of the CPa-1 protein of photosystem II: alteration of the basic residue pair 384,385R to 384,385G leads to a defect associated with the oxygen-evolving complex. Biochemistry 31:11482–11488
Putnam-Evans C, Burnap R, Wu J, Whitmarsh J, Bricker TM (1996) Site-directed mutagenesis of the CP 47 protein of photosystem II: alteration of conserved charged residues in the domain 364E-444R. Biochemistry 35:4046–4053
Qian M, Al-Khaldi SF, Putnam-Evans C, Bricker TM, Burnap RL (1997) Photoassembly of the photosystem II (Mn)4 cluster in site-directed mutants impaired in the binding of the manganese-stabilizing protein. Biochemistry 36:15244–15252
Raymond J, Blankenship RE (2004) The evolutionary development of the protein complement of photosystem 2. Biochim Biophys Acta 1655:133–139
Robinson C, Bolhuis A (2004) Tat-dependent protein targeting in prokaryotes and chloroplasts. Biochim Biophys Acta 1694:135–147
Rokka A, Suorsa M, Saleem A, Battchikova N, Aro EM (2005) Synthesis and assembly of thylakoid protein complexes: multiple assembly steps of photosystem II. Biochem J 388:159–168
Roncel M, Boussac A, Zurita JL, Bottin H, Sugiura M, Kirilovsky D, Ortega JM (2003) Redox properties of photosystem II cytochromes b559 and c550 in the cyanobacterium Thermosynechococcus elogatus. J Biol Inorg Chem 8:206–216
Roose JL, Pakrasi HB (2004) Evidence that D1 processing is required for manganese binding and extrinsic protein assembly into photosystem II. J Biol Chem 279:45417–45422
Rova EM, Mc Ewen B, Fredriksson PO, Styring S (1996) Photoactivation and photoinhibition are competing in a mutant of Chlamydomonas reinhardtii lacking the 23-kDa extrinsic subunit of photosystem II. J Biol Chem 271:28918–28924
Rova M, Franzacn L-G, Fredriksson P-O, Styring S (1994) Photosystem II in a mutant of Chlamydomonas reinhardtii lacking the 23 kDa psbP protein shows increased sensitivity to photoinhibition in the absence of chloride. Photosynth Res 39:75–83
Salter AH, Virgin I, Hagman A, Andersson B (1992) On the molecular mechanism of light-induced D1 protein degradation in photosystem II core particles. Biochemistry 31:3990–3998
Schubert M, Petersson UA, Haas BJ, Funk C, Schroder WP, Kieselbach T (2002) Proteome map of the chloroplast lumen of Arabidopsis thaliana. J Biol Chem 277:8354–8365
Seidler A (1996) The extrinsic polypeptides of Photosystem II. Biochim Biophys Acta 1277:35–60
Seidler A, Rutherford AW (1996) The role of the extrinsic 33 kDa protein in Ca2 + binding in photosystem II. Biochemistry 35:12104–12110
Shen JR, Inoue Y (1993) Binding and functional properties of two new extrinsic components, cytochrome c-550 and a 12-kDa protein, in cyanobacterial photosystem II. Biochemistry 32:1825–1832
Shen JR, Vermaas W, Inoue Y (1995) The role of cytochrome c-550 as studied through reverse genetics and mutant characterization in Synechocystis sp. PCC 6803. J Biol Chem 270:6901–6907
Shen JR, Ikeuchi M, Inoue Y (1997) Analysis of the psbU gene encoding the 12-kDa extrinsic protein of photosystem II and studies on its role by deletion mutagenesis in Synechocystis sp. PCC 6803. J Biol Chem 272:17821–17826
Shen JR, Qian M, Inoue Y, Burnap RL (1998) Functional characterization of Synechocystis sp. PCC 6803 delta psbU and delta psbV mutants reveals important roles of cytochrome c-550 in cyanobacterial oxygen evolution. Biochemistry 37:1551–1558
Spetea C, Hundal T, Lohmann F, Andersson B (1999) GTP bound to chloroplast thylakoid membranes is required for light-induced, multienzyme degradation of the photosystem II D1 protein. Proc Natl Acad Sci USA 96:6547–6552
Spetea C, Keren N, Hundal T, Doan JM, Ohad I, Andersson B (2000) GTP enhances the degradation of the photosystem II D1 protein irrespective of its conformational heterogeneity at the Q(B) site. J Biol Chem 275:7205–7211
Spetea C, Hundal T, Lundin B, Heddad M, Adamska I, Andersson B (2004) Multiple evidence for nucleotide metabolism in the chloroplast thylakoid lumen. Proc Natl Acad Sci USA 101:1409–1414
Stewart AC, Ljungberg U, Akerlund HE, Andersson B (1985) Studies on the polypeptide composition of the cyanobacterial oxygen-evolving complex. Biochim Biophys Acta 808:353–362
Stockhaus J, Hofer M, Renger G, Westhoff P, Wydrzynski T, Willmitzer L (1990) Anti-sense RNA efficiently inhibits formation of the 10 kd polypeptide of photosystem II in transgenic potato plants: analysis of the role of the 10 kd protein. EMBO J 9:3013–3021
Summerfield TC, Winter RT, Eaton-Rye JJ (2005a) Investigation of a requirement for the PsbP-like protein in Synechocystis sp. PCC 6803. Photosynth Res 84:263–268
Summerfield TC, Shand JA, Bentley FK, Eaton-Rye JJ (2005b) PsbQ (Sll1638) in Synechocystis sp. PCC 6803 is required for photosystem II activity in specific mutants and in nutrient-limiting conditions. Biochemistry 44:805–815
Suorsa M, Sirpio S, Allahverdiyeva Y, Paakkarinen V, Mamedov F, Styring S, Aro EM (2006) PsbR, a missing link in the assembly of the oxygen-evolving complex of plant photosystem II. J Biol Chem 281:145–150
Suzuki T, Ohta H, Enami I (2005) Cross-reconstitution of the extrinsic proteins and photosystem II complexes from Chlamydomonas reinhardtii and Spinacia oleracea. Photosynth Res 84:239–244
Suzuki T, Minagawa J, Tomo T, Sonoike K, Ohta H, Enami I (2003) Binding and functional properties of the extrinsic proteins in oxygen-evolving photosystem II particle from a green alga, Chlamydomonas reinhardtii having his-tagged CP47. Plant Cell Physiol 44:76–84
Swiatek M, Regel RE, Meurer J, Wanner G, Pakrasi HB, Ohad I, Herrmann RG (2003) Effects of selective inactivation of individual genes for low-molecular-mass subunits on the assembly of photosystem II, as revealed by chloroplast transformation: the psbEFLJoperon in Nicotiana tabacum. Mol Genet Genomics 268:699–710
Swiatek M, Kuras R, Sokolenko A, Higgs D, Olive J, Cinque G, Muller B, Eichacker LA, Stern DB, Bassi R, Herrmann RG, Wollman FA (2001) The chloroplast gene ycf9 encodes a photosystem II (PSII) core subunit, PsbZ, that participates in PSII supramolecular architecture. Plant Cell 13:1347–1367
Tezcan FA, Winkler JR, Gray HB (1998) Effects of Ligation and Folding on Reduction Potentials of Heme Proteins. J Am Chem Soc 120:13383–13388
Thornton LE, Roose JL, Ikeuchi M, Pakrasi HB (2005) The low molecular weight components: new insights from proteomics, genomics, and structural studies. In: Wydrzynski T, Satoh K, (eds), Photosystem II: the light-driven water:plastoquinone oxioreductase, vol. 22. Springer, Dordrecht, pp 121–137
Thornton LE, Ohkawa H, Roose JL, Kashino Y, Keren N, Pakrasi HB (2004) Homologs of plant PsbP and PsbQ proteins are necessary for regulation of photosystem II activity in the cyanobacterium Synechocystis 6803. Plant Cell 16:2164–2175
Tohri A, Dohmae N, Suzuki T, Ohta H, Inoue Y, Enami I (2004) Identification of domains on the extrinsic 23 kDa protein possibly involved in electrostatic interaction with the extrinsic 33 kDa protein in spinach photosystem II. Eur J Biochem 271:962–971
Turkina MV, Kargul J, Blanco-Rivero A, Villarejo A, Barber J, Vener AV (2006) Environmentally-modulated phosphoproteome of photosynthetic membranes in the green alga Chlamydomonas reinhardtii. Mol Cell Proteomics 5:1412–1425
Veerman J, Bentley FK, Eaton-Rye JJ, Mullineaux CW, Vasil’ev S, Bruce D (2005) The PsbU subunit of photosystem II stabilizes energy transfer and primary photochemistry in the phycobilisome-photosystem II assembly of Synechocystis sp. PCC 6803. Biochemistry 44:16939–16948
Vrettos JS, Reifler MJ, Kievet O, Lakshmi KV, de Paula JC, Brudvig GW (2001) Factors that determine the unusually low reduction potential of cytochrome c550 in cyanobacterial photosystem II. J Biol Inorg Chem 6:708–716
Webber AN, Packman LC, Gray JC (1989) A 10 kDa polypeptide associated with the oxygen-evolving complex of photosystem II has a putative C-terminal non-cleavable thylakoid transfer domain. FEBS Lett 242:435–438
Xu Q, Bricker TM (1992) Structural organization of proteins on the oxidizing side of photosystem II. Two molecules of the 33-kDa manganese-stabilizing proteins per reaction center. J Biol Chem 267:25816–25821
Yi X, Hargett SR, Frankel LK, Bricker TM (2006) The PsbQ protein is required in Arabidopsis for photosystem II assembly/stability and photoautotrophy under low light conditions. J Biol Chem 281:26260–26267
Yi X, McChargue M, Laborde S, Frankel LK, Bricker TM (2005) The manganese-stabilizing protein is required for photosystem II assembly/stability and photoautotrophy in higher plants. J Biol Chem 280:16170–16174
Yu H, Xu X, Britt RD (2006) The 33 kDa protein can be removed without affecting the association of the 23 and 17 kDa proteins with the luminal side of PS II of spinach. Biochemistry 45:3404–3411
Zouni A, Witt HT, Kern J, Fromme P, Krauss N, Saenger W, Orth P (2001) Crystal structure of photosystem II from Synechococcus elongatus at 3.8 A resolution. Nature 409:739–743
Acknowledgments
We thank members of the Pakrasi lab for helpful discussions during the preparation of this manuscript. This work was supported by funding from the National Science Foundation to HBP (MCB-0215359).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Roose, J.L., Wegener, K.M. & Pakrasi, H.B. The extrinsic proteins of Photosystem II. Photosynth Res 92, 369–387 (2007). https://doi.org/10.1007/s11120-006-9117-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11120-006-9117-1