Abstract
The differential expression profiling of the fruit-specific genes between a blood orange (Citrus sinensis cv. Tarocco) and a blonde orange (Citrus sinensis cv. twenty-first century navel orange) was investigated by fruit transcriptome sequencing. The relative expression folds for Cs6g17570 and Cs5g31400 were significantly greater in the “Tarocco” fruits, but Cs9g04810 transcripts had no significant differences between two accessions by digital gene expression profiling. In particular, R2R3-MYB, bHLH-type, and WD40-repeat proteins (encoded by Cs6g17570, Cs5g31400, and Cs9g04810) were transcription factors with each conserved domain and interacted in vitro and in vivo. Moreover, the dihydroflavonol 4-reductase (DFR), a key enzyme involved into anthocyanin biosynthesis pathway, had greater transcription levels in Tarocco fruit and also in the transgenic Arabidopsis thaliana by Cs6g17570 ectopic expression. Furthermore, Cs6g17570 and Cs5g31400 in Tarroco were upregulated by cold induction and then had similar expression profiling during cold storage. In conclusion, Cs6g17570, Cs5g31400, and Cs9g04810 encoding proteins might be involved into anthocyanin regulation in Tarocco by MYB-bHLH-WD40 complex as well as in other fruit trees. Studies on the anthocyanin regulatory mechanism will aid in development of new biotechnological tools for Citrus breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Anthocyanins are derived from flavonoid biosynthesis and responsible for the colors of various plant species. The anthocyanins are scavengers of radical species and contribute to high antioxidant activity. The blood oranges (Citrus sinensis) characterized by their unique flesh and rind colors. The major anthocyanins of the blood orange juice have been characterized as cyanidin 3-glucoside, cyanidin 3-(6ʹ-malonyglucoside) and six minor anthocyanins (Hillebrand et al. 2004). The blood orange arose from the insertion of a Copia-like retrotransposon adjacent to a gene-encoding Ruby, a MYB-type transcriptional activator of anthocyanin production (Butelli et al. 2012). In addition, the expression pattern of CsMYC2, containing a basic helix-loop-helix (bHLH) domain, is correlated with UDP-glucose flavonoid glucosyl-transferase (UFGT) activity in different tissues or cultivars, and is involved in the regulation of the flavonoid biosynthetic pathway in Citrus (Cultrone et al. 2010). Glutathione S-transferases (GST) are involved in the vacuolar import of anthocyanins, and the GST enzyme also is active against cyanidin-3-O-glucoside (Lo Piero et al. 2006). The anthocyanin contents of Citrus exposed to cold sharply increases and the structural genes of anthocyanins biosynthesis indicate it is a cold-regulated pathway (Crifo et al. 2012 Anthocyanin production is transcriptionally regulated by the MYB-bHLH-WD40 (MBW) complex, which is composed of WD40 repeat protein, one R2R3-MYB protein, as well as one bHLH protein, to activate several of the late flavonoid biosynthetic genes (LBGs) in Arabidopsis thaliana (Li 2014). Structural genes encoding the enzymes involved in the anthocyanin biosynthetic pathway are conserved among different species (Holton and Cornish 1995). Therefore, the characterization of transcription factors regulating anthocyanin biosynthesis in blood orange would be useful for understanding the pigmentation mechanism and in breeding programs.
Anthocyanin production is transcriptionally regulated by the MYB-bHLH-WD40 (MBW) transcription factors complex, which has been widely found in apple (An et al. 2012), strawberry (Schaart et al. 2013), and peach (Rahim et al. 2014), respectively. Whether the MBW complex also is involved into anthocyanin regulation in blood oranges as well as in the other fruits? Studies on the anthocyanin regulation mechanism will aid in the development of new biotechnological tools for Citrus breeding. Digital gene expression (DGE) profiling is a revolutionary approach for gene expression analysis (Tang et al. 2011). Driven by Solexa/Illumina technology, DGE creates genome-wide expression profile using the sequencing technology to produce millions of short complementary DNA (cDNA) reads for analyzing the pathway of interest (Zhang et al. 2015).
In our work, a fruit transcriptome analysis was performed to comprehensively study the differential genes expression between the blood (C. sinensis cv. Tarocco) and blonde (C. sinensis cv. twenty-first Century navel oranges). In comparison with blonde orange, genes encoding for enzymes involved in the anthocyanin biosynthesis and regulations were upregulated in the “Tarocco.” In addition, R2R3-MYB, bHLH type and WD40 repeat proteins interacted in vivo and in vitro and seemed to form a complex to regulate anthocyanin biosynthesis in the Tarocco.
Materials and Methods
Plant Materials and Fruit Quality Measurements
The fruits from blood (C. sinensis cv. Tarocco) and blonde (C. sinensis cv. twenty-first century navel orange) oranges were harvested by the end of March in 2014, respectively. The samples were frozen in liquid nitrogen and stored at −80 °C in the lab. Juice was extracted using a domestic juicer. The total acidity (TA) and total soluble solids (TSS) were determined using previously described methods (He et al. 2013) to determine fruits maturity. The anthocyanin contents of fruit juice were measured using spectrophotometry at 510 nm in a previous study (Rapisarda et al. 2000). The flesh RNA of Tarocco and twenty-first century navel orange were isolated using an RNA isolation kit (Huayueyang, Beijing, China). cDNA synthesis was performed using the PrimeScript first-strand cDNA synthesis kit (Takara, Dalian, China). The relative expression levels of chalcone synthase gene were semi-quantitatively amplified to test the RNA quality prior to transcriptome sequencing.
The RNA-seq Library Preparation and Sequencing
An aliquot (~6 μg) of the total RNA from each sample was extracted using an RNA isolation kit (Huayueyang) and purified using the Dynabeads messenger RNA (mRNA) DIRECT Micro Kit (Invitrogen, Waltham, MA, USA). Subsequently, the sequencing library was prepared using ion total RNA-Seq Kit v2.0 (Invitrogen). The transcriptome sequencing was performed by Ion Torrent (Thermofisher).
The Differential Expression Genes Identification
The reference genome and annotation of C. sinensis cv. Valencia were collected from the website http://citrus.hzau.edu.cn/orange/index.php (Xu et al. 2013). The Cuffdiff software (Pollier et al. 2013) was used to identify transcripts that had significant differences based on their relative abundance. Finally, transcripts with P value of <0.01 and fold change ≥1.8 were marked as significantly different. The differential expression genes were analyzed for gene ontology (GO) enrichment using Blast2GO software with default parameters (Conesa et al. 2005). A Venn Diagrams for quantitative comparing transcripts was produced by Venny2.1 (http://bioinfogp.cnb.csic.es/tools/venny/). Hierarchical clustering of differential expression genes that showed threshold values of ≥5 and ≤−5 fold change was generated by Multi-Experiment Viewer software (Saeed et al. 2003).
Characterization of Three Transcription Factors Regulating Anthocyanin Biosynthesis
Three transcripts from Tarocco were identified using fruit transcriptome sequencing data to be possibly involved into anthocyanin regulation pathway. The full-length open reading frames of three differentially expressed regulatory factors in ‘Tarocco’ fruits were obtained by RT-PCR with specific primer pairs (Table 1) and designated by Cs2g17570, Cs5g31400, and Cs9g04810, respectively. After adjusting the deduced amino acid sequences of Cs2g17570, Cs5g31400, and Cs9g04810 encoding proteins, WEBLOGO was used to determine the features of specific domains (Crooks et al. 2004). Then, phylogenetic trees were constructed based on the multiple sequence alignment by MEGA 5.0 and 1000 bootstrap replicates were performed using the neighbor-joining (NJ) method for each group of proteins.
Yeast Two-Hybrid and Firefly Luciferase Complementation Imaging Assays
To test in vitro interactions between Cs2g17570, Cs5g31400, and Cs9g04810 encoding proteins, the open reading frames of each gene were cloned into pENTR/D-TOPO (Invitrogen) to generate pENTRY-R2R3MYB (entry vector recombinant with full length of Cs2g17570), pENTRY-bHLH (Cs5g31400) and pENTRY-WB40 (Cs9g04810), respectively. The prey constructs of AD-R2R3MYB and AD-WD40 were generated by LR reactions between pDEST22 (Invitrogen) and pENTRY-R2R3MYB, pENTRY-WD40, respectively. The bait constructs of BD-ΔN-bHLH (N-terminal deletion of Cs5g31400 to prevent self-activation) and BD-WD40 were generated by LR reactions between pDEST32 (Invitrogen) and pENTRY-ΔN-bHLH and pENTRY-WD40, respectively. Bait and prey plasmids, as well as the blank pDEST22 (or pDEST32) were co-transformed into AH109 yeast cells. The yeast cells were selected on SD-Leu-Trp and SD-Leu-Trp-His media, respectively. In addition, the firefly luciferase complementation imaging assays was performed to verify the interaction between MYB and bHLH. The bHLH-nLuc was generated by LR reactions (Invitrogen) between nLucGW (Wei et al. 2015) and pENTRY-bHLH. Moreover, the cLuc-R2R3MYB and cLuc-WD40 were generated by LR reactions between cLucGW and pENTRY-R2R3MYB and pENTRY-WD40, respectively. These plasmids were independently transformed into Agrobacterium GV3101. Firefly luciferase complementation imaging assays were carried out as described earlier (Chen et al. 2008). Briefly, cLuc-R2R3MYB, cLuc-WD40 and nLuc-bHLH, and the blank combinations were co-transformed with pCam-P19 into the leaves of Nicotiana benthamiana. Every combination was repeated more than three times in different leaves. The plants were incubated in the dark for 24 h and then in the light for 48 h. The leaves were sprayed with 1 mM luciferin and observed under an imaging apparatus (Lumazone 1300B, Beijing, China).
Differential Expression Genes Analysis Between Blood And Blonde Orange Cultivars
Two red fleshes accessions were selected: an Italian variety (Tarocco) and a Spanish variety (“Doblefina”) as blood orange cultivars. The “Cara cara” navel orange and the twenty-first century navel orange were selected as common (blonde) orange cultivars. The relative expression folds of each gene was calculated using 2−△△Ct method. The PCR program was 95 °C for 3 min, 35 cycles of 95 °C for 15 s, 56 °C for 15 s, and 72 °C for 20 s, followed by a 2-min extension at 72 °C. All of the primers used in this study are shown in Table 1. The relative expression folds of each gene between different varieties were normalized by the transcription levels of counterparts in Doblefina and EF-1α was used as the internal standard.
Expression Profilings of the Genes Involved in Anthocyanin Biosythesis During Cold Storage
The anthocyanin content in ‘Tarocco’ fruits was measured by spectrophotometry under cold storage for 0, 28, 42, 56 and 70 day, respectively. The relative expression folds of regulatory and structural genes were determined by the protocol mentioned previously. The relative expression folds of the each gene from different cold induction periods were normalized by the transcription levels of counterparts under cold treating for 43 days and EF-1α was used as an internal standard.
Ectopic Over-Expression of Cs6g17570 in A. thaliana
The plasmid containing a 35S promoter driving CsR2R3MYB over-expression was constructed using pK2GW7 (VIB, Ghent, Belgium) and the previously obtained pENTRY-R2R3MYB by LR reaction. The destination vector was transformed into Agrobacterium GV3101 by electroporation (BIO-RAD, Hercules, USA). The 250-mL cultures of GV3101 were incubated for 12 h and then centrifuged at 3000 rpm for 10 min. The Agrobacterium pellets were re-suspended in half-strength Murashige and Skoog (1/2 MS) medium without agar. The flowers of A. thaliana were dipped in an Agrobacterium culture supplemented with 0.03 % Silwet for 15 min. After 2 weeks, A. thaliana seeds were harvested and then were grown on 1/2 MS supplemented with kanamycin (50 g/mL) for screening the transgenic plants. The kanamycin-resistant seeds were synchronized in 4 °C refrigerator for 3 days and then grown in growth chambers at a temperature of 22 ± 2 °C under long day conditions (16-h light and 8-h dark). After 7 days, the seedlings were transferred to soil and grown in a greenhouse under the same conditions as described previously. The anthocyanin contents in adult leaves of WT and transgenic lines were determined using spectrophotometry as described previously. The total RNA of WT and transgenic lines were extracted using TRIzol (Sigma-Aldrich, St. Louis, MO, USA). cDNA synthesis was performed as mentioned previously. Semi-quantitative PCR was performed. The PCR reaction conditions were 95 °C for 5 min, 27 or 30 cycles of 95 °C for 30 s, 56 °C for 30 s, and 72 °C for 50 s, followed by a 5-min extension at 72 °C. All of the primers used in this study are shown in Table 1.
Statistically Analysis of Experiments
Differences of anthocyanin contents between WT and transgenic A. thaliana were evaluated via t test analysis by SPSS (IBM, Armonk, NY, USA). The Pearson correlation coefficient was determined between the relative expression folds of each gene and anthocyanin contents by SPSS, respectively. Statistical significance was considered to be P < 0.05.
Results
Digital Gene Expression Profiling Between Two Accessions
The fruits from two varieties were harvested at a similar maturity stage (Supplement Fig. 1) to investigate the gene expression profiling. In total, 48,556,208 and 43,861,517 reads with 127 bp in mean length were selected from blood and blonde orange fruits, respectively. When mapped to the reference genome of C. sinensis cv. Valencia (Xu et al. 2013), 61.74 and 82.34 % reads uniquely anchored blood and blonde orange, respectively. The numbers of transcripts between two varieties were counted in a Venn Diagrams (Fig. 1a). Hierarchical clustering of differential expression genes that had more than 5 (log2fold change) between two accessions were produced (Fig. 1b). In total, 29,655 expressed genes were identified, out of which 1714 accounted for differential expression (P < 0.01, log2fold change ≥1.8). The structural genes involved in the anthocyanin biosynthesis pathway had significantly higher expression folds (the numbers of fragments per kilobase of transcript per million fragments mapped) in the Tarocco than the twenty-first century navel orange, including phenylalanine ammonia lyase, 4-coumarate: CoA ligase, chalcone synthase, flavanone 3-hydroxylase, dihydroflavonol 4-reductase, leucoanthocyanidin dioxygenase, and UFGT (P < 0.05, Table 2). The GO enrichment results indicated that a total of 55 differentially expressed genes were found in the nucleic acid binding transcription factors of the “molecular function” category (Fig. 1c), within which two putative transcripts were possibly involved in anthocyanin regulation. Cs2g17570 and Cs5g31400 were significantly upregulated in “Tarocco” fruits (P < 0.05, Table 2), but the transcript level of Cs9g04810 showed no significant difference between two accessions (P > 0.05).
RNA-seq analysis between two accessions. Comparative study on the number of transcripts between two accessions (a), differential expression pattern between two accessions (b), and GO distributions of the differentially expressed genes (c). T and 21C indicated Tarocco and twenty-first century navel orange, respectively. b Triangle indicated by Cs6g17570
Three Transcription Factors Found in Tarocco Fruits
Cs9g04810 (GenBank Number: KT757349), Cs6g17570 (KT757348), and Cs5g31400 (KT757350) were co-expressed in Tarocco fruits, which encoded WD40, R2R3-MYB, and bHLH type proteins, respectively. Hereinafter, Cs6g17570, Cs5g31400, and Cs9g04810 were referred to as CsR2R3MYB, CsbHLH, and CsWD40, respectively. The MYB family was clustered into two subgroups containing a conserved R2R3 domain (Fig. 2a). The representative member from a phylogenic subgroup (boxed in Fig. 2b) was AtMYB114 that regulated the anthocyanin pathway in Arabidopsis (Heppel et al. 2013). AtMYB114 shared a 66.2 % identity with the CsR2R3MYB coding protein in Tarocco at the amino acid level. In addition, CsR2R3MYB in Tarocco had 100 % identity with a Ruby transcriptional activator to produce anthocyanin in C. sinensis cv. Moro (GenBank: JN402334). However, the amino acid sequence identity was 33.6 % between the CsbHLH coding protein in Tarocco and MYC2 protein in “Moro” reported as regulator in the flavonoid biosynthetic pathway in the previous study (Cultrone et al. 2010). In Figs. 3 and 4, there was a 75.3 % identity at the amino acids level between the CsbHLH coding protein in Tarocco and peachbHLH3 in peach controlling anthocyanin production (Rahim et al. 2014), and there was a 87.1 % identity between the CsWD40 coding protein in Tarocco and PgWD40 in pomegranate involving into anthocyanin biosynthesis (Ben-Simhon et al. 2011). As a result, three transcription factors were simultaneously expressed in Tarocco fruit and had each conserved domain that functioned in anthocyanin regulation.
Three Transcription Factors Interacted In Vitro and In Vivo
A yeast two-hybrid assay indicated that the WD40 repeat protein interacted with R2R3-MYB and bHLH type proteins in vitro, respectively (Fig. 5a). But the R2R3-MYB protein and bHLH type protein failed to interact, partially because the coding sequences of CsbHLH were truncated by 231 amino acids at N-terminal to cause no binding activity with CsR2R3MYB coding protein in the yeast two-hybrid system. So, firefly luciferase complementation assay was conducted to verify interaction between R2R3-MYB and bHLH-type protein. Therefore, the stronger interaction in vivo was found between R2R3-MYB and bHLH-type proteins, but a weaker interaction was occurred between the bHLH-type and WD40 repeat proteins by firefly luciferase complementation imaging assays (Fig. 5b). Therefore, R2R3-MYB, bHLH, and WD40 repeat proteins from “Tarocco” were interacted in vitro and in vivo.
The Expression Levels of Structural and Regulatory Genes Upregulated in the Blood Orange Cultivars
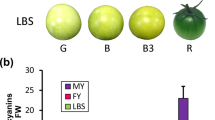
The fruits from four cultivars were harvested at a similar maturity stage (Fig. 6j). The blood orange cultivars (Tarocco and Doblefina) have pigmented fleshes and flavedo, varying from red to purple, but the blonde orange cultivars (Cara cara and twenty-first century Navel orange) has no anthocyanins at all (Fig. 6i). PAL, CHS, DFR, and UFGT involved into anthocyanin biosynthesis pathway were expressed at higher folds in the blood oranges (Fig. 6e–h). The transcription levels of PAL, CHS, and DFR were positively correlated with anthocyanin contents in blood oranges (Table 3). In addition, CsR2R3MYB, CsbHLH, and GST also had higher transcription levels in blood oranges (Fig. 6a, b, d). But CsWD40 had no differential expression between four cultivars (Fig. 6c). In comparison with the other two transcription factors, only CsbHLH transcription levels were significantly correlated with the relative expression folds of UFGT participating in to anthocyanin production (Table 3). Besides, GST transcription levels were also positively correlated with CHS and UFGT (Table 3).
The Expression Profiling of Three Transcription Factors in Tarocco Fruits During Cold Storage
The anthocyanin content in the fruit juice of Tarocco was increased rapidly during cold storage at 10 °C for 70 days. The anthocyanin content was only 8.26 mg/L after harvesting. Since then, it gradually reached the maximum level of 204.33 mg/L under cold induction for 70 days (Fig. 7a). CHS, an anthocyanin biosynthesis gene, was increased to its greatest value after 43-day cold induction and then maintained a steady transcription state (Fig. 7b). Anthocyanin accumulation in plant cells required GST for their transport into vacuoles since their cytoplasmic retention is toxic to the cell. Thus, the GST expression level was increased firstly and then decreased gradually (Fig. 7c), which might be consistent with the anthocyanin production rate in the cytoplasm by cold induction. Besides, CsR2R3MYB transcription level was significantly correlated with anthocyanin transporter-GST (Fig. 7d) during the cold storage (Pearson correlation coefficient = 0.928, P = 0.023, data not shown in this study). Therefore, CsR2R3MYB played more important roles in the anthocyanin production than other two transcription factors. In addition, CsbHLH and CsR2R3MYB had a similar expression pattern (Fig. 7e) but CsWD40 was completely different during cold storage (Fig. 7f).
The Pigmenting Transgenic A. thaliana caused by CsR2R3MYB Overexpression
The anthocyanin content in the transgenic A. thaliana was significantly greater than WT (Fig. 8a). The pigmented transgenic A. thaliana was found during the entire life at cotyledon stage (Fig. 8b), young seedling stage (Fig. 8c) and adult plant stage (Fig. 8d), respectively. In comparison with transcriptional profiling of WT, the PCR products amplified by 27 and 30 cycles showed that the late biosynthetic genes, such as DFR and LDOX, were significantly upregulated, but that the early biosynthetic genes, such as CHS and F3H, in transgenic Arabidopsis seemed to be unchanged (Fig. 8e). Moreover, the bHLH-type regulator (TT8 of Arabidopsis) was also slightly activated in the transgenic seedlings but the R2R3-MYB-type regulator and WD40-type regulator encoded by Arabidopsis were unchanged (Fig. 8e). It suggests that the anthocyanin quantity might depend on relative expression folds of transcriptional activator, such as CsR2R3MYB, to promote anthocyanin biosynthesis in the plants.
Discussion
Our investigations also have elucidated positive association between phenylalanine ammonia lyase (PAL), chalcone synthase (CHS), and dihydroflavonol 4-reductase (DFR) transcript levels and anthocyanin contents (P value <0.01, respectively), suggesting that the PAL, CHS, and DFR was produced exclusively for anthocyanin as reported in other studies (Licciardello et al. 2008; Moriguchi et al. 1999). This conclusion is also in agreement with over expressed of CHS enzyme in “Moro” flesh by LC-MS/MS approaches (Muccilli et al. 2009).
In this study, DFR participating in the anthocyanin biosynthesis were upregulated in the Tarocco fruit and also in the transgenic A.s thaliana by the CsR2R3MYB over-expression. Therefore, CsR2R3MYB played more important regulating roles in the anthocyanin biosynthesis pathway in Tarocco fruits, which was consistent with a homologous MYB-type transcriptional activator of anthocyanin production in Moro (Butelli et al. 2012). Furthermore, CsbHLH expression level increased more rapidly than CsR2R3MYB in Tarocco under cold storage for 28 days, which were in agreement with a bHLH-type regulator of Arabidopsis firstly activated in the transgenic seedling by CsR2R3MYB ectopic expression in this study.
STRING is a database of known and predicted protein interactions (Szklarczyk et al. 2015). The amino acids sequence of CsbHLH was used to predict the protein-protein interactions in Tarocco by searching in the database of A. thaliana. So that the orthologous TT8 protein was found in A. thaliana database (P value = 9e-99), which had 74 % identity with CsbHLH at amino acids level. Moreover, TTG1, MYB75, and DFR from A. thaliana were interacted by integrated interaction data (Fig. 9a) and were homologues of CsWD40, CsR2R3MYB, and Cs3g25090 (DFR of C. sinensis) from Tarocco, respectively (Fig. 9b). Therefore, three transcription factors might form a regulating complex (MYB-bHLH-WD40) to activate anthocyanin biosynthesis pathway in the C. sinensis cv. Tarocco.
The identification of genes participating in the anthocyanin modulation will be critical steps toward improving fruit quality in the blood orange breeding. The discovery of MBW regulation model may be useful for earlier identification of new varieties of blood orange. As a result, molecular markers will be developed against the three transcription factors of MBW complex for the blood orange identification. In addition, the more available primer pairs (SSR or SNP) will be generated by transcriptome sequencing, which had been reported in apricot (Dong et al. 2014), mango (Luo et al. 2015) and pummelo (Liang et al. 2015).
References
An XH, Tian Y, Chen KQ, Wang XF, Hao YJ (2012) The apple WD40 protein MdTTG1 interacts with bHLH but not MYB protein to regulate anthocyanin accumulation. J Plant Physiol 169:710–717
Ben-Simhon Z, Judeinstein S, Nadler-Hassar T, Trainin T, Bar-Ya'akov I, Borochov-Neori H, Holland D (2011) A pomegranate (Punica granatum L.) WD40-repeat gene is a functional homologue of Arabidopsis TTG1 and is involved in the regulation of anthocyanin biosynthesis during pomegranate fruit development. Planta 234(5):865–881
Butelli E, Licciardello C, Zhang Y, Liu J, Mackay S, Bailey P (2012) Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 24:1242–1255
Chen H, Zou Y, Shang Y (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146:368–376
Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M (2005) Blast2gO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Crifo T, Petrone G, Lo Cicero L, Lo Piero AR (2012) Short cold storage enhances the anthocyanin contents and level of transcripts related to their biosynthesis in blood oranges. J Agric Food Chem 60(1):476–81
Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14:1188–1190
Cultrone AA, Cotroneo PS, Recupero GR (2010) Cloning and molecular characterization of R2R3-MYB and bHLH-MYC transcription factors from Citrus sinensis. Tree Genet Genomes 6:101–112
Cotroneo PS, Russo MP, Ciuni M, Recupero GR, Lo Piero AR (2006) Quantitative real-time reverse transcriptase-PCR profiling of anthocyanin biosynthetic genes during orange fruit ripening. J Amer Soc Hort Sci 131(4):537–547
Dong S, Liu Y, Niu J, Ning Y, Lin S, Zhang Z (2014) De novo transcriptome analysis of the Siberian apricot (Prunus sibirica L.) and search for potential SSR markers by 454 pyrosequencing. Gene 544:220–227
Gonzalez A, Zhao M, Leavitt JM, Alan ML (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53:814–827
He J, Chen KL, Liu JJ, Guan B, Li HW, Wang JH (2013) Effect of liquid fertilizer by injecting into on quality of Tarocco blood orange. Southwest China J Agric Sci 26:299–303
Heppel SC, Jaffe FW, Takos AM, Schellmann S, Rausch T, Walker AR, Bogs J (2013) Identification of key amino acids for the evolution of promoter target specificity of anthocyanin and proanthocyanidin regulating MYB factors. Plant Mol Biol 82(4-5):457–71
Hillebrand S, Schwarz M, Winterhalter P (2004) Characterization of anthocyanins and pyranoanthocyanins from blood orange [Citrus sinensis (L.) osbeck] juice. J Agric Food Chem 52:7331–7338
Holton TA, Cornish EC (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7:1071–1083
Li S (2014) Transcriptional control of flavonoid biosynthesis: fine-tuning of the MYB-bHLH-WD40 (MBW) complex. Plant Signal Behav 9(1):e27522
Licciardello C, Russo MP, Valè G, Reforgiato Recupero G (2008) Identification of differentially expressed genes in the flesh of blood and common oranges. Tree Genet Genomes 4:315–31
Liang M, Yang X, Li H, Su S, Yi H, Chai L, Deng X (2015) De novo transcriptome assembly of pummelo and molecular marker development. PLoS One 10(3):e0120615. doi:10.1371/journal.pone.0120615
Lo Piero AR, Puglisi I, Rapisarda P, Petrone G (2005) Anthocyanins accumulation and related gene expression in red orange fruit induced by low temperature storage. J Agric Food Chem 53:9083–9088
Luo C, Wu HX, Yao QS, Wang SB, Xu WT (2015) Development of EST-SSR and TRAP markers from transcriptome sequencing data of the mango. Genet Mol Res 14(3):7914–7919
Lo Piero AR, Puglisi I, Petrone G (2006) Gene isolation, analysis of expression, and in vitro synthesis of glutathione s-transferase from orange fruit [Citrus sinensis L. (Osbeck)]. J Agric Food Chem 54(24):9227–9233
Moriguchi T, Kita M, Tomono Y, Endo-Inagaki T, Omura M (1999) One type of chalcone synthase gene expressed during embryogenesis regulates the flavonoid accumulation in Citrus cell cultures. Plant Cell Physiol 40:651–65
Muccilli V, Licciardello C, Fontanini D, Patrizia Russo M, Cunsolo V, Saletti R, Reforgiato Recupero G, Foti S (2009) Proteome analysis of Citrus sinensis L. (Osbeck) flesh at ripening time. J Proteomics 73(1):134–152
Pollier J, Rombauts S, Goossens A (2013) Analysis of RNA-Seq data with TopHat and Cufflinks for genome-wide expression analysis of jasmonate-treated plants and plant cultures. Methods Mol Biol 1011:305–315
Rahim MA, Busatto N, Trainotti L (2014) Regulation of anthocyanin biosynthesis in peach fruits. Planta 240(5):913–29
Rapisarda P, Fanella F, Maccarone E (2000) Reliability of analytical methods for determining anthocyanins in blood orange juices. J Agric Food Chem 48:2249–2252
Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34(2):374–378
Schaart JG, Dubos C, Fuente IRDL, Houwelingen AMML, Vos RCH, Jonker HH, Xu WJ, Routaboul JM, Lepiniec L, Bovy AG (2013) Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry fruits. New Phytologist 197:454–467
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:447–452. doi:10.1093/nar/gku1003
Tang Q, Ma X, Mo C, Wilson IW, Song C, Zhao H, Yang Y, FuW QD (2011) An efficient approach to finding Siraitia grosvenorii triterpene biosynthetic genes by RNA-seq and digital gene expression analysis. BMC Genomics 12:343
Wei BY, Zhang JZ, Pang CX, Yu H, Guo DS, Jiang H, Ding MX, Chen ZY, Tao Q, Gu HY, Qu LJ, Qin GJ (2015) The molecular mechanism of SPOROCYTELESS/NOZZLE in controlling Arabidopsis ovule development. Cell Res 25:121–134
Xu Q, Chen LL, Ruan XA, Chen DJ, Zhu AD, Chen CL, Bertrand D, Jiao WB, Hao BH, Matthew PL (2013) The draft genome of sweet orange (Citrus sinensis). Nat Genet 45(1):59–66
Zhang QJ, Tao ST, Li M, Qi XX, Wu J, Yin H, Deng JL, Zhang SL (2015) Identification of differentially expressed genes using digital gene expression profiles in Pyrus pyrifolia Nakai cv. Hosui bud release following early defoliation. Tree Genet Genomes 11:34. doi:10.1007/s11295-015-0858-x
Acknowledgement
This work was supported by the Applied and Basic Research Project of Sichuan Province.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1
Fruit quality analysis between two accessions. 1, 2 indicated Tarocco and twenty-first century navel orange, respectively. The total acidity (a), anthocyanin contents (b), and total soluble solids (c) were determined. The key genes involved into the anthocyanin biosynthesis and chalcone synthase, were expressed differentially between two accessions (d) (JPG 380 kb)
Rights and permissions
About this article
Cite this article
Wang, Jh., Liu, Jj., Chen, Kl. et al. Anthocyanin Biosynthesis Regulation in the Fruit of Citrus sinensis cv. Tarocco. Plant Mol Biol Rep 34, 1043–1055 (2016). https://doi.org/10.1007/s11105-016-0984-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-016-0984-0