Abstract
Gibberellins (GAs) are plant hormones that control many aspects of plant growth and development. Gibberellin 2-oxidase plays an important role in determining the level of bioactive GAs. In this study, we isolated three GA2ox genes (ClGA2ox1-3) from Camellia lipoensis Chang et Xu. The results of a quantitative real-time reverse transcription polymerase chain reaction analysis indicated that ClGA2ox1-3 may play a tissue-specific role in plant development. The transcript of ClGA2ox1 was more abundant in the stem and apex, ClGA2ox2 was highly expressed in mature leaves, and ClGA2ox3 was more abundant in roots. We produced transgenic plants of Nicotiana tabacum L. by overexpressing the ClGA2ox1-3 genes. Plants with overexpressed ClGA2ox1 or ClGA2ox3 genes exhibited dwarf phenotypes, including reduced growth, delayed flowering, and smaller, rounder, and darker green leaves. All of the transgenic plants overexpressing the ClGA2ox1 gene bloomed normally, but their flowers were half the size of the control plants. Plants overexpressing ClGA2ox3 could be categorized into two classes: moderately dwarfed and severely dwarfed. The ClGA2ox2 gene had little effect on the morphological characterization of transgenic plants. Quantitative real-time PCR analysis showed that the ClGA2ox3 expression level was generally correlated with the level of dwarfism. The endogenous level of bioactive GA4 and GA1 largely decreased in transgenic plants and was generally correlated with the degree of dwarfism in transgenic plants with the ClGA2ox1 or ClGA2ox3 gene. The application of GA3 rescued the dwarf phenotype of transformants, indicating that the GA signaling pathway might function normally in transgenic plants. Therefore, morphological changes in transgenic plants may result from a decrease in the endogenous level of bioactive GAs. Additionally, the possibility of molecular breeding for plant form alternation in Camellia plants by genetically engineering the GA metabolic pathway is discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dwarfism and semi-dwarfism are key traits in orchard and cereal crop breeding. In fruit trees, dwarf and semi-dwarf varieties are often preferred. These cultivars allow for dense field cultivation, facilitate mechanized maintenance, increase the efficiency of fruit collection, and allow for more precise pesticide application by reducing spray drift (Busov et al. 2003). Camellia is a genus of flowering plants in the family Theaceae. This genus is composed of evergreen shrubs or small trees that are popular around the world for a variety of uses, especially as ornamental plants. Semi-dwarf or dwarf camellias are favored in landscape applications and potted flower production.
Many studies have demonstrated that dwarfism is commonly related to deficiencies in gibberellin (GA) levels or signaling (Spielmeyer et al. 2002; Olszewski et al. 2002). GAs are a class of phytohormones that are responsible for regulating many aspects of plant growth and development throughout the life cycle of plants, including leaf expansion, stem elongation, flowering, seed germination, and the transition from vegetative growth to flowering (Yamaguchi 2008; Peters 2013). Plants defective in GA biosynthesis display typical GA-deficient phenotypes, characterized by prolonged germination dormancy; small, dark green leaves; inhibited root growth; defective flowering; reduced seed production; and male sterility (Sun and Gubler 2004; Otani et al. 2013). Therefore, it is important to maintain optimal endogenous GA levels in plants. GA levels are precisely regulated by several mechanisms, including the transcriptional regulation of genes associated with synthesis and deactivation of the bioactive GA forms. The formation of C19-GA, through the oxidation of C20-GA, and the 3β-hydroxylation of C19-GA are important steps in the biosynthesis pathway of bioactive GA; GA20-oxidase and GA3-oxidase catalyze these respective steps in GA biosynthesis (Olszewski et al. 2002; Sun 2008). The deactivation of the bioactive GA forms GA1 and GA4 and their respective precursors, GA9 and GA20, are catalyzed by GA2-oxidases (GA2ox) (Thomas et al. 1999). Many genes encoding enzymes that catalyze GA biosynthesis and catabolism have been identified (Hedden and Thomas 2012; Plackett et al. 2012; Nishii et al. 2014). However, GA biosynthetic genes have received much more attention than catabolic genes. Functional analysis of GA2ox revealed that catabolic genes are also important in the control of GA levels (Zhou et al. 2010; El-Sharkawy et al. 2012).
GA 2-oxidase in Arabidopsis, rice (Oryza sativa L.), and poplar (Populus trichocarpa Torr. & Gray ex Hook.) are encoded by small gene families (Hedden and Thomas 2012; Daviere and Achard 2013). GA2oxs were first identified by screening cDNA expression libraries for 2β-hydroxylase activity (Thomas et al. 1999; Martin et al. 1999). Five Arabidopsis GA2ox genes have been identified based on sequence homology, and their encoded protein activities have been confirmed (Hedden and Phillips 2000). The physiological functions of these genes have been studied in a variety of plant species, and it has been demonstrated that they are active against bioactive GAs and their immediate precursors. The overexpression of AtGA2ox7 and AtGA2ox8 in transgenic tobacco (Nicotiana tabacum) leads to a dwarf phenotype (Schomburg et al. 2003). Similar results have been obtained in other transgenic plants overexpressing GA 2-oxidases from rice (O. sativa) and poplar (Populus spp.) (Busov et al. 2003; Huang et al. 2010). GA2ox7 and GA2ox8 were identified from Arabidopsis, and they catabolize only the non-bioactive C20-GAs (Schomburg et al. 2003). Phylogenetic analysis has shown that the GA 2-oxidases can be divided into three classes (Yamaguchi 2008). Members of class III include GA2ox7 and GA2ox8, distinguished by their preference for C20-GAs. The biological-structural division between classes I and II remains less clear. To our knowledge, no GA2ox genes have yet been cloned from camellias, and their developmental functions and regulation during plant development are completely unknown.
In this study, we isolated three putative GA 2-oxidase genes (ClGA2ox1, ClGA2ox2, and ClGA2ox3) from Camellia lipoensis using reverse transcription PCR (RT-PCR) and the rapid amplification of cDNA ends (RACE) method. Based on the expression of ClGA2ox1-3, we suggest that the phenotypes might be largely attributed to individual GA 2-oxidases, in some cases with a specific expression pattern in plant development. To our knowledge, this is the first report of a GA 2-oxidase gene isolated from camellia plants. Moreover, the ClGA2ox1 gene not only induces dwarf phenotypes but also affects flower size, suggesting that this gene might provide new tools for research and the genetic engineering of ornamental plant stature.
Materials and Methods
Plant Material
C. lipoensis Chang et Xu is an evergreen shrub and was cultivated in a greenhouse for this study. Tobacco (N. tabacum) leaves were harvested from greenhouse-grown plants, surface sterilized by soaking in 0.1 % (w/v) HgCl2 for 5 min, and subsequently rinsed three times in sterile water. The leaves were cut into pieces through the midrib to obtain 0.5 × 0.5 cm2 rectangular explants. The tobacco explants were placed on a solid regeneration medium, based on Murashige and Skoog (MS) medium (Murashige and Skoog 1962), supplemented with 30 g/L sucrose, 6.0 g/L plant agar, 0.5 mg/L 6-benzyladenine (BA), and 0.05 mg/Lα-naphthaleneacetic acid (NAA). The pH of the medium was adjusted to 5.7 before autoclaving. In vitro shoot regeneration of tobacco was performed in 8-cm diameter Petri dishes. After 4 to 5 weeks, the tobacco shoots were harvested and transferred to a root-inducing MS medium containing half-strength MS microelements supplemented with 30 g/L sucrose and 6.0 g/L plant agar. Shoot regeneration and root induction were performed in a growth chamber at 25 °C with a 16-h photoperiod.
Agrobacterium tumefaciens Strains
A. tumefaciens strain EHA105 was used for transformation. The T-DNA region of the binary vector pCAMBIA1300 was under the control of the cauliflower mosaic virus (CaMV) 35S promoter, and the hygromycin phosphotransferase (HPT) gene was under the control of the CaMV 35S promoter.
Isolation and Sequencing of Camellia GA2ox cDNA Sequences
Total RNA was extracted from leaf of C. lipoensis using the RN38 EASYspin plus plant RNA Kit (Aidlab biotechnologies Co., Beijing, China) according to the manufacturer’s protocol. The full-length ORF of the C. lipoensis GA2-oxidase gene was cloned through reverse transcription PCR (RT-PCR), and the rapid amplification of cDNA ends (RACE). Three pairs of degenerate primers (Table S1) were designed from the conserved regions of GA2ox orthologues. The isolated fragment was cloned in the vector pGEM-T Easy (Promega, WI, USA), sequenced, and analyzed using BLAST (Altschul et al. 1997). Extension of the partial cDNA clone was carried out using a SMARTER RACE cDNA Amplification Kit (Takara). Alignment of the camellia GA2ox predicted sequence, and the neighbor-joining tree construction were performed using the ClustalW program (Thompson et al. 1994).
Vector Construction and Plant Transformation
The ClGA2ox1, ClGA2ox2, and ClGA2ox3 gene sequences were cloned separately in front of the 35S constitutive promoter within a pCAMBIA1300 vector. The T-DNA contains a hygromycin gene for selection in plants under a duplicated 35S promoter. The vector also contains an NPTII gene for bacterial selection. The modified construct was cloned in the Agrobacterium tumefaciens strain EHA105.
A bacterial suspension was prepared from a 200-μL frozen glycerol culture in 30 mL of LB medium (10 g/L bacto-tryptone, 5 g/L bacto-yeast extract, 10 g/L NaCl, pH 7.0) supplemented with 50 mg/L kanamycin and 25 mg/L rifampicin and cultured for 24 h at 28 °C. A bacterial suspension with an OD600 = 0.5 was pelleted at 5000 rpm for 10 min at 4 °C. The supernatant was discarded, and the pellet was re-suspended in 20 mL of half-strength liquid MS medium; the subsequent solution was supplemented with 200 μM acetosyringone. Young leaf explants from in vitro-grown tobacco plants were cut into 5 × 5 mm squares and submerged in the bacterial solution for 5 min. The explants were blotted dry on sterile filter paper and placed on MS medium supplemented with 100 μM acetosyringone for 2 days in the dark. Then, the explants were transferred to a shoot-inducing medium supplemented with 0.5 mg/L 6-benzylaminopurine, 0.05 mg/L NAA, 20 mg/L hygromycin, and 200 mg/L Timentin. The explants were transferred to fresh media every 2 weeks. Regenerated shoots were cut from the explants and transferred to root-inducing medium (MS, 0.05 mg/L NAA, 20 mg/L hygromycin, and 200 mg/L Timentin). After rooting, plantlets were planted in a greenhouse.
Reverse Transcription Polymerase Chain Reaction and Southern Blot
For reverse transcription polymerase chain reaction (RT-PCR) verification of ClGA2ox expression, total RNA was isolated from all transgenic lines of tobacco. The RNA was isolated from ground plant material using the RN38 EASYspin plus plant RNA Kit (Aidlab biotechnologies Co., Beijing, China) according to the manufacturer’s protocol, which also removed the genomic DNA. The RNA concentration and quality were measured using a NanoDrop 2000 (Thermo Fisher Scientific Inc., Waltham, USA). Reverse transcription was performed using the First Strand cDNA Synthesis Kit (Fermentas) with 5 μg of RNA. RT-PCRs were prepared using 10 ng of cDNA in a final volume of 25 μL containing 0.25 μM each ClGA2ox primers (Table S2), 0.15 mM each dNTP, 2 mM MgCl2, and 2 U of rTaq polymerase (Takara). The reaction consisted of 3 min at 94 °C, followed by 35 cycles of 30 s at 94 °C, 30 s at 58 °C for 30 s, and 80 s at 72 °C. A final 10-min extension step at 72 °C was applied. PCR amplification was conducted in a thermocycler (ABI PCR system 9700, USA).
Genomic DNA was isolated using the CTAB method (Stewart and Via 1993). For Southern blot hybridization, 25 μg of DNA from the control and transgenic plants was digested using 50 units of BamHI (Takara) for 24 h. The resulting DNA fragments were separated on agarose gels and transferred to a membrane (Sriskandarajah et al. 2007). A DIG-labeled (digoxigenin) probe was prepared by PCR according to the manufacturer’s protocol (Roche Applied Science Co., Germany) using the ClGA2ox1, ClGA2ox2, and ClGA2ox3 gene primer pairs (Table S4). The hybridization and post-hybridization and an estimation of the visualized fragments were also performed following the protocol.
Quantitative Real-Time Polymerase Chain Reaction Analysis
Total RNA was isolated using the RN38 EASYspin plus plant RNA kit. The gene expression levels of various organs in C. lipoensis were determined by qRT-PCR using specific primer sets (Table S3), and ClActin (actin gene of C. lipoensis, GenBank accession number: KF366912) was used as an internal standard. Transcripts of transgenes in young leaves of transgenic plants were also quantified by quantitative real-time polymerase chain reaction (qRT-PCR) analysis using primer sets (Table S3), and NicActin (actin gene of N. tabacum, GenBank accession number: AB158612) was used as an internal control.
Quantitative real-time PCR analysis was performed using the Applied Biosystems 7300 Real Time PCR System. One microgram of total RNA was reverse transcribed using a PrimeScript RT reagent kit (Takara). The qRT-PCR reaction was performed using a SYBR premix Ex Taq II kit (Takara). Each PCR was performed in three replicates under the following conditions: 30 s at 95 °C, 31 s at 60 °C, and plate read (detection of fluorescent product) for 40 cycles. To characterize the PCR products, a melting curve analysis was performed by slowly raising the temperature from 60 to 95 °C, with fluorescence data obtained at 0.5 °C intervals. Quantifications of each cDNA sample were made in triplicate to ensure consistent results from the three separately prepared RNA samples. The relative amounts of target gene transcripts were calculated using the comparative cycle threshold method, and the results were normalized to the internal control.
Phenotypic Characteristics of Transgenic Plants
Transgenic plants (generation 1) and non-transgenic plants were transplanted to pots and kept under 16 h photoperiod conditions at 24 °C/18 °C (day/night). After 2 months of cultivation, morphological characterizations (height, internode length, leaf length, and width) were performed during the flowering season. The mean plant development values of three randomly selected plants were measured.
Plant Treatments with GA3 and Paclobutrazol
The transgenic lines (generation 1) were cultivated in MS medium containing 100 μM GA3. Non-transgenic plant was treated with 2 μM of a GA biosynthesis inhibitor, paclobutrazol (PAC). Changes in growth were observed and photographed 2 weeks after treatment. Experiments were performed three times.
Analysis of the Endogenous GA Level
The procedures for extraction, purification, and quantification of endogenous GAs were performed as previously described (El-Sharkawy et al. 2012). Aliquots from apical shoots including the three youngest developed leaves from plants just before flowering were harvested and weighted for later analysis. The concentrations of GAs in the extracts were determined by high-performance liquid chromatography/mass spectrometry (HPLC/MS). All experiments were carried out in three independent replicates.
Results
Isolation of ClGA2ox1-3
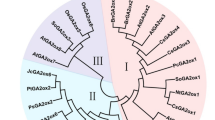
Sequence analysis of the PCR product showed that the fragment encoded a part of the GA2ox gene family. Three full-length cDNAs of GA2ox were obtained, containing open reading frames of 999, 996, and 1002 bp, and the cloned genes were subsequently named ClGA2ox1, ClGA2ox2, and ClGA2ox3, respectively (GenBank accession numbers: KJ502290, KJ502289, and KJ502291, respectively). The ClGA2ox amino acid sequences were aligned with those of other reported GA2oxs. The deduced amino acid sequences of ClGA2ox1-3 share 70, 59, and 54 % identity with P. trichocarpa PtGA2ox1 (Niu et al. 2013), A. thaliana AtGA2ox1 (Lee et al. 2014), and O. sativa OsGA2ox3 (Sakai et al. 2003), respectively. An alignment of the deduced amino acid sequences of ClGA2ox1-3 with other GA2ox sequences (Figure S1) showed that they contain characteristic sequences that are conserved within the GA 2-oxidase class, including the 2-oxoglutarate-binding and the iron-binding sites (Thomas et al. 1999), confirming that ClGA2ox1-3 belongs to this class of enzymes. A phylogenetic tree was constructed using the deduced amino acid sequences of ClGA2oxs with other plant GA2oxs (Fig. 1). The GA 2-oxidase family was distributed among three different clades.
Phylogenetic relationship of ClGA2ox1-3 with other plant GA2oxs based on the full-length amino acid sequence. The GenBank accession numbers for each protein are ClGA2ox1 (KJ502290), ClGA2ox2 (KJ502289), ClGA2ox3 (KJ502291), Populus trichocarpa PtGA2ox1 (EEE85235), PtGA2ox2 (EEE80803), PtGA2ox3 (EEE86215), PtGA2ox6 (EEE97401), PtGA2ox7 (EEE99275), Arabidopsis thaliana AtGA2ox1 (CAB41007), AtGA2ox2 (CAB41008), AtGA2ox3 (CAB41009), AtGA2ox4 (AEE32235), AtGAox6 (AEE27425), AtGAox7 (AEE32606), AtGAox8 (AEE84420), Oryza sativa OsGAox1 (BAB40934), OsGA2ox2 (BAC167451), OsGA2ox3 (BAC16752), OsGAox4 (BAF17950), OsGAox5 (BAF20604), and OsGAox6 (BAF15255). The scale value of 0.1 indicates 0.1 amino acid substitutions per site
To elucidate the roles of each GA2ox gene in various organs of C. lipoensis, total RNA was extracted from the apex, leaves, stems, roots, and seeds. To assess the function of each of the ClGA2ox1-3 genes in plant development, we analyzed their developmental expression profiles using qRT-PCR. Figure 2 shows that ClGA2ox1-3 transcripts could be detected at all developmental stages analyzed, indicating GA activity throughout plants during many developmental processes (Fleet and Sun 2005). ClGA2ox1 was the most highly expressed gene in most samples, with the exception of young and mature leaves. The expression levels of the other two genes increased with leaf development. ClGA2ox3 was dominant early in development, with high levels found in roots, and ClGA2ox2 transcripts reached the highest levels in mature leaves. This data set showed that the ClGA2ox1 transcript was more abundant in the stem and shoot apex than in other tissues, ClGA2ox2 was more abundant in mature leaves, and ClGA2ox3 was more abundant in roots. Additionally, ClGA2ox1 was the most highly expressed gene at most developmental stages.
Relative expression levels of the ClGA2ox1-3 genes measured by qRT-PCR. Expression levels in the following organs from 3-year-old Camellia lipoensis. A apex, B young leaf, C medium-aged leaf (fully elongated), D mature leaf, E internode, F roots, G seed. Relative amounts of each gene are normalized to ClActin. Values are means ± SE of three technical replicates
Southern Blot Analysis and qRT-PCR Analysis for ClGA2ox1-3 Gene Expression
RT-PCR analysis revealed that ClGA2ox1-3 was overexpressed in transgenic tobacco plants (Fig. 3). Moreover, Southern blot analysis showed that the nine transgenic lines were derived from independent plants (Fig. 4). All of the transgenic lines had one copy of the transgene. The control plants yielded no fragments. ClGA2ox1-3 transcripts were detected in all of the transgenic lines, and no transcripts were detected in non-transgenic plants according to qRT-PCR analysis (Fig. 5). Among the three transgenic lines with the ClGA2ox3 gene, two severely dwarfed lines (G3-1 and G3-3) showed relatively high expression levels, whereas the moderately dwarfed line (G3-2) showed a relatively low expression level. The transgenic lines with the ClGA2ox1 or ClGA2ox3 gene showed relatively higher expression levels than the ClGA2ox2 gene, inducing dwarfism.
RT-PCR of the transgenic lines using ClGA2ox1-3 specific sequence as the forward primer and the reverse primer (Table S2). a Transgenic lines with the ClGA2ox1 gene. b Transgenic lines with the ClGA2ox2 gene. c Transgenic lines with the ClGA2ox3 gene. M DNA molecular marker, + positive control amplifying plasmid, WT non-transgenic plants
Southern blot of transgenic plants. a Transgenic lines with the ClGA2ox1 gene. b Transgenic lines with the ClGA2ox2 gene. c Transgenic lines with the ClGA2ox3 gene. The genomic DNA was digested with BamHI, and a DIG-labeled probe was formed using the primer sets (Table S3). M DNA molecular weight marker, + positive plasmid, WT represents the non-transgenic plant
Quantitative real-time PCR analysis of ClGA2ox1-3 transcripts in young leaves of the control plant and transgenic plants. The expression levels of each gene were relative to NicActin. WT non-transgenic plant. No expression of ClGA2ox1-3 gene was detected in WT (ND not detected). G1-1~G1-3 expression level of the ClGA2ox1 gene in different transgenic lines. G2-1~G2-3 expression level of the ClGA2ox2 gene in different transgenic lines. G3-1~G3-3 expression level of the ClGA2ox3 gene in different transgenic lines
Production and Morphological Characterization of Transgenic Plants
All of the transgenic plants were morphologically distinguished from the control plants (Table 1). The transgenic plants with ClGA2ox1 or ClGA2ox3 genes exhibited dwarf phenotypes due to their decreased internode length. Both leaves and internode lengths in the transgenic lines were significantly shorter than the control, producing smaller, darker green, and rounder leaves compared with the control. Transgenic lines with the ClGA2ox3 gene could be classified into two classes according to the degree of dwarfism: one line (G3-2) was moderately dwarfed, and two lines (G3-1 and G3-3) were severely dwarfed. G3-2 produced normal flowers, but the severely dwarfed lines (G3-1 and G3-3) produced no visible flower buds. All of the transgenic lines with the ClGA2ox1 gene produced smaller flowers compared with the control (approximately half the size of the control flowers). The ClGA2ox2 gene had little effect on the morphological characterization of transgenic plants but did result in darker green leaves (Fig. 6).
The phenotypes of transgenic plants. a Phenotypes of the second generation transgenic plants grown for 4 weeks. b Leaves from transgenic plants grown for 50 days. c Flowers from transgenic plants. G1, G2, and G3 represent transgenic plants with the ClGA2ox1, ClGA2ox2, and ClGA2ox3 genes, respectively. WT represents the non-transgenic plant
Dwarfism is Reversed by GA3 Application
GA3 is a bioactive form of GA, which cannot be metabolized by GA2ox (Sakamoto et al. 2001). Therefore, if the transformants have low levels of bioactive GA due to a high rate of inactivation by GA2ox, the application of GA3 should be able to rescue the dwarf phenotype. The exogenous GA3 treatment increased the shoot length and leaf size of both transgenic lines. Two weeks after GA3 application, the phenotypes of the transgenic lines were restored, similar to the control (Fig. 7). The results demonstrated that the GA signaling pathway might function normally in transgenic plants. Therefore, morphological changes in transgenic plants may result from a decrease in the endogenous level of bioactive GA.
Effects of GA3 on plant phenotype of transgenic plants. Two-week-old plants cultivated in MS medium containing 100 μM GA3. GA 3 (+) and GA 3 (−) represent treatments with and without GA3, respectively. G1, G2, and G3 represent transgenic plants with the ClGA2ox1, ClGA2ox2, and ClGA2ox3 genes, respectively. WT represents the non-transgenic plant. Bar represents 2 cm
Analysis of the Endogenous GA Level
The level of bioactive GA4 and GA1 decreased in both transgenic plants and control plants treated with PAC (Fig. 8). The GA4 concentration in the transgenic plants overexpressing the ClGA2ox3 gene reduced to 42.9 % of the controls, similar to that of control plants treated with PAC, and the GA1 content reduced to 38.7 %. The level of GA34 increased in the transgenic plants. No metabolism of GA8 was observed in the transgenic plants. There were no significant changes in the GA3 concentration between transgenic plants and controls.
Concentration of endogenous GAs in apical shoots. WT represents the non-transgenic plant. WT-PAC represents the control plant treated with paclobutrazol. G1, G2, and G3 represent transgenic plants with the ClGA2ox1, ClGA2ox2, and ClGA2ox3 genes, respectively. Values represent the mean ± standard error of three replicates. Asterisks undetectable GAs
Discussion
In this study, we isolated three full-length cDNA clones encoding GA 2-oxidase from Camellia. The predicted proteins of these cDNAs contain the regions typically conserved in plant 2-oxoglutarate-dependent dioxygenases. The sequences are closely related to other known GA 2-oxidases and share 70 % amino acid identity with PtGA2ox1. A phylogenetic tree was constructed, and the GA 2-oxidase family was distributed among three different clades. The class I and class II enzymes catabolize C19-GAs, and class III enzymes catabolize C20-GAs (Yamaguchi 2008). The phylogenetic tree revealed that ClGA2oxs belong to class I, with AtGA2ox1, AtGA2ox2, AtGA2ox3, OsGA2ox3, and OsGA2ox4.
GA 2-oxidases are encoded by multigene families. In contrast to the 20-oxidase and 3β-hydroxylase genes, which show tissue-specific expression (Phillips et al. 1995), ClGA2ox1-3 showed different patterns of expression. The transcript abundance of the ClGA2ox1 gene in the internode and apex was higher than that of ClGA2ox2-3 genes. The ClGA2ox2 gene was found to be more abundant in mature leaves, and ClGA2ox3 was more abundant in roots. Thus, future characterization of the camellia GA2ox gene family and its regulatory mechanisms and functional roles should enable more specific manipulation of GA-related traits.
The qRT-PCR analysis showed that ClGA2ox1 was expressed at a high level in the internodes and apex, ClGA2ox2 was expressed at a high level in the mature leaves, and ClGA2ox3 was highly expressed in roots and at relatively low levels in other organs compared with ClGA2ox1-2. It can be concluded that diverse GA2oxs may function quite differently throughout a plant’s growth and development. The overexpression of a GA2ox that mainly functions in vegetative production may cause dwarf plants with normal flowering and seed production, such as with ClGA2ox1. The overexpression of a GA2ox that mainly functions in old organ metabolism may have little effect on dwarfism induction, such as with ClGA2ox2. The overexpression of a GA2ox that is expressed at a low level throughout plant growth may cause severe dwarfism, such as with ClGA2ox3.
GA is known to stimulate stem elongation in many plant species, and GA treatment causes an increase in the number of vegetative stem internodes that elongate upon bolting and in the number of flowers, resulting in an overall increase in plant height (Rieu et al. 2008). The overexpression of ClGA2ox1 or ClGA2ox3 in N. tabacum had a profound effect on plant growth and development, and resulted in reduced growth and later flowering (Table 1). GA4 and GA1 are thought to be the major bioactive GAs that existed in most species. It has been confirmed that GA2ox can catalyze GA4 and GA1 to GA34 and GA8, respectively. The analysis of the endogenous GAs showed a reduced GA4 and GA1 level in transgenic plants; the GA34 content increased compared with the wild plants. A similar severe dwarf phenotype was produced when the control tobacco was treated with the GA biosynthesis inhibitor PAC. The endogenous GA4 and GA1 level was reduced and was similar to that of transgenic plants. The GA-deficient phenotype of transgenic plants could be restored by applying GA3 (Fig. 7). This result strongly supports the hypothesis that dwarf phenotypes are a result of the deficiency of bioactive GAs. Our findings also provide a potential method for controlling the growth of transgenic plants overexpressing GA2ox genes via the exogenous application of GA3 during horticultural manipulation. For example, managers might speed early growth using GA application to reach a desired stature. Once the GA application ceases, the transgenic plant overexpressing GA2ox will resume slower growth.
Semi-dwarfism is one of the most valuable traits in horticulture breeding because semi-dwarf plants are more resistant to damage by wind and rain. The overexpression of GA 2-oxidase is an easy way to reduce the bioactive GA levels in transgenic plants; however, the ectopic expression of most GA2oxs caused severe dwarfism and seed abortion in various plant species (Singh et al. 2002; Biemelt et al. 2004; Hedden and Thomas 2012). ClGA2ox1 and ClGA2ox3 in N. tabacum caused dwarf plants to flower late and produce seeds normally. Notably, flowers from transformants overexpressing ClGA2ox1 were half the size of the controls (Fig. 6c), and transgenic plants with ClGA2ox2 or ClGA2ox3 retained normal flower size; thus, ClGA2ox1 may play a specific role in flower development. Few studies regarding GA2ox affecting flower size have been reported. This gene is very useful in horticulture breeding and not only induces dwarfism but also regulates flower development.
The manipulation of ornamental plant stature has long been a major goal in horticulture. Camellias are typically taller in stature than is desirable in potted flower production. Consequently, plant growth retardants are widely applied to inhibit plant elongation, often acting by disrupting the synthesis of GAs (Rademacher 2000). However, plant retardants require repeated applications of chemicals that are not only expensive but also hazardous to workers and the environment. Genetic manipulation may provide an alternative approach to the use of plant growth retardants because plant dwarf phenotypes can be achieved by upregulating genes encoding enzymes involved in GA catabolism (Hedden and Phillips 2000).
In conclusion, the present study shows that the overexpression of ClGA2ox1 or ClGA2ox3 in transgenic tobacco results in a dwarf phenotype. Whereas flowers from transgenic plants with 35S:ClGA2ox1 were approximately half the size of the control plants, and produced seeds normally. Flowers were not observed in transgenic lines with a high expression of ClGA2ox3, whereas the transgenic plants with lowly expressed ClGA2ox3 flowered normally. These results demonstrate that the overexpression of ClGA2ox1 may be a useful method for obtaining compact growth and small flower ornamental plants. The compact ornamental plants with normal flowers may be induced by a low expression of ClGA2ox3. The effects of transformations using constructs in which ClGA2ox3 is controlled by a weak expression activity or tissue-specific promoter will be investigated in future studies.
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Biemelt S, Tschiersch H, Sonnewald U (2004) Impact of altered gibberellin metabolism on biomass accumulation, lignin biosynthesis, and photosynthesis in transgenic tobacco plants. Plant Physiol 135:254–265
Busov VB, Meilan R, Pearce DW, Ma C, Rood SB, Strauss SH (2003) Activation tagging of a dominant gibberellin catabolism gene (GA 2-oxidase) from poplar that regulates tree stature. Plant Physiol 132:1283–1291
Daviere JM, Achard P (2013) Gibberellin signaling in plants. Development 140:1147–1151
El-Sharkawy I, El Kayal W, Prasath D, Fernandez H, Bouzayen M, Svircev AM, Jayasankar S (2012) Identification and genetic characterization of a gibberellin 2-oxidase gene that controls tree stature and reproductive growth in plum. J Exp Bot 63:1225–1239
Fleet CM, Sun T-P (2005) A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr Opin Plant Biol 8:77–85
Hedden P, Phillips AL (2000) Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci 5:523–530
Hedden P, Thomas SG (2012) Gibberellin biosynthesis and its regulation. Biochem J 444:11–25
Huang J, Tang D, Shen Y, Qin B, Hong L, You A, Li M, Wang X, Yu H, Gu M, Cheng Z (2010) Activation of gibberellin 2-oxidase 6 decreases active gibberellin levels and creates a dominant semi-dwarf phenotype in rice (Oryza sativa L.). J Genet Genomics 37:23–36
Lee D, Lee I, Kim K, Kim D, Na H, Lee I-J, Kang S-M, Jeon H-W, Le P, Ko J-H (2014) Expression of gibberellin 2-oxidase 4 from Arabidopsis under the control of a senescence-associated promoter results in a dominant semi-dwarf plant with normal flowering. J Plant Biol 57:106–116
Martin DN, Proebsting WM, Hedden P (1999) The SLENDER gene of pea encodes a gibberellin 2-oxidase. Plant Physiol 121:775–781
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–497
Nishii K, Ho M-J, Chou Y-W, Gabotti D, Wang C-N, Spada A, Möller M (2014) GA2 and GA20-oxidase expressions are associated with the meristem position in Streptocarpus rexii (Gesneriaceae). Plant Growth Regul 72:123–140
Niu S, Li Z, Yuan H, Fang P, Chen X, Li W (2013) Proper gibberellin localization in vascular tissue is required to regulate adventitious root development in tobacco. J Exp Bot 64:3411–3424
Olszewski N, Sun TP, Gubler F (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell 14:61–80
Otani M, Meguro S, Gondaira H, Hayashi M, Saito M, Han D-S, Inthima P, Supaibulwatana K, Mori S, Jikumaru Y, Kamiya Y, Li T, Niki T, Nishijima T, Koshioka M, Nakano M (2013) Overexpression of the gibberellin 2-oxidase gene from Torenia fournieri induces dwarf phenotypes in the liliaceous monocotyledon Tricyrtis sp. J Plant Physiol 170:1416–1423
Peters RJ (2013) Gibberellin phytohormone metabolism. In: Rohmer M, Bach TJ (eds) Isoprenoid synthesis in plants and microorganisms. Springer, New York, pp 233–249
Phillips AL, Ward DA, Uknes S, Appleford N, Lange T, Huttly AK, Gaskin P, Graebe JE, Hedden P (1995) Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol 108:1049–1057
Plackett ARG, Powers SJ, Fernandez-Garcia N, Urbanova T, Takebayashi Y, Seo M, Jikumaru Y, Benlloch R, Nilsson O, Ruiz-Rivero O, Phillips AL, Wilson ZA, Thomas SG, Hedden P (2012) Analysis of the developmental roles of the Arabidopsis gibberellin 20-oxidases demonstrates that GA20ox1, −2, and −3 are the dominant paralogs. Plant Cell 24:941–960
Rademacher W (2000) Growth retardants: effects on gibberellin biosynthesis and other metabolic pathways. Annu Rev Plant Biol 51:501–531
Rieu I, Ruiz-Rivero O, Fernandez-Garcia N, Griffiths J, Powers SJ, Gong F, Linhartova T, Eriksson S, Nilsson O, Thomas SG, Phillips AL, Hedden P (2008) The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J 53:488–504
Sakai M, Sakamoto T, Saito T, Matsuoka M, Tanaka H, Kobayashi M (2003) Expression of novel rice gibberellin 2-oxidase gene is under homeostatic regulation by biologically active gibberellins. J Plant Res 116:161–164
Sakamoto T, Kobayashi M, Itoh H, Tagiri A, Kayano T, Tanaka H, Iwahori S, Matsuoka M (2001) Expression of a gibberellin 2-oxidase gene around the shoot apex is related to phase transition in rice. Plant Physiol 125:1508–1516
Schomburg FM, Bizzell CM, Lee DJ, Zeevaart JAD, Amasino RM (2003) Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 15:151–163
Singh DP, Jermakow AM, Swain SM (2002) Gibberellins are required for seed development and pollen tube growth in Arabidopsis. Plant Cell 14:3133–3147
Spielmeyer W, Ellis MH, Chandler PM (2002) Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc Natl Acad Sci U S A 99:9043–9048
Sriskandarajah S, Mibus H, Serek M (2007) Transgenic Campanula carpatica plants with reduced ethylene sensitivity. Plant Cell Rep 26:805–813
Stewart CN Jr, Via LE (1993) A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. BioTechniques 14:748–750
Sun T-P (2008) Gibberellin metabolism, perception and signaling pathways in Arabidopsis. The Arabidopsis Book/American Society of Plant Biologists 6:e0103
Sun T-P, Gubler F (2004) Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol 55:197–223
Thomas SG, Phillips AL, Hedden P (1999) Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc Natl Acad Sci 96:4698–4703
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Yamaguchi S (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59:225–251
Zhou P, Ren B, Zhang XM, Wang Y, Wei CH, Li Y (2010) Stable expression of rice dwarf virus Pns10 suppresses the post-transcriptional gene silencing in transgenic Nicotiana benthamiana plants. Acta Virol 54:99–104
Acknowledgments
This work was supported by the National Science & Technology Pillar Program during the 12th Five-year Plan Period (No. 2012BA01B0703) and International Cooperation in Science and Technology Project (No. 2011DFA30490). We also acknowledge Breeding New Flower Varieties Program of Zhejiang Province (2012C12909-6) and State Forestry Administration of the People’s Republic of China, Project 948 (2014-4-16).
Author information
Authors and Affiliations
Corresponding author
Additional information
Zheng Xiao and Ruipeng Fu contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1
Comparison of the deduced amino acid sequences of ClGA2ox1-3 with other GA2oxs. The closed triangles indicate the three amino acid residues forming the iron-binding site. The asterisks show the putative 2-oxoglutarate binding site. The GenBank accession numbers for each protein are ClGA2ox1 (KJ502290), ClGA2ox2 (KJ502289), ClGA2ox3 (KJ502291), AtGA2ox1 (CAB41007), OsGA2ox3 (BAC16752) and PtGA2ox1 (EEE85235). (GIF 182 kb)
Table S1
Primers used for amplifying GA2ox gene fragment (DOC 30 kb)
Table S2
Primers used for RT-PCR analysis (DOC 31 kb)
Table S3
Primers used for quantitative real-time PCR (DOC 29 kb)
Table S4
Detective primers for southern blot (DOC 28 kb)
Rights and permissions
About this article
Cite this article
Xiao, Z., Fu, R., Li, J. et al. Overexpression of the Gibberellin 2-Oxidase Gene from Camellia lipoensis Induces Dwarfism and Smaller Flowers in Nicotiana tabacum . Plant Mol Biol Rep 34, 182–191 (2016). https://doi.org/10.1007/s11105-015-0917-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-015-0917-3