Abstract
Assessment of genetic diversity is an essential component in germplasm characterization and utilization. Molecular markers serve as a valuable tool to assess the genetic variation and germplasm identification, which play a key role for faba (Vicia faba L.) bean breeding. In this study, we analyzed the genetic diversity of faba bean accessions based on simple sequence repeats (SSRs) and morphological traits. Forty-six faba bean accessions, originating from different countries and from the ICARDA breeding program, were evaluated by using 15 morphological and agronomic traits and 17 simple sequence repeat (SSR) loci. Significant differences among accessions for the 15 morphological descriptors were observed. Analysis by SSR markers showed a high genetic diversity among the accessions: All SSRs showed polymorphism, and 101 alleles were revealed for all accessions. The number of alleles at each locus ranged from 2 to 10, with an average of 5.94 alleles per marker, and the polymorphic information content (PIC) values ranged from 0.38 to 0.84 with a mean of 0.69. Expected heterozygosity (He), observed heterozygosity (Ho), unbiased expected heterozygosity (UHe), and Shannon’s information index (I) showed existence of high genetic variation between accessions from different pedigree. Analyses of genetic distance of the accessions separated the accessions into two groups and seven and five subgroups according to morphological and SSR analysis, respectively. Principal component analysis (PCA) of the SSR markers showed that the first two principal components (PCs) explained a total of 43.90 % of the genetic variation and allowed to distinguish three groups of accessions. Regardless of the method of analysis, Tunisian cultivars are grouped together. For the rest of the accessions, the geographical origin did not seem to be the main factor for structuring the variability of the studied accessions. Elite accessions from the ICARDA faba bean program differed from others and clustered together. The results obtained suggested that the faba bean microsatellite markers can be used to efficiently distinguish faba bean genotypes and to estimate their genetic diversity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Faba bean (Vicia faba L.) is one of the ancient cultivated crops widely believed to have originated in the Mediterranean-West Asia region (Tanno and Willcox 2006). In the Mediterranean countries, this crop is used as source of protein in both human and animal nutrition because of the high nutritional value of its seeds (Mejri et al. 2012). Faba bean includes 13–14 % of cell wall slightly lignified, 3 % of soluble sugars, 6.5 % of oligosaccharides, 27–34 % of proteins, and around 45 % of starch of total dry matter (Ayed 2011). Besides being an important food crop, it contributes to feed and fodder supply for livestock and affects positively the soil productivity for the cereal crops grown in rotation through biological nitrogen fixation (Duc 1997). Faba bean is a diploid with 2n = 2x = 12 chromosomes, possesses one of the largest genomes among crop legumes (~13,000 Mb), and is partially cross-pollinated ranging from 4 to 84 % (Ellwood et al. 2008). Despite the partial allogamous status of this crop, Perrino et al. (1991) suggested that the genetic variability of faba bean species is quite large. Although most researchers refer to four botanical varieties, namely, Vicia faba major, Vicia faba equina, Vicia faba minor, and Vicia faba paucijuga, in practice, a continuous variation for most seed, pod, plant, floral biology, and chemical and physiological traits, and also tolerance to several biotic and abiotic stresses, has been observed (Duc et al. 2010). North Africa is one of the top-producing regions of the faba bean in the world (Aouar-sadli et al. 2008). The average seed yield of faba bean in Africa (1.3 t/ha) is below world average (1.5 t/ha), while the average yields obtained in Asia (1.7 t/ha) and Europe (2.2 t/ha) are higher. Exceptionally high yields are obtained in Egypt and Sudan where the crop is irrigated (3.0 and 2.3 t/ha, respectively), whereas the national average yield of faba bean in Tunisia is low (0.7–0.8 t/ha) and characterized by wide fluctuations (Kharrat et al. 1991). The instability of the yield is mainly due to the lack of suitable genetic material with stable yield and reasonable resistance/tolerance to the prevalent diseases, pests, and abiotic stress. High yield and resistance/tolerance to both biotic and abiotic stresses are the prime objectives across faba bean breeding programs. It is therefore imperative to develop new cultivars that are able to overcome the biotic and abiotic factors responsible for low faba bean productivity in Tunisia. The introduction of new genetic resources into breeding programs may further improve agronomic traits and increase genetic variability and thereby lead to the development of high-yield cultivars (Hwang et al. 2008). In this context, a proper characterization of the genetic variation in the available local and Mediterranean faba bean germplasm resources is necessary for further improvement of crop yield and to impart tolerance to biotic and abiotic stress. Using morphological and agronomic traits for studying genetic diversity was previously achieved for many plants including faba bean (Mustafa 2007). Terzopoulos and Bebeli (2008) suggested that using only morphological/agronomic traits is not adequate for the development of gene pools since either these traits are influenced by environmental factors and stage of plant development or they reveal only limited variation. The rapid development of molecular markers has provided new tools for evaluating germplasm and assessing the genetic diversity and population structure of plant species (Baranski et al. 2012; Boczkowska et al. 2012; Cao et al. 2012; Mir et al. 2012; Roy et al. 2012; Sardos et al. 2012; Wang and Li 2012). Different types of markers such as isozymes, random amplification of polymorphic DNAs (RAPDs), restriction fragment length polymorphisms (RFLPs), target region amplification polymorphisms (TRAPs), sequence-specific amplification polymorphisms (SSAPs), and amplified fragment length polymorphisms (AFLPs) have already been used for the assessment of genetic variability and diversity and to perform fingerprinting of Vicia species and V. faba L. accessions (Van de Ven et al. 1990; Przybylska et al. 1998; Gresta et al. 2010; Kwon et al. 2010; Liu and Hou 2010; Ouji et al. 2012; Waly et al. 2012).
Simple sequence repeats (SSRs), also called microsatellites, which correspond to tandemly repeated DNA sequences with a very short repeat unit, have been introduced as powerful genetic markers in plant improvement studies (Smith et al. 1997). Moreover, SSRs have been used as DNA markers for genomic fingerprinting and phylogenetic analysis, because they are economical to score, highly abundant in the genome, and co-dominant, they have high allelic diversity, and they are usually selectively neutral (Achtar et al. 2010). Some SSRs, derived from transcripts (known as expressed sequence tag (EST)-SSR markers), have become a powerful tool in plant genetic analysis and breeding research. Moreover, these markers are more closely connected with known function genes and possess higher transferability to related species (Ellis and Burke 2007). SSRs and EST-SSRs have been isolated, characterized, and reported in various species, including V. faba L. (Zeid et al. 2009; Gong et al. 2010; Ma et al. 2011).
In Mediterranean countries such as Tunisia, V. faba represents a crucial source of protein for foods, especially V. faba var. minor for animal feed. Most diversity studies on cultivated V. faba published by now have focused on Tunisian V. faba var. major germplasm, but only a few genotypes of V. faba var. minor have been represented among germplasm studied (Ouji et al. 2012; Yahia et al. 2012).
In order to develop V. faba var. minor cultivars, a preliminary characterization and evaluation of the genetic variability among local and cultivated V. faba var. minor accessions originating from Mediterranean countries and a comparison with widely grown commercial varieties could be useful for the exploitation of this genetic diversity in breeding programs.
The aim of this work was to assess the genotypic variability and relationship among local V. faba var. minor genetic resources distributed in Tunisia, commercial cultivars and cultivated and improved genotypes from different geographic origins based on agronomic and morphological characters, and SSR molecular markers previously developed in V. faba. In addition, the results of this study provide guidelines and a scientific base to better explore V. faba L.var. minor germplasm resources and facilitate local cultivar improvement.
Materials and Methods
Plant Materials
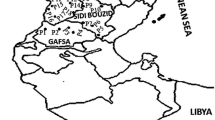
Plant materials of faba bean used in this study totaled 46 accessions and were received from the International Center for Agricultural Research in the Dry Areas (ICARDA), Aleppo, Syria, or directly collected by the authors (Table 1). This set includes ICARDA elite faba bean accessions used in the ICARDA faba bean breeding program and samples from eight countries, encompassing three different geographical regions (Fig. 1) such as Northern Africa (Algeria, Tunisia, Libya, and Egypt), Eastern Africa (Ethiopia, and Sudan), and Near East (Syria, and Turkey). Eight seeds per accession were germinated, and seedlings were transferred to individual pots into a greenhouse. Leaves were collected after about 3 weeks on the eight individual plants, pooled to represent a bulk DNA, frozen in liquid nitrogen, and then stored at −80 °C prior to DNA extraction.
Morphological Characterization
The 46 accessions were evaluated in greenhouse for their morphological and agronomic traits. Morphological characteristics from eight plants per accession were recorded using 15 descriptors including qualitative characters (seed color, flower color, seed texture, seed shape, standard anthocyanin coloration, and standard extent of anthocyanin coloration) and quantitative plant and seed traits (100-seed weight, number of pod per node, number of seeds per pod, seed length, seed width, number of leaflets per leaf, plant height, pod length, and duration (number of days from sowing to 50 % of flowering)) chosen from faba bean descriptors of the International Board for Plant Genetic Resources (IBPGR 1985).
Molecular Characterization
DNA Extraction
Total genomic DNA was extracted from the young leaves (0.5 g) which were powdered in liquid nitrogen, mixed with 700 μl CTAB extraction buffer and 20 mg PVP 40000, and incubated at 65 °C for 30 min with slow shaking every 5–10 min. The mixture was treated two times with 700 μl chloroform-isoamyl alcohol (24:1). DNA was precipitated by adding (1/10) volume acetate sodium (3 M, pH 5.2) and (2/3) volume ice-cold isopropanol. The tubes were placed on ice for 20 min, after which the samples were centrifuged for 15 min at 13,800 rpm. The DNA pellet was washed in 70 % ethanol and dissolved in 50 μl sterilized water. The quality of the DNA was estimated on an agarose gel (0.8 %) stained with ethidium bromide.
PCR Amplification and SSR Analysis
Twenty-one primer pairs were tested. These standard loci have been used in characterization of faba bean genetic diversity in several germplasm collections (Gong et al. 2011; Ma et al. 2011). After optimization of the PCR conditions, 17 primer pairs were selected on the basis of the reproducibility and the polymorphism of the generated bands (Table 3). PCRs were performed in a final volume of 25 μl containing 20 ng of faba bean DNA template, 1× PCR buffer (10 mM Tris–HCl (pH 8.5), 50 mM KCl, 2.5 mM MgCl2), 0.2 mM of each dNTP, 0.2 μM of each primer, and 1 U of Taq polymerase (Vivantis, USA). Reactions were performed in a Minicycler (Bio-Rad iCycler) thermal cycler with the following conditions: 94 °C for 5 min, followed by 35 cycles at 94 °C for 30 s, 55–60 °C depending on the primer pair for 30 s, and 72 °C for 1 min. The last step was 72 °C for 5 min. A volume of 5 μl of stop solution (10 mM NaOH, 95 % formamide, 0.05 % bromophenol blue, 0.05 % xylene cyanol) was added to the product of the PCR amplification before denaturation for a period of 5 min at 94 °C. Four microliters of the reaction mixture was loaded onto denaturing gels of polyacrylamide at 6 % (w/v; 19:1 acrylamide-bisacrylamide) with 8 M urea. Electrophoresis was performed at 75-W constant power for about 3 h (Sequi-Gen GT sequencing cell, Bio-Rad). The products of the amplification were visualized by silver staining according to the protocol of Benbouza et al. (2006). Gels were dried at room temperature and scanned. The reproducibility of PCR experiments was confirmed by repeating the procedure at least two times.
Data Analysis
For quantitative morphological characteristics, an ANOVA analysis was performed; homogeneous groups (Newman and Keuls) were determined by using SYSTAT 8.0 software. The data of morphological characteristics were scored according to Table 2 and used to determine distance matrices for morphological characteristics based on Gower general similarity coefficient (Gower 1971). Unweighted Pair-Group Method using Arithmetic average (UPGMA) analysis was performed with the program MVSP 3.1 (Multi-Variate Statistical Package; Kovach 2007).
For molecular analysis by SSR markers, allele sizes were estimated using gel_analyzer software. The summary statistics including the number of alleles per locus (Na), number of effective alleles (Ne), unbiased expected heterozygosity (UHe), observed heterozygosity (Ho), expected heterozygosity (He), Shannon’s information index (I), and fixation index (F st ) were determined using GenAlEx6 software (Peakall and Smouse 2006). The polymorphism information content (PIC) value was calculated following the formula described by Botstein et al. (1980).
For genetic distance analysis based on SSR results, the allelic size data were transformed to binary data: presence (1) versus absence (0) of each allele. A similarity matrix was generated using the Nei’s genetic distance (Nei and Li 1979). Additionally, a matrix of genetic distance (Nei 1972) was calculated from the three main geographical groups and ICARDA elite faba bean accessions included in this study. Similarity data were processed through the UPGMA cluster analysis conducted using the MVSP 3.1 program. To assess further the genetic relationships of faba bean accessions as individual plants, a principal component analysis (PCA) was conducted based on the SSR variation patterns using MVSP 3.1 software program.
Results
Morphological Characterization
Morphological comparison was performed between 46 faba bean accessions (Table 1) from different geographic origins. Among these accessions, nine originated from Tunisia such as Saber 02, Mahasen, Hara, and Beja which represent the major faba bean accessions currently grown in various regions of the country. All genotypes did flower under our trial conditions, and their morphological characterization could be completed. The results are summarized in Table 2. Each quantitative characteristic was compared among accessions by using ANOVA. Data show significant differences (P ˂ 0.05) between accessions for the nine quantitative agronomic characters considered.
The 46 accessions showed significant difference in days to 50 % flowering. Five different mean groups were found according to the Newman and Keuls comparison denoted by alphabets from a to e (Table 2). Triple white, FLIP 06-003FB, and F.390 had the earliest flowering date, whereas NEB 397, FLIP 03-76FB, and local cultivar (Beja) had the latest. There was a significant difference between accessions for hundred-seed weight. Using the Newman and Keuls procedure, the mean result allowed separation into eight statistically different groups. Accessions NEB 404, Malti, Giza 3, FLIP 03-52FB, FLIP 03-67FB, FLIP 03-82FB, FLIP 05-03FB, FLIP 05-05FB, FLIP 06-001FB, and FLIP 06-003FB had the highest hundred-seed weight, whereas VIR 2037, NEB 102, NEB 118, VIR 2040, Saber 02, and FLIP 03-35FB had the lower hundred-seed weight.
Surprisingly, the four currently cultivated genotypes in Tunisia showed late flowering dates, and lower hundred-seed weight compared to the average of all accessions (41.46 and 72.81, respectively) and have a higher number of seeds per pod, and a higher number of pods per node compared to the average of all accessions (2.14 and 1.22, respectively).
The qualitative and quantitative data were used to evaluate genetic distances among accessions and to construct a dendrogram (Fig. 2). The 46 analyzed accessions were classified according to the 15 selected descriptors, indicating two main groups. At a coefficient of 0.76, accessions of the first group was divided into three subgroups in which the first subgroup (cluster I) comprised only the currently four cultivated genotypes in Tunisia. The second group was divided into four subgroups (clusters IV to VII). The cluster IV comprised only one accession (NEB 397) while the cluster VII comprised the majority of the accessions from the ICARDA elite faba bean breeding programs. The cluster V comprised two accessions NEB 99 and one from ICARDA elite accessions (FLIP 03-67FB). Although this accession share similar qualitative morphological traits with other accessions (particularly with those from the ICARDA elite faba been breeding program), it differed significantly for quantitative traits such as seed number per pod (SN/P) from most of the other ICARDA elite accessions.
Molecular Characterization
Seventeen SSR markers were used for the characterization and genetic diversity analysis of the 46 faba bean accessions (Table 3). A total of 101 alleles were detected; the number of alleles per locus ranged from 2 (M43) to 10 (M41), with an average of 5.94 alleles across the 17 loci. The polymorphism information content (PIC) values ranged from 0.38 (M43) to 0.84 (P121), with an average of 0.69. The frequency of the most common allele at each locus ranged from 21 % (P121) to 64 % (M66). On average, 35.35 % of the 46 faba bean accessions shared a common major allele at any given locus. The He mean was 0.73, ranging from 0.49 in M43 to 0.85 in P121, and Ho ranged from 45 % of heterozygous individuals in M14 to 100 % in M41. The Wright’s fixation index (Fst) compares He with Ho, estimating the degree of allelic fixation, and ranged from 0.75 in M43 to 0.39 in M14 with an average value of 0.14. Finally, Shannon-Weaver’s information indices ranged from 0.68 to 1.98 and averaged at 1.5. The results showed an abundant genetic diversity among the 46 accessions, and the most polymorphic loci, P121 and M41, revealed a high level of genetic variation.
Cluster Analysis
In order to understand the genetic relationships of faba bean accessions from different sources and origins, the Nei’s genetic distances were calculated. UPGMA cluster analysis of the similarity matrix obtained from 17 SSR markers resulted in a dendrogram of genetic relationships (Fig. 3) producing two major clusters. The first group included 11 among the 19 accessions originating from ICARDA elite faba bean accessions. The second cluster was subdivided into four subclusters, one of which included currently cultivated Tunisian varieties along with some genotypes originate from Egypt, Libya, and Ethiopia such as VIR2163, VIR 2128, NEB 482, F.390, and NEB 453. The second subcluster comprised accessions of different countries of origins. The third subcluster included five accessions originating from ICARDA elite faba bean accessions, and finally, the fourth subcluster comprised of six NEB accessions and three accessions originating from ICARDA elite faba bean accessions that the register data available assigned tentatively to an ICARDA improved genotypes origin probably derived from NEB. Interestingly, a number of traditional and improved genotypes originating from ICARDA elite faba bean accessions did not form a well and defined group. This dendrogram shows that the members of the same geographic origin were not assigned to the same group. However, the four currently cultivated Tunisian cultivars belonged to the same group. The cluster analysis performed using the similarity data between groups including accessions that belong to each geographical area (Fig. 4) showed that North Africa and Eastern Africa accessions were very similar (D = 0.82). Near East accessions seemed to be as closely related to the Eastern Africa as to the Northern Africa group (D = 0.74). Finally, accessions originating from ICARDA elite faba bean accessions were clustered separately from the other geographical area. The cophenetic correlation value (r = 0.93) for this analysis was also statistically significant.
Dendrogram of the 46 faba bean accessions included in this study generated by UPGMA cluster analysis from the similarity matrix obtained using Nei’s genetic distance (Nei and Li 1979) based on the results of 17 SSR markers
Dendrogram of the four main originate groups included in this study generated by UPGMA cluster analysis from the similarity matrix obtained using Nei’s unbaised genetic distance coefficients (Nei 1972)
Principal Component Analysis
Associations among 46 accessions were investigated by the PCA method. The localization of individuals was defined by the first principal component (PC1), and the second principal component (PC2). In this analysis, PC1 and PC2 explained 26.49 and 17.41 %, respectively, of the total variability. The PCA provides an alternative view of the genetic distances among faba bean accessions compared to the UPGMA dendogram (Fig. 5). Most individuals were grouped more closely according to their pedigree. Indeed, the 19 individuals from ICARDA elite faba bean accessions were quite distant from the other genotypes but cover a very large area in the plot, suggesting a high level of genetic diversity. The eight individuals from VIR germplasm were located quite distant from the remaining individuals. Moreover, PCA analysis showed that the commercial and local Tunisian cultivars could constitute one group. The NEB genotypes clustered together with the remaining genotypes. PCA also revealed that the genetic distance does not correlate with the geographical origin in the 46 accessions studied.
Two-dimensional principal component analysis (PCA) based on SSR polymorphisms in the 46 accessions. Codes for the faba bean accessions refer to those in Table 1
Discussion
The knowledge about the genetic relationships of genotypes provides useful information to address problems in breeding programs and germplasm resource management (Salem et al. 2008). DNA fingerprinting techniques such as RAPD, AFLP, and ISSR have been largely utilized to characterize the diversity of genetic of faba bean (Link et al. 1995; Zied et al. 2003; Terzopoulos and Bebeli 2008), while SSRs markers are not yet universally used in this species. In this study, morphological data analysis was coupled with SSR molecular analyses to investigate the genetic relationships among 46 faba bean accessions. The accessions showed diverse morphological traits and distinct SSR marker patterns.
Using morphological data, the genetic diversity of faba bean germplasm was studied in different countries such as Egypt, Spain, Italy, Greece, Iraq, Iran, and Lebanon (Perrino et al. 1991; Polignano et al. 1999; Terzopoulos et al. 2003; Terzopoulos et al. 2004). Recently, Yahia et al. (2012) described a substantial phenotypic variability in the Tunisian faba bean germplasm using morphological descriptors. However, the morphological variability could be inflated by environmental conditions if the data were recorded for different accessions in different regions and ecosystems. Our study provides a morphological characterization of accessions from different origins tested in a same location. This morphological analysis showed a high morphological diversity among the material analyzed at within-species level. This morphological diversity is higher between accessions of three different geographic origins than in elite faba bean accessions used in breeding programs. Indeed, most ICARDA elite faba bean accessions were grouped together suggesting that it is likely that these accessions were selected on the basis of identical morphological characters. The local and commercial Tunisian genotypes were clearly discriminated from other genotypes and belonged to a distinct subgroup. This suggested that these genotypes are adapted to similar environmental conditions and have undergone selection.
Many types of molecular markers, particularly SSR markers, have been successfully used to assess genetic diversity and characterize crop resources (Keneni et al. 2012; McClean et al. 2012; Suresh et al. 2013).
In this study, we utilized SSR analysis to characterize genetic diversity in 46 faba bean accessions. The average number of alleles per locus (5.94) was higher compared to AFLP (1.45) and SSAP markers (1.68) (Liu and Hou 2010; Ouji et al. 2012). These suggest that SSR markers are a very suitable tool for assessing genetic diversity of faba bean.
The molecular analysis was used to tentatively classify the 46 accessions by different ways: UPGMA cluster analysis and principal components analysis.
Based on the UPGMA cluster analysis, the 46 accessions were divided into two major groups (Fig. 3). Group 1 included 11 among 19 ICARDA elite faba bean accessions. The remaining eight ICARDA elite faba bean accessions were clustered in group 2. Within this group, four subgroups were observed. Interestingly all ICARDA elite faba bean accessions observed in group 1 and group 2 were clustered together. Moreover, PCA analysis showed that all ICARDA elite faba bean accessions were clustered separately from the genotypes of other origin but simultaneously formed three subgroups of points. These data suggested that ICARDA elite faba bean accessions differ from other accessions and grouped together regardless of the type of analysis. This probably means that they were selected according to the same characters. Interestingly, SSR provide more information than morphological analysis distinguishing some subgroups that morphological analysis did not distinguish. Moreover, subgroups observed in ICARDA elite faba bean accessions could suggest a common pedigree. According to the dendrogram analysis (Fig. 3), six accessions—NEB 97, NEB 404, NEB 99, NEB 102, NEB 397, and NEB 118—from Egypt, Turkey, Syria, Tunisia, and Ethiopia, respectively, were classified with ICARDA elite faba bean accessions into group 2. This may suggest that accessions from these regions could have been used as a basis for the development of the ICARDA elite faba bean accessions. NEB accessions have been utilized in faba bean breeding programs, especially at ICARDA, in order to introduce genes to confer tolerance to abiotic and biotic stress, and these faba bean germplasms have been successfully introgresed into faba bean cultivars (Stoddard et al. 1999).
Duc et al. (2010) described main ecogeographical groups; three of them were represented in this study (ICARDA elite faba bean accessions, Eastern Africa, and Northern Africa), and we performed a cluster analysis on this base using the similarity data between groups including accessions that belong to each geographical area and ICARDA elite faba bean accessions. This cluster analysis showed the most diverging set of accessions to be from ICARDA elite faba bean accessions (Fig. 4) and showed a lower level of diversity between the other regions. The second most distinct set was from Near East (Fig. 4). On the other hand, Fig. 3 shows that accessions were clustered into two groups but that the groups do not correspond to the ecogeographic origin of the accessions. This clustering may reflect the pedigree relations and commonality of important agricultural traits in accessions from different regions. Moreover, the observed genetic diversity of SSR loci was subjected to principal component analysis. The multivariate analyses classified the 46 accessions into groups consisting of ICARDA elite faba bean accessions, VIR genotypes, and other accessions from different pedigree. Figure 5 allows assessment of the genetic relationship among accessions. There is no obvious relationship between geographical origin and grouping based on genetic similarities. Both UPGMA cluster analysis and PCA showed relatively distant relationship of ICARDA elite faba bean accessions with other accessions from different origin. On the other hand, the correlation between genetic and geographical distances among faba bean accessions needs to be further studied using more accessions across grater geographic.
SSR and morphological analyses showed significant differences between Tunisian local and commercial faba bean cultivars (Mahasen, Hara, local Beja, and Saber 02) and the remaining accessions. The Tunisian accessions are characterized by late flowering dates, weak 100-seed weight, higher number of seeds per pod, and a higher number of pods per node compared to the average of all accessions. The molecular and morphological differentiation of the Tunisian cultivars suggests a common genetic pool as well as an adaptation to local agro-climatic conditions.
Elite faba bean cultivars were clearly discriminated from accessions originating from different geographical origin. Indeed, the genetic distances among some elite faba bean accessions were relatively high suggesting the existence of an interesting diversity among these elite accessions. On the other hand, we found that the genetic diversity between the Tunisian faba bean genotypes is narrow and that the Tunisian faba bean genotypes are closer to the other faba bean germplasm sources than to the ICARDA elite faba bean accessions. Therefore, we suggest the importance of introducing elite faba bean accessions to enrich the genetic foundation of breeding of faba bean in Tunisia. Similar results were approved by Liu and Hou (2010) which suggested that introducing elite foreign germplasm to enrich genetic foundation of breeding of faba bean in Qinghai should be necessary.
Although the 46 accessions could not be obviously divided into several unattached groups, some valuable information about the genetic relationship of some accessions was obtained from the dendrogram based on SSRs. Indeed, breeders might introduce some heterogeneity in their material from other breeding centers (Li et al. 2008). It is interesting to note that PCA analysis placed all VIR accessions in one group showing a high level of genetic relatedness and close parentage. This analysis suggests that the collection for the in situ conservation of VIR accessions was, may be, made on the basis of specific criteria which allowed us to distinguish them from most of the other accessions.
Our study confirmed the high potential of SSRs as excellent molecular markers for faba bean genotype identification, differentiation, and evaluation of genetic variation.
Conclusion
In conclusion, our data showed significant variation in morphological traits and microsatellite DNA polymorphisms among 46 faba bean accessions. The relatively high average number of alleles per locus and allelic diversity observed in the set of genotypes analyzed indicate that the genetic base of the material is relatively wide. The allelic richness of the total accessions suggested the presence of potential unique and rare alleles. ICARDA elite faba bean accessions had clustered together versus the other accessions from different origins suggesting a common pedigree and the similarity of the selective traits. Morphological and molecular analysis showed that local and commercial Tunisian genotypes seem to be different from all the other accessions, suggesting an adaptation to local conditions but also a narrow genetic base. Results from this study show that SSRs markers are a useful tool for faba bean diversity studies, which is a key in conducting faba bean breeding programs. Indeed, knowledge of the genetic diversity of a species is important for the choice of crossing parents in hybrid breeding (Tams et al. 2004). To ensure sustaining breeding progress in the future, the introduction of new germplasm into national faba bean breeding programs, especially by the aid of molecular markers, is recommended.
References
Achtar S, Moualla MY, Kalhout A, Röder MS, MirAli N (2010) Assessment of genetic diversity among Syrian durum (Triticum ssp. durum) and bread wheat (Triticum aestivum L.) using SSR markers. Russ J Genet 46:1320–1326
Aouar-sadli M, Louadi K, Doumandji S (2008) Pollination of the broad bean (Vicia faba L. var. major) (Fabaceae) by wild bees and honey bees (Hymenoptera: Apoidea) and its impact on the seed production in the Tizi-Ouzou area (Algeria). Afr J Agric Res 3:266–272
Ayed MH (2011) Effect of partially replacing soybean by broad beans “vicia faba var minor” on digestibility, growth performances and carcass characteristics in broiler chickens. Adv Environ Biol 5:2332–2335
Baranski R, Maksylewicz-Kaul A, Nothnagel T, Cavagnaro PF, Simon PW, Grzebelus D (2012) Genetic diversity of carrot (Daucus carota L.) cultivars revealed by analysis of SSR loci. Genet Resour Crop Evol 59:163–170
Benbouza H, Jacquemin JM, Baudoin JP, Mergeai G (2006) Optimization of a reliable, fast, cheap and sensitive silver staining method to detect SSR markers in polyacrylamide gels. Biotechnol Agron Soc Environ 10:77–81
Boczkowska M, Bulińska-Radomska Z, Nowosielski J (2012) AFLP analysis of genetic diversity in five accessions of Polish runner bean (Phaseolus coccineus L.). Genet Resour Crop Evol 59:473–478
Botstein D, White RL, Skalnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphism. Am J Hum Genet 32:314–331
Cao Y, Tian L, Gao Y, Liu F (2012) Genetic diversity of cultivated and wild Ussurian Pear (Pyrus ussuriensis Maxim.) in China evaluated with M13-tailed SSR markers. Genet Resour Crop Evol 59:9–17
Duc G (1997) Faba bean (Vicia faba L.). Field Crop Res 53:99–109
Duc G, Bao S, Baum M, Redden B, Sadiki M, Suso MJ, Vishniakova M, Zong X (2010) Diversity maintenance and use of Vicia faba L. genetic resources. Field Crop Res 115:270–278
Ellis JR, Burke JM (2007) EST-SSRs as a resource for population genetic analyses. Heredity 99:125–132
Ellwood SR, Phan HTT, Jordan M, Hane J, Torres AM, Avila CM, Cruz-Izquierdo S, Oliver RP (2008) Construction of a comparative genetic map in faba bean (Vicia faba L.); conservation of genome structure with Lens culinaris. BMC Genomics 9:380–391
Gong YM, Xu SC, Mao WH, Hu QZ, Zhang GW, Ding J, Li ZY (2010) Generation and characterization of 11 novel EST derived microsatellites from Vicia faba (Fabaceae). Am J Bot 97:69–71
Gong YM, Xu SH, Mao WH, Li ZY, Hu QZ, Zhang GW, Ding J (2011) Genetic diversity analysis of faba bean (Vicia faba L.) based on EST-SSR markers. Agric Sci China 10:838–844
Gower JC (1971) A general coefficient of similarity and some of its properties. Biometrics 27:857–874
Gresta F, Avola G, Albertini E, Raggi L, Abbate V (2010) A study of variability in the Sicilian faba bean landrace ‘Larga di Leonforte’. Genet Resour Crop Evol 57:523–531
Hwang TY, Nakamoto Y, Kono I, Enoki H, Funatsuki H, Kitamura K, Ishimoto M (2008) Genetic diversity of cultivated and wild soybean including Japanese elite cultivars as revealed by length polymorphism of SSR markers. Breed Sci 58:315–323
International Board for Plant Genetic Resources (IBPGR) (1985) Faba bean descriptors. IBPGR, Rome (31 pp)
Keneni G, Bekele E, Imtiaz M, Dagne K, Getu E, Assefia F (2012) Genetic diversity and population structure of Ethiopian chickpea (Cicer arietinum L.) germplasm accessions from different geographical origins as revealed by microsatellite markers. Plant Mol Biol Rep 30:654–665
Kharrat M, Ben Saleh H, Halila HM (1991) Faba bean status and prospects in Tunisia. CIHEAM Options Mediterr 10:169–172
Kovach WL (2007) MVSP - A MultiVariate Statistical Package for Windows, ver. 3.1. Kovach Computing Services, Pentraeth, Wales, U.K
Kwon SJ, Hu J, Coyne CJ (2010) Genetic diversity and relationship among faba bean (Vicia faba L.) germplasm entries as revealed by TRAP markers. Plant Genet Resour Characterization Util 8:204–213
Li Z, Wang X, Zhang Y, Zhang G, Wu L, Chi J, Ma Z (2008) Assessment of genetic diversity in glandless cotton germplasm resources by using agronomic traits and molecular markers. Front Agric China 2:245–252
Link W, Dixkens C, Singh M, Schwall M (1995) Genetic diversity in European and Mediterranean faba bean germplasm revealed by RAPD markers. Theor Appl Genet 90:27–32
Liu Y, Hou W (2010) Genetic diversity of faba bean germplasms in qinghai and core germplasm identified based on AFLP analysis. Legume Genomics Genet 1:1–6
Ma Y, Yang T, Guan J, Wang S, Wang H, Sun X, Zong X (2011) Developpment and characterization of 21 EST-derived microsatellite markers in Vicia faba (faba bean). Am J Bot 98:22–24
McClean PE, Terpstra J, McConnell M, White C, Lee R, Mamidi S (2012) Population structure and genetic differentiation among the USDA common bean (Phaseolus vulgaris L.) core collection. Genet Resour Crop Evol 59:499–515
Mejri S, Mabrouk Y, Voisin M, Delavault P, Simier P, Saidi M, Belhadj O (2012) Variation in quantitative characters of faba bean after seed irradiation and associated molecular changes. Afr J Biotechnol 11:8383–8390
Mir RR, Kumar J, Balyan HS, Gupta PK (2012) A study of genetic diversity among Indian bread wheat (Triticum aestivum L.) cultivars released during last 100 years. Genet Resour Crop Evol 59:717–726
Mustafa AZMA (2007) Genetic variation among Egyptian cultivars of Vicia faba L. Pak J Biol Sci 10:4204–4210
Nei M (1972) genetic distance between populations. Am Nat 106:283–291
Nei M, Li WH (1979) mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Soc USA 76:5269–5273
Ouji A, El Bok S, Syed NH, Abdellaoui R, Rouaissi M, Flavell AJ, El Gazzah M (2012) Genetic diversity of faba bean (Vicia faba L.) populations revealed by sequence specific amplified polymorphism (SSAP) markers. Afr J Biotechnol 11:2162–2168
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel, population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Perrino P, Robertson LD, Silh MB (1991) Maintenance, evaluation and use of faba bean germplasm collections: problems and prospects. CIHEAM Options Méditerr 10:21–31
Polignano GB, Alba E, Ugentil P, Scippa G (1999) Geographical patterns of variation in bari faba bean germplasm collection. Genet Res Crop Evol 46:183–192
Przybylska J, Zimniak-Przybylska Z, Krajewski P (1998) Isoenzyme variation in the wild relatives of Vicia faba (Fabaceae). Plant Syst Evol 213:173–186
Roy A, Bal SS, Fergany M, Kaur S, Singh H, Malik AA, Singh J, Monforte AJ, Dhillon NPS (2012) Wild melon diversity in India (Punjab State). Genet Resour Crop Evol 59:755–767
Salem KFM, El-Zanaty AM, Esmail RM (2008) Assessing wheat (Triticum aestivum L.) genetic diversity using morphological characters and microsatellite markers. Word J Agric Sci 5:538–544
Sardos J, Noyer JL, Malapa R, Bouchet S, Lebot V (2012) Genetic diversity of taro (Colocasia esculenta (L.) Schott) in Vanuatu (Oceania): an appraisal of the distribution of allelic diversity (DAD) with SSR markers. Genet Resour Crop Evol 59:805–820
Smith JSC, Chin ECL, Shu H, Smith OS, Wall SJ et al (1997) An evaluation of the utility of SSR loci as molecular markers in maize (Zea mays L.): comparisons with data from RFLPs and pedigree. Theor Appl Genet 95:163–173
Stoddard FL, Kohpina S, Knight R (1999) Variability of Ascochyta fabae in South Australia. Aust J Agr Res 50:1475–1481
Suresh S, Park JH, Cho GT, Lee HS, Baek HJ, Lee SY, Chung JW (2013) Development and molecular characterization of 55 novel polymorphic cDNA-SSR markers in faba bean (vicia faba L.) using 454 pyrosequencing. Molecules 18:1844–1856
Tams SH, Bauer E, Oettler G, Melchinger AE (2004) Genetic diversity in European winter triticale determined with SSR markers and coancestry coefficient. Theor Appl Genet 108:1385–1391
Tanno K, Willcox G (2006) The origins of cultivation of Cicer arietinum L. and Vicia faba L.: early finds from Tell el-Kerkh, north-west Syria, late10th millennium B.P. Veget Hist Archaeobot 15:197–204
Terzopoulos PJ, Kaltsikes PJ, Bebeli PJ (2003) Collection, evaluation and classification of Greek population of faba bean (Vicia faba L.). Genet Res Crop Evol 50:373–381
Terzopoulos PJ, Kaltsikes PJ, Bebeli PJ (2004) Characterization of Greek populations of faba bean (Vicia faba L.) and their evaluation using a new parameter. Genet Res Crop Evol 51:655–662
Terzopoulos PJ, Bebeli PJ (2008) Genetic diversity analysis of Mediterranean faba bean (Vicia faba L.) with ISSR markers. Field Crop Res 108:39–44
Van de Ven M, Powell W, Ramsay G, Waugh R (1990) Restriction fragment length polymorphisms as genetic markers in Vicia. Heredity 65:329–342
Waly EA, Farghali MA, Abbas HS, Mosselhy DS (2012) Identification of the genetic differences between some faba bean genotypes by finger print characters. J Appl Sci Res 8:17–24
Wang KJ, Li XH (2012) Genetic diversity and geographical peculiarity of Tibetan wild soybean (Glycine soja). Genet Resour Crop Evol 59:479–490
Yahia Y, Guetat A, Elfalleh W, Ferchichi A, Yahia H, Loumerm M (2012) Analysis of agromorphological diversity of southern Tunisia faba bean (Vicia faba L.) germplasm. Afr J Biotechnol 11(56):11913–11924
Zied M, Schon CC, Link W (2003) Genetic diversity in recent elite faba bean lines using AFLP markers. Theor Appl Genet 107:1304–1314
Zeid M, Mitchell S, Link W, Carter M, Nawar A, Fulton T, Kresovich S (2009) Simple sequence repeats (SSRs) in faba bean: new loci from Orobanche-resistant cultivar ‘Giza 402’. Plant Breed 128:149–155
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Abid, G., Mingeot, D., Udupa, S.M. et al. Genetic Relationship and Diversity Analysis of Faba Bean (Vicia Faba L. var. Minor) Genetic Resources Using Morphological and Microsatellite Molecular Markers. Plant Mol Biol Rep 33, 1755–1767 (2015). https://doi.org/10.1007/s11105-015-0871-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-015-0871-0