Abstract
Abiotic stresses such as drought and high salinity inhibit plant growth and productivity. NAC transcription factors play a variety of important roles in plant development and abiotic stress responses. To date, the transformation of NAC genes into poplar plants has not been reported. In this study, we introduced the CarNAC3 and CarNAC6 salinity and drought tolerant genes from Cicer arietinum (chickpea) into Populus deltoides × Populus euramericana ‘Nanlin895’ (poplar) plants using Agrobacterium tumefaciens-mediated transformation. We verified the integration of the two genes into the poplar genome using polymerase chain reaction (PCR) and their stable expression was confirmed at the transcript level using real-time (RT)-PCR. The growth rates of roots and shoots and the clawed root rate increased in transformed plants grown in agar medium or in soil and increased in response to drought and salt stress conditions. Proline and photoprotectant pigment accumulation and antioxidant enzyme activities increased in response to abiotic stress, but the accumulation of photosynthetic pigments and malondialdehyde (MDA) decreased in transgenic lines relative to wild-type control plants. The CarNAC3 transgene was expressed at higher levels than the CarNAC6 transgene. Our results showed that the CarNAC3 and CarNAC6 genes enhanced the capacity for osmotic adjustment and increased antioxidant enzyme activity and suggest that these genes could play a significant role in improving drought and salt tolerance when expressed in poplar plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants must adjust to environmental conditions to coordinate their growth and development (Santner and Estelle 2009). Drought and high salinity are two important abiotic stresses that reduce plant growth and productivity (Nakashima et al. 2012). Many abiotic stress-responsive genes have been identified in plants using molecular techniques such as DNA microarrays (Fowler and Thomashow 2002; Rabbani et al. 2003; Yamaguchi-Shinozaki and Shinozaki 2006; Nakashima et al. 2009). NAC domain proteins are transcription factors (TFs) whose functions are related to plant development or biotic and abiotic stresses (Nakashima et al. 2012). Many NAC proteins have been reported to be involved in shoot apical meristem development (Souer et al. 1996), clawed ratio and lateral root (He et al. 2005), senescence (Guo and Gan 2006; Uauy et al. 2006), secondary cell wall formation (Mitsuda et al. 2007), and in response to fungal infection and to drought and high-salinity stress (Hu et al. 2006; Zheng et al. 2009). Previous research on NAC proteins focused on model plant species such as Arabidopsis thaliana (Arabidopsis) and rice (Olsen et al. 2005). The RESPONSIVE TO DEHYRATION 26 (RD26) gene from Arabidopsis was the first reported gene encoding a NAC protein (Yamaguchi-Shinozaki et al. 1992). The NAC domain was identified based on consensus sequences present in the petunia NAM and Arabidopsis AFAF1/2 and CUC2 protein sequences (Aida et al. 1997; Nakashima et al. 2012). Some NAC genes are upregulated by wounding and bacterial infection (Collinge and Boller 2001; Mysore et al. 2002; Hegedus et al. 2003) whereas others mediate viral resistance (Xie et al. 1999).
Hui et al. (2009) reported that the CarNAC3 protein from Cicer arietinum (chickpea) was 285 amino acids (aa) in length and included one conserved domain. The sequence responsible for the transcriptional activity of this protein was located in the C-terminal region. Under stress conditions, expression of the CarNAC3 gene in poplar increased proline and photosynthetic pigment levels and antioxidant enzyme activities. Furthermore, its expression decreased the concentration of malondialdehyde (MDA) relative to that of wild-type (WT) control plants (Movahedi et al. 2014a).
The CarNAC6 gene from chickpea encodes 308 amino acids and regulates transcription factors (Nakashima et al. 2012; Movahedi et al. 2014b). The CarNAC6 gene belongs to the NAC domain protein family (NAM, ATAF1, 2, and CUC2), which enhances plant resistance to biotic and abiotic stresses and plays an important role in plant development (Movahedi et al. 2014b).
Sophisticated mechanisms have evolved in plants to protect against severe conditions through the synergistic actions of antioxidants and osmoprotectants (Al-Whaibi et al. 2011). Drought and high salinity stresses cause oxidative stress in plants (Ahmad et al. 2010; Barba-Espin et al. 2011). Plants respond to abiotic stress by increasing the formation of reactive oxygen species (ROS) including H2O2, Ō2, and hydroxyl radicals, which impair electron transport (Mishra et al. 2011; Nounjan et al. 2012). In response to increased ROS levels, plants produce an array of enzymatic and non-enzymatic antioxidant molecules to scavenge ROS (Krantev et al. 2008; Zhou et al. 2014). Superoxide dismutase (SOD), guaiacol peroxidase (POD), and catalase (CAT) activities and MDA levels are induced by cell membrane damage (Abdullahil et al. 2010; Dong et al. 2010; Mishra et al. 2011). Proline, a soluble osmolyte, plays an important role in adjusting plant osmylotic functions and protecting cell structures from abiotic stress (Abraham et al. 2010; Hou et al. 2013). Increased proline accumulation in transgenic poplar plants enhanced drought and salt tolerance (Ben et al. 2010; Su et al. 2011). Cho et al. (2012) reported that increasing the activity of antioxidant enzyme proline, influenced by abiotic stresses, cause to increase tolerance in plant. To date, the transformation of NAC genes into poplar plants has not been reported. In this study, we introduced the CarNAC3 and CarNAC6 genes into the poplar hybrid clone ‘Nanlin895’ (Populus deltoides × Populus euramericana ‘Nanlin895’) using Agrobacterium tumefaciens-mediated transformation. We compared the physiological and biochemical responses of transformed and wild-type poplar plants to abiotic stress. In addition, we investigated the effect of CarNAC3 and CarNAC6 gene expression in transgenic poplar plants on the antioxidant system and on osmotic protection against drought and high salinity stresses.
Materials and Methods
Plant Materials and Genetic Transformation
We transformed poplar using the LBA404 A. tumefaciens strain resistant to rifampicin (Hoekema et al. 1983) and kanamycin. Two binary vectors containing either the CarNAC3 gene (GenBank accession number FJ356671.1) or the CarNAC6 gene (GenBank accession number FJ477887.1) isolated from C. arietinum (chickpea) cultivar 209 (PBI121/CarNAC3 and PBI121/CarNAC6, respectively) were prepared by the State Key Laboratory of Crop Genetics and Germplasm Enhancement of the National Center for Soybean Improvement at Nanjing Agricultural University, China (Hui et al. 2009). The CarNAC3 gene was 1160 bp in length and contained a single open reading frame (ORF). The CarNAC6 gene was 1154 bp in length and contained two opposing ORFs. The PBI121/CarNAC3 plasmid was digested with XbaI to isolate an 858-nt fragment of CarNAC3 and the PBI121/CarNAC6 plasmid was digested with XbaI and BamHI to isolate a 924-nt fragment of CarNAC6 for preparing transformant Agrobacteria. Expression of the CarNAC3 and CarNAC6 genes in these constructs was driven by the cauliflower mosaic virus (CaMV) 35S promoter, and both genes contained the nopaline synthase (NOS) gene terminator. Agrobacterium cultures were grown at 28 °C for 48 h in liquid Luria-Bertani medium supplemented with 50 mg/l rifampicin and 50 mg/l kanamycin (Horsch et al. 1988) with gentle shaking until an optical density (OD) of 0.7 was reached. After centrifugation of the cultures, the bacterial pellets were re-suspended in liquid Murashige and Skoog (MS) medium containing 5 % sucrose to prepare Agrobacterium infective suspensions which were cold-treated at 4 °C (Logemann et al. 2006). Leaves from P. deltoides × P. euramericana ‘Nanlin895’ (poplar) plants were pre-cultured for 2 days and then cut in 0.5 × 0.5 cm for inoculation by immersion into the Agrobacterium infective suspensions for 120 min at 28 °C with constant agitation at 220 rpm. MS medium supplemented with 0.5 mg/l 6-benzylaminopurine (6-BA), 0.004 mg/l thidiazuron (TDZ), 4.0 g/l agar (semi-solid medium), 25.0 g/l sucrose, and 200 μM acetosyringone (AS) was used to co-cultivate the inoculated explants with Agrobacterium for 2 days at 28 °C without light.
Explants from 45 putative transformed lines for each of the two genes were transferred to MS selection medium containing 0.5 mg/l 6-BA, 0.004 mg/l TDZ, 6.0 g/l agar, and 25.0 g/l sucrose supplemented with 400 mg/l rifampicin and 50 mg/l kanamycin and grown at 23 ± 1 °C and pH 5.8 under a 16/8-h photoperiod and 50–60 % relative humidity. The shoot tips (~5 cm) of regenerated shoots were excised from 42 CarNAC3 to 43 CarNAC6 lines and transferred to half-strength MS medium containing no added hormones for rooting under phytotron conditions. Forty well-developed poplar lines for each transgene were recovered to use in comparisons with untransformed WT poplar plants. Eight lines for each transgene were transferred to MS agar medium containing various salt concentrations. Thirty transgenic poplar lines for each gene and eight WT lines were transferred to the greenhouse for complementary experiments.
PCR and Real-Time-PCR
Total genomic DNA was extracted from young leaves of putative transformants using the CTAB method (Porebski et al. 1997) and DNA at concentrations of 900–1000 ng/μl determined using a BioDrop spectrophotometer (UK). Transformations were confirmed by the amplification of an 858-bp fragment from the CarNAC3 gene by polymerase chain reaction (PCR) using the primers CarNAC3 F (5′-ATGAATGGAAGAACAAG-3′) and CarNAC3 R (5′-ATATTCTCTATGGTCATATAC-3′) and the amplification of a 924-bp fragment from the CarNAC6 gene using the primers CarNAC6 F (5′-ATACATGGCATCAATGGA-3′) and CarNAC6 R (5′-ATAAAAATTGAGGGTGTTGGAATTAG-3′). The PCR reactions were carried out under the following conditions: 94.5 °C for 5 min; 35 cycles of 94 °C for 35 s, 56 °C for 40 s, and 72 °C for 75 s, followed by 72 °C for 10 min. The PCR products were visualized on a 1 % agarose gel.
Total RNA was extracted from young leaves of transformed poplar plants using TRIzol (Tiangen Biotech, Beijing, China) according to the manufacturer's instructions with RNA concentrations of 1100–1300 ng/μl determined using a BioDrop spectrophotometer. Real-time (RT)-PCR) reactions containing 4.0 μg of total RNA and an oligo-dT primer were carried out to synthesize first-strand complementary DNA (cDNA) using a PrimeScript One Step RT-PCR Ver. 2 kit (Takara Biotechnology, Dalian, China) according to the manufacturer’s instructions. The RT-PCR products were re-amplified in 50-μl PCR reactions using the same PCR primers described above to confirm their identities. Absolute quantification of CarNAC3 and CarNAC6 transgene copy numbers was performed using RT-PCR and relative expression levels were determined by the ΔΔCt method using an Applied Biosystems RT-PCR system (Applied Biosystems, USA) and Fast Start Universal SYBR Green Master (Rox; No. 04913914001: Roche, USA). Amplification was performed on 1000-, 500-, 250-, 125-, and 62.5-fold cDNA dilutions to create a standard curve using the primers CarNAC3 messenger RNA (mRNA) F (5′-GTTTCAGATTTCACCCAACTGATGA-3′) and CarNAC3 mRNA R (5′-ATCAACTTCAGGGATGATAGATGCA-3′) to amplify a 101-nt fragment from the CarNAC3 cDNA and the primers CarNAC6 mRNA F (5′-ATTTCACCCCACAGAAGAAGAACTA-3′) and CarNAC6 mRNA R (5′-ATACCACTCTCTTTCTCCAACCTTT-3′) to amplify a 157-nt fragment from the CarNAC6 cDNA. Expression of the housekeeping gene β-actin (NCBI Reference Sequence: XM-006370951.1) was used for normalization of expression. The primers β-actin F (5′-GACCTTCAATGTGCCTGCAA-3′) and β-actin R (5′-ACCATCACCAGAATCCAGCA-3′) were used to amplify a 100-nt fragment from the β-actin mRNA.
Plantlet Transplantation and Phenotypic Analysis
To analyze the phenotypic effects of CarNAC3 and CarNAC6 transgene expression, 40 putative transformed poplars lines were selected for each gene. Plants with three to four expanded leaves on the top bud were transplanted to fresh agar medium incorporating various NaCl concentrations for phenotypic analysis. The growth rates of roots and shoots of plantlets grown on NaCl concentrations of 0, 50, 100, 150, and 200 mM were measured over a period of 4 weeks. Each treatment was applied to eight putative transformed poplar lines for each gene.
To determine the levels of protective compounds and antioxidant enzyme activities, 15 each of CarNAC3 and CarNAC6 transgenic poplar lines were transferred to the greenhouse. After 3 days of acclimatization, the plantlets were transplanted to soil consisting of a 2:1 mixture of sterilized peat and perlite. During the first 2 weeks, humidity was maintained by spraying with water. After 2 weeks, the 15 lines for each transgene were irrigated with a series of increasing salt concentrations over a 4-week period. Eight untransformed WT lines served as controls. The plants were watered with 0 mM NaCl solution every 2 days during the first week. The NaCl concentration was then increased by 50 mM every week to a maximum of 150 mM. The amount of water supplied was kept constant and trays under the pots maintained the salt content of the soil. To impose drought stress on both the transgenic and WT lines, watering was then discontinued for 15 days. Shoot heights and the concentrations of photosynthetic pigments, proline, and malondialdehyde (MDA), and the activities of superoxide dismutase (SOD) and guaiacol peroxidase (POD) were measured and compared between the transgenic and WT poplar lines.
In a complementary experiment, the clawed root rate was calculated using the formula: percent clawed roots = number of main roots/(main roots + lateral roots) × 100 to compare the effect of transgene expression on the clawed root phenotype in transgenic and WT lines grown for 4 weeks in medium containing 0 or 25 mM NaCl.
Determination of Chlorophyll, Proline, and MDA Concentrations and SOD and POD Activities
A 10-ml volume of 80 % acetone was added to 10 ml of young leaf tissue previously homogenized in liquid nitrogen and incubated for 24 h at 4 °C. The supernatant was collected by centrifugation at 10,000 × g for 10 min at 4 °C. A microplate reader spectrophotometer (Bio-Rad) set to measure absorbance at 663.8, 646.8, and 470.0 nm was used to determine the absorbance of chlorophyll a (C a ), chlorophyll b (C b ), total chlorophyll (C a + b ), and total carotenoids (C x + c ) according to Lichtenthaler (1987). The SOD (EC1.15.1.1) activity and MDA concentration were determined according to the methods of Satoh (1978), Stewart and Bewley (1980), and Wang et al. (2012). The POD (E.C. 1.11.1.7) activity and proline content were measured using the methods of Pagariya et al. (2012) and Lei et al. (2007), respectively.
Statistical Analysis
All data analysis was carried out in triplicate using ANOVA with mean separations determined by Duncan's test calculated using SPSS version 16 (SPSS Inc., Chicago, IL, USA) and Excel 2013 software (Microsoft, Redmond, WA, USA). Differences were considered statistically significant when the confidence intervals showed no overlap of the mean values with an error value of 0.05.
Results
Analysis of CarNAC3 and CarNAC6 Nucleotide Sequences
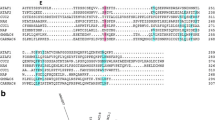
Schematic diagrams of the CarNAC3 and CarNAC6 full-length cDNA sequences contained in the PBI121 binary vector constructs used for transformation of poplar are shown in Fig. 1. Based on analysis using the CLC Genomics Workbench 3 software (CLC, Taipei, Taiwan), the CarNAC3 gene contained an 858-nt open reading frame (ORF), a 64-bp 5′-untranslated region (UTR), and a 238-bp 3′-UTR. The predicted CarNAC3 protein was 286 aa in length with an approximate molecular weight of 277 kDa, contained seven predicted α-helices, and the N-terminal region between amino acids 9 and 161 contained five subdomains according to Ooka et al. (2003). The CarNAC6 nucleotide sequence contained two opposing ORFs that were 924 and 309 nt in length with 145-bp 5′- and 85-bp 3′-UTRs and 332-bp 5′- and 513-bp 3′-UTRs, respectively. The predicted positive-strand CarNAC6 protein was 307 aa in length with an approximate molecular weight of 299 kDa and contained seven predicted α-helices. The predicted complementary-strand CarNAC6 protein was 102 aa in length with an approximate molecular weight of 99 kDa and without predicted α-helix. Alignments of the CarNAC3 and CarNAC6 protein sequences were generated using the Clustal program in the CLC Genomics Workbench 3 software to detect conserved sequence motifs and to determine homology scores (Supplemental Data 1).
Confirmation of Transformation, Transgene Copy Number, and Relative Expression Levels
We confirmed the transformation of eight (each gene four lines) putative transgenic poplar lines that grew on MS selection medium supplemented with kanamycin using PCR followed by electrophoresis of the reaction products with DL2000 DNA Markers on a 1.2 % agarose gel. The production of bright gel bands from all of the transgenic lines corresponding to the relevant positive control bands produced from the PBI121/CarNAC3 and PBI121/CarNAC6 plasmid DNAs demonstrated that both transgenes were integrated into the poplar genome (Supplemental Data 2).
We determined CarNAC3 and CarNAC6 transgene copy numbers using RT-PCR. Based on the formula, X = Y − intercept/slope degree where X = copy number and Y = C t , the value average of four lines of CarNAC3 gene copy number was 6.668 with a slope of −3.35 and an R 2 value of 1.0 (Supplemental Data 3c). The value average of four lines of CarNAC6 gene copy number was 5.933 with a slope of −3.557 and an R 2 value of 0.999 (Supplemental Data 3f).
The C t values for the CarNAC3 and CarNAC6 genes during the exponential phase of amplification were 1.6 and 1.5, respectively (Supplemental Data 3a, d). Comparative quantitative analysis using the ΔΔCt method with expression of the β-actin housekeeping gene serving as an internal reference showed that CarNAC3 expression was approximately fivefold higher than that of CarNAC6, taking into account the similar gene copy numbers (Supplemental Data 4c).
Effects of Transgenic CarNAC3 and CarNAC6 Expression on Growth Phenotypes of Poplar Plants
We examined the effects of CarNAC3 and CarNAC6 gene expression on salt tolerance in transgenic poplar plants as indicated by root and shoot growth rates and the clawed root rate in plants grown on MS agar medium containing various concentrations of NaCl. Both transgenic and WT poplar plants exhibited normal root growth in medium lacking NaCl over a 4-week period, but the transgenic poplar plants exhibited a higher rate of root growth than the WT plants (Fig. 2a). Under mild salt stress (50 mM NaCl), the leaves of WT plants turned yellow and their root growth rates were lower than those of the transgenic poplar lines (Fig. 2b). These results showed that, under mild salt stress, the transgenic lines maintained normal root growth rates while the WT lines did not.
Under moderate salt stress (100 mM NaCl), most leaves of the WT plants appeared withered and yellow and root growth ceased, whereas the transgenic plants exhibited normal root growth. Under this condition, the CarNAC3 transgenic plants started to root during the second week (Fig. 2c). Under high salt stress (150 mM NaCl), the WT plants did not survive, but all of the transgenic lines remained unaffected until the fourth week. Root growth ceased in some of the transgenic lines during the first and second weeks. Under severe salt stress (200 mM NaCl), root growth continued in all of the transgenic lines but with different rates and the CarNAC6 transgenic plants exhibited greater root growth than the CarNAC3 transgenic plants, consistent with the higher expression of CarNAC6 than CarNAC3 in roots (Fig. 2d, e).
In contrast, the transgenic and WT plants maintained normal shoot growth rates during the 4-week period in the absence of salt stress (0 mM NaCl), with the WT plants exhibiting higher shoot growth rates than the transgenic lines (Fig. 3a). Increasing the NaCl concentration to mild or moderate levels of salt stress (50 and 100 mM NaCl) decreased the shoot growth rates in WT plants but had no effect on the transgenic plants, which maintained approximately constant growth rates with a higher rate in the CarNAC3 plants than in the CarNAC6 plants (Fig. 3b, c).
Shoot growth stopped during the fourth week in the WT plants subjected to 150 mM NaCl, but the transgenic lines were unaffected (Fig. 3d). Under severe salt stress (200 mM NaCl), all of the transgenic lines maintained normal shoot growth rates, but some of the WT plants withered and none maintained normal shoot growth (Fig. 3e).
The effects of the transgenes on the clawed root rate were high in both types of transgenic lines and increased significantly under mild salt-stress conditions (Fig. 4a, b). The CarNAC6 transgenic plants exhibited a higher clawed root rate than the CarNAC3 transgenic plants (Fig. 4c, d).
Comparison of clawed root rates (%) in WT and transgenic poplar plants after 4 weeks of growth. a Clawed root rate (%) in the absence of stress (0 mM NaCl). b Clawed root rate under mild salt stress (25 mM NaCl). c A representative CarNAC3 transgenic poplar plant after the third week of treatment with 25 mM NaCl. d A representative CarNAC6 transgenic poplar plant after the third week of treatment with 25 mM NaCl. P-3 CarNAC3 plants, P-6 CarNAC6 plants. Scale bar 1 cm
Evaluation of Drought and Salt Tolerance of Transgenic CarNAC3 and CarNAC6 Poplar Plants Under Greenhouse Conditions
When grown in soil in the greenhouse without salt stress, the WT plants exhibited higher growth rates after 3 weeks than the transgenic plants and the transgenic CarNAC6 plants grew at higher rates than the transgenic CarNAC3 plants (Fig. 5a). To compare the growth of the transgenic CarNAC3 and CarNAC6 plants with that of the WT plants under drought and salt stress conditions, growth continued to be monitored after commencing irrigation with NaCl concentrations of 0, 50, 100, and 150 mM at the start of the fifth week. For all treatments, the plants were irrigated at intervals of 3 days for 2 weeks. Each treatment included three clones of the transformed poplar lines with three replications and the averages of the data from all of the clones were used for statistical analysis.
Comparison of shoot heights between WT and transgenic poplar plants with standard error bars. a Representative WT and transgenic CarNAC3 and CarNAC6 poplar plants after 3 weeks. b Shoot height before and after salt stress with various NaCl concentrations. c Shoot height before and after drought stress for various lengths of time. P-3 CarNAC3 plants, P-6 CarNAC6 plants. White scale bar, 1 cm
The growth of the WT plants in soil watered with 50 mM NaCl continued, but at a much lower rate than when watered without NaCl and was suppressed by 100 mM NaCl, while the transgenic lines maintained normal growth rates at both NaCl concentrations (Fig. 5b). These results indicated that the growth of the transgenic CarNAC3 and CarNAC6 plants was enhanced under salt stress conditions relative to that of the WT plants. The growth rates of the CarNAC3 plants in 100 and 150 mM NaCl were significantly higher than those of the CarNAC6 plants, possibly due to the much higher expression of the CarNAC3 transgene (Fig. 5c). These results indicated that the growth of the transgenic CarNAC3 and CarNAC6 poplar plants was superior to that of the WT plants under salt-stress conditions. These results were also consistent with the much higher expression level of the CarNAC3 transgene under conditions of severe drought and salt stress, as determined by RT-PCR.
Proline, MDA, Chlorophyll, and Carotenoid Amounts and SOD and POD Activities
Proline, one of the most important osmoprotectants in plants, protects cells from damage caused by drought and salt stresses. The amounts of proline were significantly higher in the CarNAC3 and CarNAC6 plants than in the WT control plants under drought- and salt-stress conditions (Fig. 6a, b). SOD activity did not increase significantly in WT plants in response to drought or salt stress, but did increase significantly in all of the transgenic lines (Fig. 6c, d). In addition, POD activity was significantly higher in the transgenic lines than in the WT plants under all treatment conditions (Fig. 6e, f).
Antioxidant activities in WT and transgenic poplar plants under drought- and salt-stress conditions. a, b Proline accumulation in transgenic CarNAC3 and CarNAC6 transgenic plants and WT control plants under drought- and salt-stress conditions. c, d Superoxide dismutase (SOD) enzymatic activity in CarNAC3 and CarNAC6 transgenic and WT control plants under drought- and salt-stress conditions. e, f Guaiacol peroxidase (POD) enzymatic activity in CarNAC3 and CarNAC6 transgenic plants and WT control plants under drought- and salt-stress conditions. g, h Malondialdehyde (MDH) accumulation in response to drought and salt stresses in CarNAC3 and CarNAC6 transgenic plants and WT plants. P-3 CarNAC3 plants, P-6 CarNAC6 plants
In the absence of salt stress, the MDA concentrations in the expressing CarNAC3 and CarNAC6 plants were 1.1- and 1.2-fold lower respectively than WT plants mathematically (Fig. 6g, h). After salt treatment for 2 weeks, the MDA concentrations increase significantly in the CarNAC3 and CarNAC6 transgenic lines.
In the absence of salt stress, the chlorophyll and carotenoid concentrations in the transgenic lines were similar to those in the WT plants (Fig. 7a, b). Under drought- and salt-stress conditions, the chlorophyll concentrations decreased significantly in both the transgenic and WT plants in contrast to the carotenoid levels, which increased. Unlike the transgenic plants, the decrease in the chlorophyll content was greater in response to salt stress than to drought stress in the WT plants. The increases in the carotenoid levels in the WT and transgenic plants were similar but were greater the transgenic plants. These results demonstrated that the effects of transgenic expression of the CarNAC3 and CarNAC6 genes were greater under salt stress than under drought stress. The CarNAC3 lines were more tolerant of both stress conditions than the CarNAC6 lines in all experiments.
Comparison of photosynthetic pigment levels in CarNAC3 and CarNAC6 transgenic plants and WT plants under drought- and salt-stress conditions. a Effects of various levels of salt stress on chlorophyll and carotenoid contents. b Effects of drought stress on chlorophyll and carotenoid contents. P-3 CarNAC3 plants, P-6 CarNAC6 plants
Discussion
The presence of related domains in different proteins may indicate that the proteins have similar biological functions. A phylogenetic analysis of the CarNAC3 protein sequence placed it in the NAC-LIKE ACTIVATED BY AP3/PI (NAP) subgroup containing the NAP, AtNAM, AtNAC2, ANAC047, NAM-B1, GmNAC1, and GhNAC5 proteins (Hui et al. 2009). NAC-type TFs play important roles in the control of responses that confer tolerance to environmental stresses such as drought and salt stress. Overexpression of stress-responsive Arabidopsis SNAC-A genes such as RD26 and ATAF1 and rice SNAC-A genes such as SNAC1, OsNAC6/SNAC2, and OsNAC5 enhanced drought and salt stress tolerance (Hu et al. 2006; Nakashima et al. 2007; Takasaki et al. 2010). Transgenic plants overexpressing the OsNAC6 gene exhibited less growth than transgenic plants overexpressing the SNAC1 or OsNAC5 genes, which exhibited growth similar to that of control plants (Hu et al. 2006; Nakashima et al. 2007; Takasaki et al. 2010). The CarNAC3 gene was expressed primarily in blooming flowers, similar to GmNAC1 and NAP (Sablowski and Meyerowitz 1998; Meng et al. 2007). Moreover, a close homolog of the CarNAC3 and AtNAC2 proteins was shown to be a transcription factor that acts downstream of the auxin signaling pathway to promote lateral root formation (He et al. 2005). Based on these prior results, we investigated the role of CarNAC3 in lateral root development. In this study, we transformed the CarNAC3 and CarANC6 genes into poplar to investigate the effects of their expression in comparison with WT poplar plants. We confirmed the integration of these genes into the poplar genome and their expression using PCR and RT-PCR.
Proline accumulation can affect various physiological processes of plants, such as osmotic adjustment, which is an important mechanism that confers tolerance to drought and salt stress conditions. Proline plays a critical role as an osmoprotectant in the regulation of gene expression in plants subjected to salt stress (Bursy et al. 2007). Proline accumulation could increase the capacity for osmotic adjustment and increase the level of abiotic stress tolerance. In our study, the proline levels in WT and transgenic lines were approximately similar in the absence of stress, but drought and salt treatments caused increased CarNAC3 and CarNAC6 expression in transformed plants and an associated increase in proline accumulation (Fig. 6a, b). Therefore, the osmotic adjustment capacity of the transgenic lines was enhanced relative to the control plants. The improved osmotic adjustment capacity resulting from CarNAC3 and CarNAC6 expression may enable the maintenance of chlorophyll and carotenoid levels to preserve plant growth while subjected to drought and salt stress conditions (Fig. 7a, b). SOD and POD activities and MDA concentration are commonly used indicators of the magnitude of a plant response to abiotic stress (Mittova et al. 2004). Transgenic expression of the CarNAC3 and CarNAC6 genes was shown to increase SOD and POD activities significantly under abiotic stress conditions and to decrease MDA concentrations markedly (Song et al. 2006; Bhagat et al. 2011). In this study, the SOD and POD activities in transgenic lines increased markedly in response to drought and salt stress conditions (Fig. 6c–f) and the MDA concentrations decreased significantly in response to drought stress and severe salt stress relative to the control plants (Fig. 6g, h).
These results suggest that CarNAC3 and CarNAC6 gene expression in poplar reduces oxidative damage to membranes and increases salinity and drought tolerance against stresses. Furthermore, the CarNAC3 gene may play a central role in this resistance due to its higher level of expression than that of the CarNAC6 gene. In the absence of stress, all of the CarNAC3 and CarNAC6 transgenic lines exhibited SOD and POD activity levels similar to those of the WT plants, but under abiotic stress conditions, the CarNAC3 transgenic lines exhibited higher activity levels than the CarNAC6 transgenic lines. These results suggest that CarNAC3 and CarNAC6 perform different functions in oxidative stress responses.
In summary, we introduced the CarNAC3 and CarNAC6 genes into poplar and confirmed by molecular analyses integration of both into the poplar genome and their stable expression at the transcript level. CarNAC3 gene expression mediated osmotic protection and induced antioxidant enzyme systems to higher levels than did CarNAC6 gene expression resulting in improved drought and salt tolerance.
Abbreviations
- 6-BA:
-
N-6-benzyladenine
- TDZ:
-
Thidiazuron
- AS:
-
Acetosyringone
- NPTII :
-
Neomycin phosphotransferase
- MS:
-
Murashige and Skoog
- ORF:
-
Open reading frame
- PCR:
-
Polymerase chain reaction
- RT-PCR:
-
Reverse transcription polymerase chain reaction
- MDA:
-
Malondialdehyde
- POD:
-
Guaiacol peroxidase
- SOD:
-
Superoxide dismutase
- ROS:
-
Reactive oxygen species
References
Abdullahil BM, Lee EJ, Paek KY (2010) Medium salt strength induced changes in growth, physiology and secondary metabolite content in adventitious roots of Morinda citrifolia: the role of antioxidant enzymes and phenylalanine ammonia lyase. Plant Cell Rep 29:685–694
Abraham E, Hourton-Cabassa C, Erdei L, Szabados L (2010) Methods for determination of proline in plants. Methods Mol Biol 639:317–331
Ahmad MSA, Ashraf M, Ali Q (2010) Soil salinity as a selection pressure is a key determinant for the evolution of salt tolerance in blue panic grass (Panicum antidotale Retz.). Flora 205:37–45
Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9:841–857
Al-Whaibi MH, Siddiqui MH, Basalah MO (2011) Salicylic acid and calcium-induced protection of wheat against salinity. Protoplasma 249:769–778
Barba-Espin G, Clemente-Moreno MJ, Alvarez S, Garcia-Legaz MF, Hernandez JA, Diaz-Vivancos P (2011) Salicylic acid negatively affects the response to salt stress in pea plants. Plant Biol (Stuttg) 13:909–917
Ben AC, Ben RB, Sensoy S, Boukhriss M, Ben AF (2010) Exogenous proline effects on photosynthetic performance and antioxidant defense system of young olive tree. J Agric Food Chem 58:4216–4222
Bhagat SS, Ghone RA, Suryakar AN, Hundekar PS (2011) Lipid peroxidation and antioxidant vitamin status in colorectal cancer patients. Indian J Physiol Pharmacol 55:72–76
Bursy J, Pierik AJ, Pica N, Bremer E (2007) Osmotically induced synthesis of the compatible solute hydroxyectoine is mediated by an evolutionarily conserved ectoine hydroxylase. J Biol Chem 282:31147–31155
Cho K, Kim YC, Woo JC, Rakwal R, Agrawal GK, Yoeun S, Han O (2012) Transgenic expression of dual positional maize lipoxygenase-1 leads to the regulation of defense-related signaling molecules and activation of the antioxidative enzyme system in rice. Plant Sci 185–186:238–245
Collinge M, Boller T (2001) Differential induction of two potato genes, Stprx2 and StNAC, in response to infection by Phytophthora infestans and to wounding. Plant Mol Biol 46:521–529
Dong J, Wan G, Liang Z (2010) Accumulation of salicylic acid-induced phenolic compounds and raised activities of secondary metabolic and antioxidative enzymes in Salvia miltiorrhiza cell culture. J Biotechnol 148:99–104
Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14:1675–1690
Guo YF, Gan SS (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46:601–612
He XJ, Mu RL, Cao WH, Zhang ZG, Zhang JS, Chen SY (2005) AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J 44:903–916
Hegedus D, Yu M, Baldwin D, Gruber M, Sharpe A, Parkin I, Whitwill S, Lydiate D (2003) Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Mol Biol 53:383–397
Hoekema A, Hirsch PR, Hooykaas PJJ, Schilperoort RA (1983) A binary plant vector strategy based on separation of vir- and T-region of the Agrobacterium tumefaciens Ti plasmid. Nature 303:179–180
Horsch RB, Fry J, Hoffmann N, Neidermeyer J, Rogers SG, Fraley RT (1988) Leaf disc transformation: plant molecular biology manual. Kluwer Academic Publishers, Dordrecht, pp 1–9
Hou XN, Liang YZ, He XL, Shen YZ, Huang ZJ (2013) A novel ABA responsive TaSRHP gene from wheat contributes to enhanced resistance to salt stress in Arabidopsis thaliana. Plant Mol Biol Rep 31:791–801
Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci U S A 103:12987–12992
Hui P, Cheng HY, Chen C, Yu XW, Yang JN, Gao WR, Shi QH, Zhang H, Li JG, Ma H (2009) A NAC transcription factor gene of chickpea (Cicer arietinum), CarNAC3, is involved in drought stress response and various developmental processes. J Plant Physiol 166:1934–1945
Krantev A, Yordanova R, Janda T, Szalai G, Popova L (2008) Treatment with salicylic acid decreases the effect of cadmium on photosynthesis in maize plants. J Plant Physiol 165:920–931
Lei Y, Korpelainen H, Li C (2007) Physiological and biochemical responses to high Mn concentrations in two contrasting Populus cathayana populations. Chemosphere 68:686–694
Lichtenthaler HK (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148:350–382
Logemann E, Birkenbih RP, Ülker B, Somssich IE (2006) An improved method for preparing Agrobacterium cells that simplifies the Arabidopsis transformation protocol. Plant Methods 2:16. doi:10.1186/1746-4811-2-16
Meng QC, Zhang CH, Gai JY, Yu DY (2007) Molecular cloning, sequence characterization and tissue-specific expression of six NAC-like genes in soybean (Glycine max (L.) Merr.). J Plant Physiol 51:617–30
Mishra P, Bhoomika K, Dubey RS (2011) Differential responses of antioxidative defense system to prolonged salinity stress in salt tolerant and salt-sensitive Indica rice (Oryza sativa L.) seedlings. Protoplasma. doi:10.1007/s00709-011-0365-3
Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagi M (2007) NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19:270–280
Mittova V, Guy M, Tal M, Volokita M (2004) Salinity up-regulates the antioxidative system in root mitochondria and peroxisomes of the wild salt-tolerant tomato species Lycopersicon pennellii. J Exp Bot 55:1105–1113
Movahedi A, Zhang J, Gao P, Yang Y, Wang L, Yin T, Kadkhodaei S, Ebrahimi M, Zhuge Q (2014a) Expression of the chickpea CarNAC3 gene enhances salinity and drought tolerance in transgenic poplars. Plant Cell Tiss Organ Cult. doi:10.1007/s11240-014-0588-z
Movahedi A, Zhang J, Amirian R, Zhuge Q (2014b) An efficient Agrobacterium-mediated transformation system for poplar. Int J Mol Sci 15(6):10780–10793
Mysore KS, Crasta OR, Tuori RP, Folkerts O, Swirsky PB, Martin GB (2002) Comprehensive transcript profiling of Pto- and Prf-mediated host defense responses to infection by Pseudomonas syringae pv. tomato. Plant J 32:299–315
Nakashima K, Tran LS, Van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K (2007) Functional analysis of a NAC type transcription factor OsNAC6 involved in abiotic and biotic stress responsive gene expression in rice. Plant J 51:617–630
Nakashima K, Ito Y, Yamaguchi-Shinozaki K (2009) Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol 149:88–95
Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) NAC transcription factors in plant abiotic stress responses. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms 1819:97–103
Nounjan N, Nghia PT, Theerakulpisut P (2012) Exogenous proline and trehalose promote recovery of rice seedlings from salt-stress and differentially modulate antioxidant enzymes and expression of related genes. J Plant Physiol 169:596–604
Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10:79–87
Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K et al (2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 10:239–47
Pagariya MC, Devarumath RM, Kawar PG (2012) Biochemical characterization and identification of differentially expressed candidate genes in salt-stressed sugarcane. Plant Sci 184:1–13
Porebski L, Bailey L, Baum B (1997) Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep 15:8–15
Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol 133:1755–1767
Sablowski RW, Meyerowitz EM (1998) A homolog of No Apical Meristem is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92:93–103
Santner A, Estelle M (2009) Recent advances and emerging trends in plant hormone signalling. Nature 459:1071–1078
Satoh K (1978) Serum lipid peroxide in cerebrovascular disorders determined by a new colorimetric method. Clin Chim Acta 90:37–43
Song S, Liu W, Guo S, Shang Q, Zhang Z (2006) Salt resistance and its mechanism of cucumber under effects of exogenous chemical activators. Ying Yong Sheng Tai Xue Bao 17:1871–1876
Souer E, Van AH, Kloos D, Mol J, Koes R (1996) The No Apical Meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85:159–170
Stewart RR, Bewley JD (1980) Lipid peroxidation associated with accelerated aging of soybean axes. Plant Physiol 65:245–248
Su X, Chu Y, Li H, Hou Y, Zhang B, Huang Q, Hu Z, Huang R, Tian Y (2011) Expression of multiple resistance genes enhances tolerance to environmental stressors in transgenic poplar (Populus x euramericana ‘Guariento’). PLoS ONE 6:e24614
Takasaki H, Maruyama K, Kidokoro S, Ito Y, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K, Nakashima K (2010) The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol Genet Genomics 284:173–183
Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J (2006) A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314:1298–301
Wang N, Hua H, Egrinya Eneji A, Li Z, Duan L, Tian X (2012) Genotypic variations in photosynthetic and physiological adjustment to potassium deficiency in cotton (Gossypium hirsutum). J Photochem Photobiol B 110:1–8
Xie Q, Sanz-Burgos AP, Guo H, García JA, Gutiérrez C (1999) GRAB proteins, novel members of the NAC domain family, isolated by their interaction with a Gemini virus protein. Plant Mol Biol 39:647–656
Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57:781–803
Yamaguchi-Shinozaki K, Koizumi M, Urao S, Shinozaki K (1992) Molecular cloning and characterization of 9 cDNAs for genes that are responsive to desiccation in Arabidopsis thaliana: sequence analysis of one cDNA clone that encodes a putative transmembrane channel protein. Plant Cell Physiol 33:217–224
Zheng XN, Chen B, Lu GJ, Han B (2009) Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochem Biophys Res Commun 379:985–989
Zhou J, Wang J, Bi Y, Wang L, Tang L, Yu X, Ohtani M, Demura T, Zhuge Q (2014) Overexpression of PtSOS2 enhances salt tolerance in transgenic poplars. Plant Mol Biol Rep 32:185–197
Acknowledgments
We thank Prof. Hao Ma (Nanjing Agricultural University, China) for providing the CarNAC3 and CarNAC6 plasmids. This work was supported by grants from the Forestry Public Benefic Research Program (No. 201304102) and the International Science and Technology Cooperation Program of China (No. 2014DFG32440), Priority Academic Program Development of Jiangsu Higher Education Institution, and the Program for Innovative Research Team at the University of Educational Department and Jiangsu Province, China.
Conflict of Interest
The authors do not have conflicts of interest to declare.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Data 1
(DOC 681 kb)
Supplemental Data 2
(DOC 69 kb)
Supplemental Data 3
(DOC 906 kb)
Supplemental Data 4
(DOC 589 kb)
Rights and permissions
About this article
Cite this article
Movahedi, A., Zhang, J., Yin, T. et al. Functional Analysis of Two Orthologous NAC Genes, CarNAC3, and CarNAC6 from Cicer arietinum, Involved in Abiotic Stresses in Poplar. Plant Mol Biol Rep 33, 1539–1551 (2015). https://doi.org/10.1007/s11105-015-0855-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-015-0855-0