Abstract
RNA-binding proteins are critical to RNA metabolism in cells and, thus, play important roles in diverse biological processes. In the present study, we identified the YTH domain-containing RNA-binding protein (RBP) family in Arabidopsis thaliana and rice at the molecular and biochemical levels. A total of 13 and 12 genes were found to encode YTH domain-containing RBPs in Arabidopsis and rice and named as AtYTH01–13 and OsYTH01–12, respectively. The phylogeny, chromosomal location, and structures of genes and proteins were analyzed. Electrophoretic mobility shift assays demonstrated that recombinant AtYTH05 protein could bind to single-stranded RNA in vitro, demonstrating that the YTH proteins have RNA-binding activity. Analyses of publicly available microarray data, gene expression by qRT-PCR, and AtYTH05 promoter activity indicate that the Arabidopsis AtYTHs and rice OsYTHs genes have distinct and diverse expression patterns in different tissues and developmental stages, showing tissue- and developmental-specific expression patterns. Furthermore, analyses of publicly available microarray data also indicate that many of the Arabidopsis AtYTHs and rice OsYTHs genes might be involved in responses to various abiotic and biotic stresses as well as in response to hormones. Our data demonstrate that the YTH family proteins are a novel group of RBPs and provide useful clues to define their biological functions of this RBP family in plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The regulation of gene expression at the posttranscriptional level is important for plant growth, development, and stress responses. Posttranscriptional regulation includes pre-messenger RNA (mRNA) splicing, capping, polyadenylation, mRNA stability, and mRNA transport (Glisovic et al. 2008; Morris et al. 2010; Mittal et al. 2011; Stower 2014). In these processes, regulation is mainly achieved either directly by RNA-binding proteins (RBPs) or indirectly, whereby RBPs modulate the function of other regulatory factors (Janga and Mittal 2011). In eukaryotes, RBPs participate in synthesizing, processing, editing, modifying, transporting, and exporting RNA molecules from the nucleus (Ambrosone et al. 2012; Huh and Paek 2013; Pallas and Gomez 2013) and, thus, are involved in all aspects of translating mRNAs, as well as in storing them while they are not being translated (Johnstone and Lasko 2001; Zhong et al. 2001). RBPs also regulate and maintain the stability of mRNA in cells (Glisovic et al. 2008; Ambrosone et al. 2012). In addition to their involvement in these very central aspects of decoding the information stored in DNA, RBPs are also involved in certain aspects of chromosome structuring and coarse regulation such as telomere maintenance (Chen and Lingner 2012).
All aspects of RNA metabolism are accompanied by the activities of a myriad of RBPs (Glisovic et al. 2008). Most of the RBPs are built-in versatile modular structures with multiple repeats of a few conserved domains arranged in a variety of ways to fulfil their diverse functional requirements (Lunde et al. 2007). RBPs are characterized by the presence of one or more conserved RNA-binding domains such as RNA recognition motif (RRM), heterogeneous nuclear ribonucleoprotein K homology domain, Piwi Argonaut and Zwille domain, and the double-stranded RNA-binding domains (Ambrosone et al. 2012). It was estimated that there are more than 200 putative RBP genes in Arabidopsis thaliana genome (Lorkovic 2009), whereas about 250 RBP genes have been identified in Oryza sativa (Cook et al. 2011). Many RBPs seem to be unique to plants, suggesting that they might serve specific functions in plants (Ambrosone et al. 2012).
The YTH (YT521-B homology) domain, comprising 100–150 amino acids, is highly conserved and was identified by comparing some known protein sequences with the splicing factor YT521-B (Stoilov et al. 2002). The first member of the YTH-containing proteins is the YT521-B, which is a nuclear protein interacting with other proteins (e.g., Sam68/p62, rSLM-1, rSLM-2, hnRNPG, rSAF-B, and emerin) and is implicated in RNA metabolism (Imai et al. 1998; Hartmann et al. 1999; Stoss et al. 2004). The human splicing factor YT521 (YTHDC1) is characterized by alternatively spliced isoforms with regulatory impact on cancer-associated gene expression (Zhang et al. 2010a; Hirschfeld et al. 2014). Recent studies also found that the human YTH family members YTHDF1/2/3 selectively bind to N6-methyladenosine and thus regulate the stability of mRNA (Dominissini et al. 2012; Wang et al. 2014). In yeast, Mmi1 (meiotic mRNA interception), a YTH protein, was found to be a part of the mRNA-destruction system that eliminates meiotic-specific mRNAs (Harigaya et al. 2006; McPheeters et al. 2009), whereas Ydr374c (Pho92), the homologue of human YTHDF2, was shown to participate in cellular phosphate metabolism through the regulation of PHO4 mRNA stability (Kang et al. 2014).
It was speculated that the YTH domain is especially abundant in plants (Stoilov et al. 2002). However, information on the YTH genes or proteins in plants is very limited. In the present study, we performed genome-wide identification, biochemical characterization, and expression patterns of the YTH genes in Arabidopsis and rice. Gene expression analyses based on publicly available microarray data, qRT-PCR, and AtYTH05 promoter activity indicate that members of the YTH family in Arabidopsis and rice should have yet-unknown functions in growth and development as well as in stress responses. Our studies demonstrate that the YTH family proteins are a novel group of RBPs and provide clues for studying their biological functions in the future.
Materials and Methods
Plant Materials and Growth Conditions
Arabidopsis ecotype Columbia-0 was used in all experiments. Plants were grown in soil in a growth room under a 10-h light (100 μmol s−1 m−2 photons m−2 s−1 of intensity) and 14-h dark cycle at 22 ± 2 °C with 60 % relative humidity. Samples for the analysis of gene expression patterns were collected from 6-week-old soil-grown plants and stored at −80 °C until use. Nicotiana benthamiana plants used for subcellular localization study were grown in the same growth room under same conditions.
Identification and Analysis of the YTH Genes in Arabidopsis and Rice
Two rounds of multiple database searches were performed to identify YTH genes in Arabidopsis and rice. In the first round, the rice (http://rice.plantbiology.msu.edu/, version 6.1) and Arabidopsis (http://www.arabidopsis.org/, TAIR10) genome databases were searched using the default settings with the YTH domain (Pfam PF04146, http://pfam.sanger.ac.uk/family/PF04146) sequence as a query. The protein and predicted complementary DNA (cDNA) and genomic sequences for putative YTH genes were downloaded from http://rice.plantbiology.msu.edu and http://www.arabidopsis.org websites, respectively. In the second round, the predicted protein and cDNA sequences of the YTH genes were used as queries for blastp or blastn searches against several nucleic acid and protein databases including TIGR (http://www.tigr.org/tdb/agi/), DDBJ database (http://www.ddbj.nig.ac.jp/E-mail/homology.html), and GenBank (http://www.ncbi.nlm.nih.gov/) to identify other possible YTH genes and retrieve full-length cDNA sequences. Intron/exon organization for each YTH gene was analyzed at Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/) by comparing the predicted cDNA, available full-length cDNA, and genomic DNA sequences. Searches for putative conserved domains in the YTH proteins were performed using Scanprosite tool at http://prosite.expasy.org/scanprosite/. Molecular weight calculations and pI prediction were carried out using Compute pI/Mw tool at http://us.expasy.org/tools/pi_tool.html (Bjellqvist et al. 1994). The putative secondary structure of the YTH domains was determined using the PHD program (Rost 1996). Further domains or motifs in the YTH proteins were searched using SMART (Letunic et al. 2012; Schultz et al. 1998).
Phylogenetic Analysis
The conserved YTH domain sequences for the identified Arabidopsis and rice YTH proteins were aligned using ClustalX (version 2.0.8) followed by manual adjustment. A phylogenetic tree was generated by the neighbor-joining (NJ) algorithm with p-distance method and pairwise deletion of gaps using MEGA version 6.05 with default parameters (Tamura et al. 2013). A bootstrap statistical analysis was performed with 1,000 replicates to test the phylogeny.
Subcellular Localization of AtYTH05 and AtYTH07 Proteins
Total RNA was extracted from the leaves of 2-week-old Arabidopsis seedlings, and first-strand cDNAs were synthesized using AMV reverse transcriptase following the manufacturer’s recommendation (TaKaRa, Dalian, China). Coding sequences of AtYTH05 and AtYTH07 were amplified using specific primers AtYTH05-GFP-F (5′-GGA AGA TCT ATG GAT TCG AAT GGT CAA GTT C-3′, a BglII site underlined)/AtYTH05-GFP-R (5′-TCC CCC GGG TTA CAT GGT AGA ATC AAC AGT-3′; a SmaI site underlined) and AtYTH07-GFP-F (5′-CGC GGA TCC ATG GCT GGA GCC GCT TCT-3′, a BamHI site underlined)/AtYTH07-GFP-R (5′-TGC TCT AGA TCA ACT CAA GTT AAT TGT TTC-3′, a XbaI site underlined) and cloned into pFGC-Egfp. The resulting pFGC-Egfp-AtYTH05 and pFGC-Egfp-AtYTH07 were transformed into Agrobacterium tumefaciens strain GV3101 by the electrotransformation method. Agrobacterium cells were grown at 28 °C overnight, collected by centrifugation and resuspended in MES buffer (10 mM MgCl2, 10 mM MES, pH 5.6, and 150 mM acetosyringone) to OD600 = 0.6. The Agrobacterium suspension was infiltrated into fully expanded leaves of N. benthamiana plants (Goodin et al. 2002). GFP-expressing cells were detected with a confocal laser scanning microscope (Zeiss LSM 510 META; argon laser exCitation wavelength, 488 nm).
Purification of the Recombinant AtYTH05 Protein and RNA-Binding Assay
The coding region of AtYTH05 was amplified with primers AtYTH05-pRSET-1F (5′-CC CTC GAG ATG GAT TCG AAT GGT CAA GTT C-3′, a XhoI site underlined) and AtYTH05-pRSET-1R (5′-GG GGT ACC TTA CAT GGT AGA ATC AAC AGT-3′, a KpnI site underlined). The amplified fragment was digested with XhoI/KpnI and cloned into pRSET-A (Invitrogen, Carlsbad, CA, USA). The resulting plasmid, pRSET(A)-AtYTH05, was confirmed by sequencing and introduced into Escherichia coli BL21(DE3) plus cells. Purification of the recombinant protein from induced and uninduced bacteria pRSET(A)-AtYTH05 was carried out with the His-Bind kit (NovaGen, Madison, WI, USA) following the manufacturer’s recommendations. Protein concentration was determined with the Bio-Rad protein assay kit (Bio-Rad, Hercules, CA, USA).
RNA-binding activity of the recombinant AtYTH05 protein was analyzed by electrophoretic mobility shift assay. All reactions were performed on ice in 20 μl binding buffer (20 mM HEPES, pH 8.0, 50 mM KCl, 1 mM DTT, and 5 % glycerol). RNA substrate used for RNA-binding activity assays was prepared as described (Iost et al. 1999). A 58-nt single-strand RNA substrate (5′-UGC AUG CCU GCA GGU CGA CUC UAG AGG AUC CCC GGG UAC CGA GCU CGA AUU CGU AUU C-3′), labeled with [α-32P]-dCTP, was generated by in vitro transcription from pGEM-4Z (Promega, Madison, WI, USA) using the MAXIscript system according to the manufacturer’s instructions (Ambion, Austin, TX, USA). The synthesized transcript was purified by native polyacrylamide gel electrophoresis. Binding reactions contained 1 nM labeled RNA and 1 or 2 μg of protein, with or without cold competitor RNA, incubated on ice for 1 h, and run on a 6 % nondenaturing acrylamide gel (Mini Protean II, Bio-Rad, Hercules, CA, USA) at 100 V for 45 min at 4 °C. Gels were dried, exposed to a storage phosphor screen for 6–12 h, and autoradiographed by exposure to storage phosphor screens (GE Healthcare, Pittsburgh, PA, USA) for 4 h.
Generation of AtYTH05 pro ::GUS Transgenic Lines and GUS Staining
A 1.5-kb fragment upstream of the AtYTH05 gene was amplified from genomic DNA using specific primers AtYTH05p-1F (5′-GC GTC GAC CAA AGA CAG ACA AAA TTC AGC AC-3′, a SalI site underlined) and AtYTH05p-1R (5′-AT GAA TTC AGC TCC ATA CTG GTA AAC ACC-3′, a EcoRI site underlined). The PCR product was cloned into pMD19-T vector (Takara, Dalian, China) and sequenced for confirmation. The entire 1.5-kb promoter fragment was cloned into the pCAMBIA1301 to create AtYTH05p-GUS fusion construct and transformed into A. tumefaciens strain GV3101. The floral dip method was used for Arabidopsis transformation (Clough and Bent 1998). Independent AtYTH05p-GUS transgenic lines were carried to T3 homozygous and stained for GUS activity as described previously (Jefferson et al. 1987).
Expression Analyses of the AtYTHs and OsYTHs Genes
Microarray expression data from various datasets were obtained using GENEVESTIGATOR (https://www.genevestigator.com/gv/) with the Arabidopsis and rice Gene Chip platform. The web site provides a web-based search interface to search for probes of genes on the Arabidopsis and rice GeneChip by using keywords as well as probe ID numbers and GO terms as query terms. The expression data for each gene in different development stages, organs, and under different abiotic and biotic stress conditions were mined. Results are given as heatmaps in different color coding that reflects absolute signal values. Only the expression data from Arabidopsis ecotype Columbia-0 were mined, while the expression data from different rice cultivars were extracted for analyses. The Arabidopsis microarray expression analyses were obtained from samples infected by Botrytis cinerea (Bc), Blumeria graminis f. sp. hordei (Bgh), cabbage leaf curl virus (CaLCuV), Golovinomyces cichoracearum (Gc), Golovinomyces orontii (Go), Hyaloperonospora arabidopsidis (Ha), Meloidogyne incognita (Mi), Phytophthora infestans (Pi), Phytophthora parasitica (Pp), Pseudomonas syringae pv. maculicola (Psm), P. syringae pv. phaseolicola (Psp), P. syringae pv. tomato (Pst), or turnip mosaic virus (TuMV). The rice microarray expression analyses were performed on samples infected with Magnaporthe grisea (Mg), Magnaporthe oryzae (Mo), Xanthomonas oryzae pv. oryzae (Xoo), or X. oryzae pv. oryzicola (Xoc). Elicitors and their concentrations used for microarray expression analysis in Arabidopsis are as follows: elongation factor Tu (EF-Tu, 1 μM), Flg22 (10 μM for seedlings, 100 nM for leaves), necrosis-inducing Phytophthora Protein 1 (NPP, 1 μM), HarpinZ (HrpZ, 10 μM), LPS (1 μM), oligogalacturonides (OGs, degree of polymerization 9–16; 50 μg/ml), or Pep2 (1 μM). Abiotic stress treatments in Arabidopsis microarray expression analyses include cold (4 °C), drought (withholding water), heat (38 °C for seedlings, 37 °C for leaf and whole plants), hypoxia (0.1 % oxygen), osmotic (300 mM mannitol), oxidative (10 μM methyl viologen), salt (150 mM NaCl for leaf and 140 mM for root), or submergence (placing plants into water-filled tanks in darkness). Rice abiotic stress treatments include anaerobic (seeds submerged at 30 °C in darkness and continuously bubbled with nitrogen), cold (4 °C), drought (slowly draining to relative water content of 65–75 % in leaves), heat (42 °C), salt (200 mM NaCl), or submergence (submerged in deionized water). Hormones and their concentrations used in Arabidopsis microarray expression analyses are as follows: abscisic acid (ABA, 10 μM for seedlings, 100 μM for leaf and whole plants, or 50 μM for guard cells), 1-aminocyclopropane-1-carboxylic acid (ACC, 10 μM), brassinolide (BL, 10 μM), ethylene (5 ppm), gibberellic acid (GA3, 1 μM), methyl jasmonate (MeJA, 10 μM), 1-naphthalene acetic acid (NAA, 1 μg/ml), salicylic acid (SA, 2 mM for seedlings and 0.3 mM for leaf), 1-N-naphthylphthalamic acid (NPA, 10 μM), or Zeatin (20 μM). Hormones and their concentrations used in rice microarray expression analyses include 6-benzylaminopurine (BAP, 50 μM), GA3 (100 μM), indole-3-acetic acid (IAA, 50 μM), kinetin (KT, 100 μM), NAA (100 μM), or trans-zeatin (5 μM).
For qRT-PCR analysis of gene expression, total RNA was extracted from different plant tissues with TRIzol (Invitrogen, Carlsbad, CA, USA) and then treated with RNase-free DNase (TaKaRa, Dalian, China). First-strand cDNA was synthesized from 0.6 μg total RNA using AMV reverse transcriptase (TaKaRa, Dalian, China) as instructed. Each qPCR reaction contained 12.5 μl 2× SYBR Premix Ex Taq (TaKaRa, Dalian, China), 0.1 μg cDNA, and 7.5 pmol of each gene-specific primer (Table 1) in a final volume of 25 μl and had three independent biological replicates. AtActin2 expression was used as an internal control. The qPCR was performed in a CFX96 real-time PCR detection system (Bio-Rad, Hercules, CA, USA). Relative gene expression levels were calculated using the 2−ΔΔCT or 2−ΔCT method as described (Livak and Schmittgen 2001).
Results
Identification of the Arabidopsis and Rice YTH Gene Family
Considering that most of the plant YTH genes or proteins have not been studied for their biological functions, we therefore performed genome-wide identification, biochemical characterization, and comparative expression analysis of the YTH genes in Arabidopsis and rice. Using the YTH domain sequence as a query, database searches against the Arabidopsis and rice genomic sequences identified totals of 13 and 12 genes encoding putative YTH domain-containing proteins in Arabidopsis and rice, respectively (Table 2). No more putative YTH genes in Arabidopsis and rice were identified in further searches against other databases using the obtained YTH genes as queries, suggesting that there are 13 and 12 genes encoding for the YTH domain-containing proteins in Arabidopsis and rice genomes, respectively. For convenience, we assigned names to these YTH genes as AtYTH01–13 (Arabidopsis thaliana YTHs) and OsYTH01–12 (Oryza sativa YTHs) according to the chromosomal positions. Information of the gene loci, molecular weights, pIs, and chromosomal locations for AtYTH01–13 and OsYTH01–12 are listed in Table 2. Full-length cDNAs were identified in the GenBank database for most of the Arabidopsis and rice YTH genes except for AtYTH03, AtYTH11, and OsYTH09, indicating that the YTH genes are normally expressed in Arabidopsis and rice plants. The YTH family members show an uneven distribution on the rice and Arabidopsis chromosomes. The AtYTHs genes are distributed on chromosomes I, III, IV, and V, and three pairs AtYTH02/AtYTH03, AtYTH08/AtYTH09, and AtYTH12/AtYTH13 are located closely (interval distance ~1,000 kB) on chromosomes I, III, and V, respectively (Table 2). By contrast, the OsYTHs genes separately distribute on chromosomes I, III, IV, V, VI, VII, and VIII (Table 2). According to their exon and intron organizations, the Arabidopsis and rice YTH genes can be divided into different groups with similar exon and intron arrangements within each group (Fig. 1). Interestingly, none of the members in each group shows identical gene structure with respect to the size and arrangement of the exons and introns (Fig. 1). The features in gene structure and exon/intron organization might indicate that the members of the Arabidopsis and rice YTH family might evolve independently.
Characterization of the Arabidopsis and Rice YTH Proteins and Phylogenetic Analysis
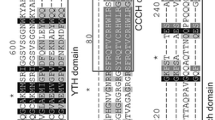
The Arabidopsis and rice YTH proteins comprise 360 ~ 717 amino acids (Table 2). Each of the predicted AtYTHs and OsYTHs protein contains an YTH domain as confirmed by blastp searches at GenBank and the YTH domain is usually located near the C-terminal except the AtYTH03, AtYTH11, OsYTH04, and OsYTH09, whose YTH domains are located at the middle regions in the proteins (Fig. 2). The YTH domains in the AtYTHs and OsYTHs proteins are highly conserved with 14 invariant and 19 highly conserved residues and are predicted to have a mixed αβ-fold secondary structure with four α-helices and six β-strands (Fig. 3). A notable feature of the YTH domain is the presence of highly conserved aromatic residues located in the β-sheet (Fig. 3). Overall, the sequences outside the conserved YTH domain in the AtYTHs and OsYTHs proteins vary dramatically without significant similarity. The SMART and PFAM databases were searched for additional known domains probably existing in the AtYTHs and OsYTHs proteins, and these searches identified a conserved CCCH-type zinc finger domain in AtYTH03 and OsYTH09 proteins (Fig. 2). No other conserved domain and motifs were found in all the rest of the AtYTHs and OsYTHs proteins (Fig. 2).
To investigate the molecular evolution and phylogenetic relationships among the YTH proteins in Arabidopsis and rice, a phylogenetic tree was constructed by alignment of the YTH domain sequences because the sequences outside the YTH domain in the OsYTHs and AtYTHs proteins showed very low levels of similarity (Fig. 4). This phylogenetic tree analysis identified roughly four main groups and most of the AtYTHs and OsYTHs fall into one of these four groups except that OsYTH01 could cluster into any group (Fig. 4). Members of groups I, II, and III only contain one conserved YTH domain without any other conserved domain or motif. AtYTH03 and OsYTH09 in group IV have two conserved domains: one YTH domain and one CCCH zinc finger domain. By contrast, AtYTH11 in this group, most similar to AtYTH03 in blast searching, contains only one YTH domain and does not have other conserved domain or motif. These data indicate that the Arabidopsis and rice YTH proteins might be structurally divergent for their different biological and biochemical functions.
Phylogenetic tree of the AtYTHs and OsYTHs proteins. An unrooted neighbor-joining tree was generated from multiple sequence alignment of the YTH domains in AtYTHs and OsYTHs proteins. Sequence alignment was done using ClustalX 2.0.8 and the tree was generated using MEGA6. The AtYTHs and OsYTHs proteins are grouped based on the bootstrap supporting value ≧50 %. Scale bar represents 0.05 amino acid substitutions per site
Subcellular Localization of AtYTH05 and AtYTH07 Proteins
The AtYTH05 and AtYTH07 were clustered in the same group III with the rice OsYTH03 (Fig. 4), whose gene was found to be differentially expressed gene in rice response to M. grisea in another study. Therefore, we chose these two AtYTHs proteins for further biochemical analysis. To examine the subcellular localization of the YTH proteins, we fused the coding sequences of AtYTH05 and AtYTH05 to GFP at the N-terminals under the control of CaMV 35S promoter and transiently expressed in N. benthamiana leaves. Transient expression assays reveal that GFP signals in the GFP-AtYTH05- and GFP-AtYTH07-expressed N. benthamiana leaves were observed in the cytoplasm, membrane, and nucleus of the cells, similar to the pattern as seen for GFP alone (Fig. 5a). These results indicate that the AtYTH05 and AtYTH07 proteins do not target specific compartments and are likely to distribute ubiquitously in cells.
Subcellular localization of AtYTH05 and AtYTH07 and RNA-binding activity of AtYTH05. a Subcellular localization analysis. The AtYTH05::GFP, AtYTH07::GFP, and GFP alone constructs were transiently expressed through agroinfiltration in Nicotiana benthamiana leaves and green fluorescence of the GFP was viewed using confocal laser microscopy (left row). The same cells were also viewed by transmission microscopy (middle row) and both images were merged (right row). b Purification of recombinant AtYTH05 protein from E. coli. M, marker for protein molecular weight; lane 1, total cell proteins from un-induced bacteria carrying pRSET(A)-AtYTH05; lane 2, total cell protein from induced bacteria carrying pRSET(A)-AtYTH05; lane 3, purified protein from uninduced bacteria carrying pRSET(A)-AtYTH05; lane 4, purified AtYTH05 protein. c RNA-binding activity of the recombinant AtYTH05 protein. Lane 1, BSA alone as the negative control; lane 2, [α-32P]-labeled ssRNA probe; lane 3, competition assay with unlabeled ssRNA probe; lanes 4 and 5, 2 ng [α-32P]-labeled ssRNA probe and 1 μg (lane 4) or 2 μg (lane 5) AtYTH05 protein, respectively
Recombinant AtYTH05 Protein Showed RNA-Binding Activity
Recent studies showed that splicing factor YT521-B, a typical YTH domain-containing protein, can bind to a short, degenerated, single-stranded RNA sequence (Zhang et al. 2010b). To determine whether the plant YTH proteins have RNA-binding activity, we expressed the AtYTH05 in E. coli, purified the recombinant AtYTH05 protein, and examined its RNA-binding activity in vitro. In SDS-PAGE gel, the recombinant AtYTH05 protein from induced bacteria carrying pRSET(A)-AtYTH05 showed a single band with the calculated molecular weight similar to the predicted amino acid sequence (Fig. 5b). In gel mobility shift assays, the negative controls, in the presence of BSA only or absence of the AtYTH05 protein in the binding reactions containing the ssRNA probe, did not show any signal (Fig. 5c, lanes 1, 2, and 3). The recombinant AtYTH05 protein bound to the ssRNA probe, and this binding activity was in competition with an unlabeled wild-type probe (Fig. 5c, lanes 4 and 5). These results demonstrate that the AtYTH05 protein can bind to single-strand RNA molecules.
Development-Specific Expression Patterns of the YTH Genes
To gain insights into the possible functions of the YTH genes in Arabidopsis and rice growth and development, we took advantage of the publicly available microarray data and analyzed the expression patterns using the GENEVESTIGATOR tool. Expression patterns of the AtYTHs and OsYTHs genes over different developmental stages of Arabidopsis and rice plants are shown in Fig. 5. The data showed that expression of the AtYTHs and OsYTHs genes was highly developmentally regulated in Arabidopsis and rice plants. Most of the AtYTHs genes except AtYTH08 and AtYTH10 have high expression potentials during senescence stage. AtYTH09 and AtYTH13 showed relatively high levels of expression potentials in vegetative and senescence stages and relatively low potentials in the flowering stage (Fig. 6a). Relatively high levels of expression potentials were detected for AtYTH02 and AtYTH04 in mature siliques and for AtYTH06 in rosettes (Fig. 6a). Most of the AtYTHs genes show expression potentials during different developmental stages, but the expression potential of AtYTH12 was primarily limited to the late stages of silique development and senescence process (Fig. 6a). AtYTH01, AtYTH03, AtYTH08, and AtYTH10 showed low expression potentials in developmental stages including germinated seed, seedlings, and rosettes (Fig. 6a). In rice, OsYTH01, OsYTH06, and OsYTH08 showed high expression potentials in the flowering stage, while OsYTH02, OsYTH03, OsYTH04, OsYTH05, OsYTH07, OsYTH10, and OsYTH12 had low potentials of expression in the flowering stage (Fig. 6b). OsYTH02, OsYTH03, OsYTH05, OsYTH07, and OsYTH10 exhibited relatively high expression potentials in the early stages of rice development (Fig. 6b). Notably, expression of OsYTH01 was largely restricted to the late stages such as flowering, milking, and dough stages, whereas only OsYTH10 showed an upregulated expression during seed germination (Fig. 6b).
Expression patterns of AtYTHs (a) and OsYTHs (b) in different developmental stages. The color scheme represents log2 ratio of expression levels with the color deep red indicating relatively high expression and the scale representing the relative signal values is shown. The developmental stages and numbers of samples used for analysis are shown on the top
To confirm the expression patterns established by analysis of the public microarray data, we examined by qRT-PCR analysis the expression patterns of the AtYTHs genes using total RNA extracted from the roots, leaves, stems, flowers, and siliques of soil-grown 6-week-old plants (Fig. 7a). Distinct expression patterns of the AtYTHs genes were observed. In the roots, most of the AtYTHs genes showed high levels of expression except AtYTH11 and AtYTH12. Higher levels of expression were detected in the stems for AtYTH01 ~ 04, AtYTH08 ~ 11, and AtYTH13, especially for AtYTH04 and AtYTH13, whereas in the same samples, the expression levels of the rest of the AtYTHs genes were low (Fig. 7a). AtYTH04 ~ 06, AtYTH08, AtYTH09, and AtYTH13 showed high levels of expression in the leaves. In contrast, high levels of expression were detected for AtYTH05, AtYTH06, and AtYTH07 in the flowers and siliques (Fig. 7a). AtYTH06 showed high levels of expression in the leaves, flowers, and siliques but low levels in the roots. However, expression of AtYTH13 was detected in all organs examined with relatively high levels of expression in the roots, stems, leaves, and siliques and modest levels in the flowers. On the contrary, AtYTH12 showed relatively low expression level in all the organs examined. Overall, the expression patterns established by qRT-PCR analysis of the AtYTH01–13 genes in different developmental stages were similar or comparable to the patterns obtained from the microarray data.
Expression patterns of the AtYTHs genes in different tissues of Arabidopsis plants. a qRT-PCR analyses of AtYTHs expressions in different tissues. b GUS staining-based analysis of AtYTH05 expression in AtYTH05 pro ::GUS transgenic plants. a Emerging leaves and shoot apex in seedlings; b leaf from 4-week-old transgenic plant; c flower buds, a mature flower, and stem; d a developing silique
The expression pattern of AtYTH05 was further confirmed by analysis of its promoter activity in transgenic plants. A 1.5-kb promoter sequence upstream of the AtYTH05 coding region was fused to the GUS reporter gene, and stable transgenic AtYTH05 pro ::GUS lines were generated. In 2-week-old AtYTH05 pro ::GUS seedlings, no significant GUS staining could be detected in the entire cotyledons including petioles, distal part of the first leaves, vascular tissues of the hypocotyls, and maturation and elongation zones of the roots (data not shown). GUS staining could not be detected until the AtYTH05 pro ::GUS plants started into the flowering stage. When the AtYTH05 pro ::GUS plants began flowering, GUS staining was detected in the margin of mature leaves, stem, style of the mature flowers, and developing siliques (Fig. 7b). These data further confirm that the AtYTH01–13 genes, at least for AtYTH05, are regulated developmentally.
Tissue-Specific Expression Patterns of the YTH Genes
To gain further information on the possible involvement of the YTHs genes in growth and development, we analyzed the tissue-specific expression patterns of AtYTHs and OsYTHs genes in Arabidopsis and rice plants. Generally, no identical expression pattern for each of the AtYTHs or OsYTHs gene in different tissues examined was observed (Fig. 8). However, most of the AtYTHs or OsYTHs genes showed overlapping expression patterns at least in two or more different tissues (Fig. 8). The AtYTHs genes except AtYTH06 showed relatively high expression potentials in seeds such as the embryo, endosperm, and testa and in the shoot, especially for AtYTH03, AtYTH07, AtYTH08, AtYTH10, and AtYTH12 in seeds and for AtYTH13 in axillary shoot (Fig. 8a). The AtYTHs genes except AtYTH01, AtYTH02, and AtYTH11 showed high expression potentials in the root tip (Fig. 8a). AtYTH01–04, AtYTH08, AtYTH09, and AtYTH11-13 but not AtYTH05-07 and AtYTH10 exhibited moderate expression potentials in raceme such as flowers and pedicel (Fig. 8a). However, the AtYTHs genes except AtYTH09 and AtYTH13 showed low expression potentials in inflorescence, rosettes (stem, leaf, and bud), and pericarp (Fig. 8a). Notably, AtYTH12 in pedicel, AtYTH06 in embryo and pericarp, and AtYTH01 and AtYTH03 in stem exhibited very low expression potentials (Fig. 8a). In rice, more diverse patterns of expression potentials for OsYTHs genes in different tissues of rice plants were observed as compared with those for AtYTHs in Arabidopsis plants. Most of the OsYTHs genes showed differential expression potentials in almost all the examined tissues in rice plants (Fig. 8b). Surprisingly, OsYTH02 and OsYTH06 exhibited high expression potentials in the stele of internodes (Fig. 8b). OsYTH09 in coleptile, OsYTH05 in node, and OsYTH04 in collar and crown tissues exhibited very low expression potentials (Fig. 8b). Taken together, these data demonstrate that the AtYTHs and OsYTHs genes have tissue-specific expression potentials in Arabidopsis and rice plants.
Expression patterns of AtYTHs (a) and OsYTHs (b) in different tissues. The color scheme represents log2 ratio of expression levels with the color deep red indicating relatively high expression and the scale representing the relative signal values is shown. Different tissues used for analysis are shown and numbers of samples used are listed in the middle
Expression Patterns of the YTH Genes in Response to Pathogen Infection and Elicitor Treatment
To gain insights into the possible involvement of the Arabidopsis and rice YTH genes in response to various stresses, we analyzed the expression patterns of AtYTHs and OsYTHs in Arabidopsis and rice plants, respectively, after infections with different types of pathogens. Differential expression patterns of AtYTHs were detected in Arabidopsis plants after pathogen infections (Fig. 8a). Expression of AtYTH06 and AtYTH10 in the leaf and root tissues was reduced in response to infection by G. cichoracearum and G. orontii (powdery mildew), H. arabidopsidis (downy mildew), P. parasitica (phytophthora blight), P. syringae pv. maculicola (leaf spot), or M. incognita (root knot) (Fig. 9a). Notably, expression of AtYTH06 in the roots of plants infected with P. parasitica or M. incognita and expression of AtYTH10 in the leaves of plants infected with G. cichoracearum or P. syringae pv. maculicola were significantly reduced (Fig. 9a). Infection by M. incognita could upregulate the expression of AtYTH01, AtYTH05, and AtYTH11 in the roots of the plants grown on MS medium. Slight increases in expression levels for AtYTH05 and AtYTH02 in the leaves infected by a nonadapted pathogen B. graminis f. sp. hordei, AtYTH02 and AtYTH12 in the leaves infected with P. infestans (late blight), and AtYTH06 in the leaves infected by P. syringae pv. maculicola, P. syringae pv. tomato, or B. cinerea were detected (Fig. 9a). Infection by cabbage leaf curl virus or turnip mosaic virus also resulted in changes of expression levels for some of the AtYTH genes. These results imply that the Arabidopsis YTH genes show differentially expression patterns upon infection by different types of pathogens including fungi, oomycetes, bacteria, viruses, and nematodes.
Expression patterns of the AtYTHs (a) and OsYTHs (b) genes in response to pathogen infection and elicitor treatment. The red or green shading represents an upregulated or downregulated expression level, respectively. The color scale with heatmap is given in log2 ratio values. a Expression patterns of the AtYTHs genes in response to pathogen infection. b Expression patterns of the OsYTHs genes in response to pathogen infection. NP Nipponbare, 309 Taipei 309. c Expression patterns of the AtYTHs genes in response to elicitor treatment. Time points of sampling are indicated as days post-inoculation (dpi) or hours post-inoculation (hpi), and tissues sampled for analyses are also indicated as root, leaf, or seedlings (sdlg)
We also analyzed the expression patterns of OsYTHs genes in the leaves or roots of different rice cultivar plants after infection by M. oryzae (M. grisea) (rice blast), X. oryzae pv. oryzae (leaf blight) or X. oryzae pv. oryzicola (leaf streak). OsYTH12 showed decreased expression levels in the leaves of cv. Nipponbare and cv. IR24 plants infected by M. grisea strain FR13 or X. oryzae pv. oryzae strain PXO99A but exhibited an increased expression level in the leaves of cv. Nipponbare plants infected by X. oryzae pv. oryzae strain PXO99A (Fig. 9b). Increased expression levels for OsYTH01, OsYTH03, and OsYTH10 were also detected in the leaves of cv. Nipponbare and cv. Taipei 309 plants infected with M. oryzae or in the leaves of cv. IR24 plants infected by X. oryzae pv. oryzae strain PXO99A or X. oryzae pv. oryzicola strain BLS303 (Fig. 9b). These data indicate that the members of the rice YTH genes respond differentially to different fungal and bacterial pathogens.
We further examined whether the AtYTHs genes could be induced by some known effectors or elicitors from pathogenic fungi and bacteria. Expression of AtYTH06 was reduced in the seedlings after treatments with effectors including EF-Tu (Zipfel et al. 2006), Flg22 (Felix et al. 1999), or Pep2 (Albert 2013) (Fig. 9c). Treatments with EF-Tu or Pep2 also affected the expression of AtYTH01, AtYTH02, and AtYTH10 in seedlings (Fig. 9c). However, treatments with other elicitors including NPP1 (Fellbrich et al. 2002), HrpZ (Alfano et al. 1996), LPS (Newman et al. 2007), or OGs (Denoux et al. 2008) did not significantly affect the expression of the AtYTH genes (Fig. 9c). These data indicate that some of the Arabidopsis YTH genes can also respond to pathogen-derived effectors or elicitors.
Expression Patterns of the YTH Genes in Response to Abiotic Stresses
We next analyzed the expression patterns of the AtYTHs and OsYTHs genes in Arabidopsis and rice plants after treatments with different abiotic stresses. In Arabidopsis, expression of AtYTH06 in the leaves, rosettes, and seedlings was significantly upregulated during cold, drought, osmotic, salt, and heat stress conditions but was markedly downregulated in bolting tissues and rosettes under hypoxia and submergence stress (Fig. 10a). Expression of AtYTH07 and AtYTH10 in seedlings and flowers was also dramatically induced at early stages after heat treatment (Fig. 10a). Increased expression levels for AtYTH05 under cold stress, AtYTH02 under hypoxia stress, and AtYTH01 and AtYTH02 under submergence stress were also detected (Fig. 10a). In contrast to its upregulated expression during heat stress, expression levels of AtYTH10 were generally decreased under cold, drought, salt, and osmotic stress, especially in rosettes during the late stage of osmotic stress (Fig. 10a). AtYTH08 also showed decreased expression at 30 h after treatment with heat stress (Fig. 10a). Expression of other AtYTHs genes was affected by most of the stresses examined leading to upregulated or downregulated levels; however, oxidative stress did not affect the expression of almost all AtYTHs genes (Fig. 10a).
Expression patterns of the AtYTHs (a) and OsYTHs (b) genes in response to abiotic stress treatment. The red or green shading represents an upregulated or downregulated expression level, respectively. The color scale with heatmap is given in log2 ratio values. a Expression patterns of the AtYTHs genes in response to abiotic stress. b Expression patterns of the OsYTHs genes in response to abiotic stress. Am cv. Amaroo, ZH11 cv. Zhonghua 11, LTH Lijiangxintuanheigu, Submerg. submergence stress. Time points of sampling are indicated as days post-treatment (dpt) or hours post-treatment (hpt) and tissues sampled for analyses are also indicated as root, leaf, or seedlings (sdlg)
In rice, different expression patterns of OsYTHs genes in response to abiotic stress were detected. Expression levels of OsYTH10 were decreased significantly in the shoots of cv. C418 and cv. K354 plants during cold stress and in panicle leaves of cv. DK151 plants under drought stress, but increased significantly in seedlings by a temperature shift from cold (4 °C) to normal condition (29 °C) (Fig. 10b). Expression of OsYTH05, OsYTH06, OsYTH07, and OsYTH09 was also downregulated in the shoot of rice plants under cold stress (Fig. 10b). Rice plants under anaerobic stress showed decreased expression levels of OsYTH01, OsYTH02, OsYTH05, OsYTH09, and OsYTH12 but exhibited an increased expression of OsYTH11 (Fig. 10b). Decreased expressions of OsYTH08 in seedlings by a temperature shift of 4 to 29 °C, OsYTH11 and OsYTH12 in seedlings under heat stress, and OsYTH11 in the roots of tillering plants under drought stress were also observed (Fig. 10b). Meanwhile, expression of OsYTH03 in rice plants under submergence stress and expression of OsYTH08 in seedlings under heat stress was upregulated markedly (Fig. 10b). Drought stress treatment increased the expression of OsYTH01, OsYTH02, OsYTH03, OsYTH05, and OsYTH12 in the root and leaf tissues, while a temperature shift of 4 to 29 °C also increased the expression of OsYTH12 in seedlings (Fig. 10b). Notably, none of the OsYTHs genes showed an altered expression pattern in rice plants under salt stress, whereas OsYTH01, OsYTH02, OsYTH03, OsYTH04, and OsYTH08 also did not exhibit altered expression patterns in response to cold stress (Fig. 10b).
Expression Patterns of the YTH Genes in Response to Hormones
Considering that the AtYTHs and OsYTHs genes were differentially regulated in responses to various abiotic and biotic stresses and that different hormones are involved in the regulation of growth/development and the responses to abiotic and biotic stresses, we therefore analyzed the expression patterns of the AtYTHs and OsYTHs genes after treatments with different hormones. In Arabidopsis, expression of AtYTH06 was significantly upregulated in seedlings, suspension cells, leaves, and guard cells after treatments with abscisic acid (ABA) or brassinolide (BL); however, AtYTH01, AtYTH02, AtYTH10, and AtYTH13 showed decreased expression in ABA-treated leaves (Fig. 11a). Expression of AtYTH05 and AtYTH13 in the root and seedlings after treatment with 1-naphthalene acetic acid (NAA) or zeatin and expression of AtYTH08 in suspension cells after treatment with BL were upregulated (Fig. 11a). Interestingly, treatments with 1-aminocyclopropane-1-carboxylic acid (ACC, precursor of ET), ethylene (ET), methyl jasmonic acid (MeJA), or salicylic acid (SA), well-known defense hormones involved in plant defense response, and treatments with indole-3-acetic acid (IAA) and gibberellic acid (GA3) did not affect the expression patterns of all AtYTHs genes in Arabidopsis plants (Fig. 11a). In rice, decreased expression of OsYTH08 in seedlings or leaves after treatments with GA3, kinetin (KT), or zeatin was detected (Fig. 11b). However, expression of OsYTH01 and OsYTH09 was upregulated in the leaves or seedlings after treatments with KT, NAA, or trans-zeatin (Fig. 11b). These data indicate that the AtYTHs and OsYTHs genes respond differentially to hormones in different tissues or developmental stages of Arabidopsis and rice plants.
Expression patterns of the Arabidopsis (a) and rice (b) YTH genes in response to hormones. The red or green shading represents an upregulated or downregulated expression level, respectively. The color scale with heatmap is given in log2 ratio values. a Expression patterns of the AtYTHs genes in response to different hormones. b Expression patterns of the OsYTHs genes in response to different hormones. MH63 cv. Minghui 63, ZS97 cv. Zhenshan 97. Time points of sampling are indicated as days post treatment (dpt), hours post-treatment (hpt), or minutes post-treatment (mpt), and tissues sampled for analyses are also indicated as root, leaf, or seedlings (sdlg)
Discussion
The YTH domain in diverse proteins was recognized as a structurally conserved domain involved in the binding of RNA molecules (Glisovic et al. 2008; Janga and Mittal 2011). Information on the YTH proteins and their biological functions in plants is limited. In the present study, we performed a comprehensive genome-wide identification, biochemical characterization, and comparative expression analyses of the YTH gene family in Arabidopsis and rice. Our results demonstrated that the YTH proteins in plants have RNA-binding activity in vitro, and expression of the YTH genes in Arabidopsis and rice is regulated in response to developmental cues as well as in response to abiotic and biotic stresses. Therefore, our study not only characterizes systematically the YTH gene family in Arabidopsis and rice at genomic and biochemical levels but also provides new information on the possible biological functions of the YTH genes in plant development and stress response.
Our genome-wide searches using the conserved YTH domain sequence as a query identified a total of 13 and 12 genes encoding YTH domain-containing proteins in Arabidopsis and rice genomes, respectively. Ok et al. (2005) identified 11 ECTs for their interacting with Calcineurin B-Like-Interacting Protein Kinase1 (CIPK1) through yeast two-hybrid screening, and all these ECTs contain the conserved YTH domains in their C-terminals. The genes encoding for these 11 ECTs are the same with AtYTH01, AtYTH02, AtYTH04-10, AtYTH12, and AtYTH13 (Table 2). In this study, we identified two more Arabidopsis genes, AtYTH03 and AtYTH11, which encode YTH domain-containing proteins (Table 2), and thus, we considered that they should be members of the YTH gene family in Arabidopsis. AtYTH03 was also identified as a gene encoding AtCPSF30 (cleavage and polyadenylation specificity factor) that is involved in alternative polyadenylation of mRNA (Delaney et al. 2006; Thomas et al. 2012). Structures such as the feature of exon/intron organization of the AtYTHs and OsYTHs genes are quite different between each other (Fig. 1), implying that the YTH gene family in Arabidopsis and rice may evolve independently. Although most of the AtYTHs genes separately distribute on different chromosomal locations, one pair of the AtYTHs genes, AtYTH08/AtYTH09, locates within 25-kb distance on chromosome III (Table 2), indicating that some members of the Arabidopsis YTH gene family may result from tandem duplication. Thus, it is likely that both tandem duplication and segmental duplication events are involved in the evolution of the AtYTHs gene family in Arabidopsis. By contrast, the rice OsYTHs genes do not form clusters and are separately distributed on different chromosomes, indicating that tandem duplication is not involved in the evolution of OsYTHs gene family in rice. Indeed, the rice OsYTHs genes show scattered distribution on the chromosomes and any two of the OsYTHs genes have quite large physical distances on the chromosomes. It is thus speculated that segmental duplication events may be the main source for the evolution of the OsYTHs gene family in rice. This is similar to the observations on the evolution of the maize ZmMPKs gene family (Liu et al. 2013).
The biochemical characteristics have been studied in some of the YTH proteins. In our study, we found that the recombinant AtYTH05 could bind to single-stranded RNA molecules, demonstrating that the AtYTH05 protein has RNA-binding activity in vitro (Fig. 5c). This is similar to YT521-B that was previously shown to bind to single-strand RNA sequences (Zhang et al. 2010b). The YTH proteins generally only have conserved YTH domains but do not contain any other well-known RNA-binding domains or motifs such as RRM, pseudouridine synthase and archaeosine transglycosylase (PUA), and oligonucleotide/oligosaccharide binding fold (OB-fold) (Stoilov et al. 2002). It was recently found that the YTH domain is required for RNA-binding activity as deletion of the YTH domain abolished the RNA-binding activity in YT521-B (Zhang et al. 2010b). Structurally, the β-strands in the YTH domain contain highly conserved aromatic residues, which are often present in RRM, PUA, and OB-fold (Stoilov et al. 2002) and are crucial for RNA binding (Hoffman et al. 1991). These structural features and experimental evidence demonstrate that the YTH domain is a novel RNA-binding domain (Zhang et al. 2010b). However, AtYTH03 (AtCPSF30), showing less sequence similarity to other AtYTHs proteins, contains an additional CCCH zinc finger domain except a YTH domain and was shown that the CCCH domain is necessary for its RNA-binding activity (Addepalli and Hunt 2007). On the other hand, it was shown that the yeast Mmi1 and the human YT521-B are localized in dynamic nuclear foci (Rafalska et al. 2004; Harigaya et al. 2006), especially that the YT521-B is localized in a specific nuclear compartment, the YT body (Rafalska et al. 2004). By contrast, results from our experiments revealed that, when transiently expressed in the leaves of N. benthamiana, AtYTH05 and AtYTH07 were not localized in a specific compartment of the cells (Fig. 5a). This is in agreement with the observation that the human YTH protein family members reside both in the nucleus and cytosol (Zhang et al. 2010b). It is thus likely that the YTH domain is not a cellular targeting signal and the YTH proteins may function in different aspects of cellular RNA metabolism (Zhang et al. 2010b).
Generally speaking, a gene expressed abundantly or increasingly in a tissue, a developmental stage, or a stress condition may indicate its function related to developmental and stress response. Different approaches including analyses of publicly available microarray data, gene expression by qRT-PCR, and promoter activity in transgenic plants were taken to explore possible involvement of the AtYTHs and OsYTHs genes in development and stress response. The facts that the expression patterns of the AtYTHs and OsYTHs genes are quite divergent and no two AtYTHs or OsYTHs genes show similar or same expression patterns in different tissues and developmental stages (Figs. 6 and 8) suggest that each member of the Arabidopsis and rice YTH gene family may have specific functions in growth and development. This is partially supported by the observations that expression of some of the AtYTHs and OsYTHs genes could be affected by exogenous treatments with growth/development-related hormones such as NAA, BL, KT, and zeatin (Fig. 11). The existence of homozygous lines for each of the AtYTHs gene (the SALK T-DNA insertion population at http://signal.salk.edu/cgi-bin/tdnaexpress?; Zhang et al. 2008 for AtYTH03) indicates that mutations in the AtYTHs genes do not result in a lethal effect. Surprisingly, most of the AtYTHs genes, especially for AtYTH01 and AtYTH05, showed higher expression potentials during senescence stage (Fig. 6a), indicating a relationship of the AtYTHs genes to senescence, probably having specific and most important functions. Notably, AtYTH05, AtYTH06, and AtYTH07 were expressed at relatively higher levels in flowers and siliques than those in other tissues (Fig. 7a), and the high level of AtYTH05 expression in flowers and siliques was further confirmed by the AtYTH05 promoter activity in AtYTH05 Pro ::GUS transgenic plants (Fig. 7b), indicating that these three AtYTHs genes might be associated with their functions in the reproduction process. This is supported by the tissue-specific expression patterns of the AtYTHs genes, which show relatively high levels of expression potentials in seeds such as embryo, endosperm, and testa than those in inflorescence and rosettes (Fig. 8a). Interestingly, the AtYTHs genes expressed abundantly in the root, stem, or leaves showed relatively low expression in the flowers and siliques, and vice versa (Fig. 7a). Such opposite expression patterns indicate that the AtYTHs genes have specific functions during different developmental stages. For the rice OsYTHs genes, it is interesting that two genes, OsYTH02 and OsYTH06, have the highest levels of expression potentials in internode and stele tissues, suggesting that both of the OsYTH02 and OsYTH06 have functions in the regulation of stem development in rice.
Several lines of evidence supporting the involvement of the YTH family in response to abiotic and biotic stresses came from our comparative analyses of the expression patterns of the AtYTHs and OsYTHs genes under stress conditions. Expression of AtYTH06 and AtYTH10 was reduced significantly after infection by different types of pathogens including fungi, oomycetes, bacteria, and nematode and by treatment with well-known pathogen-associated molecular patterns (PAMPs) such as EF-Tu and Pep2 (Zipfel 2008) (Figs. 9a and 11a), indicating that AtYTH06 and AtYTH10 should have a function in response to pathogen infection. In rice, expression of the OsYTH08 gene showed different patterns in response to infection by M. oryzae or X. oryzae pv. oryzae (Fig. 9b), indicating that OsYTH08 is involved in the interactions between rice and its pathogens. Surprisingly, however, exogenous treatments with SA, JA, ET, or ACC, which are well-known defense signaling hormones (Pieterse et al. 2012), did not significantly affect the expression of the AtYTHs genes in Arabidopsis plants (Fig. 11a). Thus, the involvement and function (if any) of the YTH family in response to biotic stress need to be further examined experimentally. On the other hand, the involvement of the YTH family in response to various abiotic stress is supported by the altered expression of AtYTH06, AtYTH07, AtYTH10, OsYTH04, and OsYTH10 (Fig. 10) under abiotic stress conditions and by the interactions of some AtYTHs proteins (also known as ECTs) with CIPK1 that is involved in the regulation of abiotic stress response (Ok et al. 2005). ABA is well documented to play important roles in signaling pathways involved in the regulation of abiotic and biotic stress responses (Chinnusamy et al. 2008; Cao et al. 2011; Kim 2012; Lee and Luan 2012; Nakashima and Yamaguchi-Shinozaki 2013; Roychoudhury et al. 2013). Similar expression patterns of AtYTH06 in Arabidopsis after treatments with cold, drought, heat, and osmotic stress and by treatment with ABA (Figs. 10a and 11a) indicate that AtYTH06 has diverse functions in responses to multiple abiotic stresses. Surprisingly, none of the AtYTHs genes including AtYTH03 responded with altered expression levels at early and late stages after treatment with oxidative stress (Fig. 10a). However, it was found that mutation in AtYTH03 (also known as AtCPSF30) led to enhanced tolerance against oxidative stress (Zhang et al. 2008; Addepalli et al. 2010). Interestingly, the AtYTHs and OsYTHs genes exhibited different expression patterns in response to same abiotic stress, e.g., induced expression of AtYTH06 vs. reduced expression of OsYTH10 in cold stress and reduced expression of AtYTH06 vs. induced expression of OsYTH03 in submergence stress (Fig. 10). Therefore, it is likely that members of the YTH family function with different modes of action in response to various abiotic stresses in Arabidopsis and rice plants.
In summary, this study presents a comprehensive characterization of the YTH gene/protein family in Arabidopsis and rice and comparative analyses of expression of the AtYTHs and OsYTHs genes in different tissues or developmental stages as well as in response to abiotic and biotic stresses. Further studies are absolutely required to elucidate the biological function for each member of the YTH family in Arabidopsis and rice using functional genomics approaches by phenotypic analyses of transgenic lines with altered expression (knockout/knockdown and overexpression).
References
Addepalli B, Hunt AG (2007) A novel endonuclease activity associated with the Arabidopsis ortholog of the 30-kDa subunit of cleavage and polyadenylation specificity factor. Nucleic Acids Res 35:4453–4463
Addepalli B, Limbach PA, Hunt AG (2010) A disulfide linkage in a CCCH zinc finger motif of an Arabidopsis CPSF30 ortholog. FEBS Lett 584:4408–4412
Albert M (2013) Peptides as triggers of plant defence. J Exp Bot 64:5269–5279
Alfano JR, Bauer DW, Milos TM, Collmer A (1996) Analysis of the role of the Pseudomonas syringae pv. syringae HrpZ harpin in elicitation of the hypersensitive response in tobacco using functionally non-polar hrpZ deletion mutations, truncated HrpZ fragments, and hrmA mutations. Mol Microbiol 19:715–728
Ambrosone A, Costa A, Leone A, Grillo S (2012) Beyond transcription: RNA-binding proteins as emerging regulators of plant response to environmental constraints. Plant Sci 182:12–18
Bjellqvist B, Basse B, Olsen E, Celis JE (1994) Reference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. Electrophoresis 15:529–539
Cao FY, Yoshioka K, Desveaux D (2011) The roles of ABA in plant-pathogen interactions. J Plant Res 124:489–499
Chen LY, Lingner J (2012) AUF1/HnRNP D RNA binding protein functions in telomere maintenance. Mol Cell 47:1–2
Chinnusamy V, Gong Z, Zhu JK (2008) Abscisic acid-mediated epigenetic processes in plant development and stress responses. J Integr Plant Biol 50:1187–1195
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Cook KB, Kazan H, Zuberi K, Morris Q, Hughes TR (2011) RBPDB: a database of RNA-binding specificities. Nucleic Acids Res 39:D301–D308
Delaney KJ, Xu R, Zhang J, Li QQ, Yun KY, Falcone DL, Hunt AG (2006) Calmodulin interacts with and regulates the RNA-binding activity of an Arabidopsis polyadenylation factor subunit. Plant Physiol 140:1507–1521
Denoux C, Galletti R, Mammarella N, Gopalan S, Werck D, De Lorenzo G, Ferrari S, Ausubel FM, Dewdney J (2008) Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol Plant 1:423–445
Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, Sorek R, Rechavi G (2012) Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485:201–206
Felix G, Duran JD, Volko S, Boller T (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18:265–276
Fellbrich G, Romanski A, Varet A, Blume B, Brunner F, Engelhardt S, Felix G, Kemmerling B, Krzymowska M, Nurnberger T (2002) NPP1, a Phytophthora-associated trigger of plant defense in parsley and Arabidopsis. Plant J 32:375–390
Glisovic T, Bachorik JL, Yong J, Dreyfuss G (2008) RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 582:1977–1986
Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO (2002) pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J 31:375–383
Harigaya Y, Tanaka H, Yamanaka S, Tanaka K, Watanabe Y, Tsutsumi C, Chikashige Y, Hiraoka Y, Yamashita A, Yamamoto M (2006) Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature 442:45–50
Hartmann AM, Nayler O, Schwaiger FW, Obermeier A, Stamm S (1999) The interaction and colocalization of Sam68 with the splicing-associated factor YT521-B in nuclear dots is regulated by the Src family kinase p59fyn. Mol Biol Cell 10:3909–3926
Hirschfeld M, Zhang B, Jaeger M, Stamm S, Erbes T, Mayer S, Tong X, Stickeler E (2014) Hypoxia-dependent mRNA expression pattern of splicing factor YT521 and its impact on oncological important target gene expression. Mol Carcinog. doi:10.1002/mc.22045
Hoffman DW, Query CC, Golden BL, White SW, Keene JD (1991) RNA-binding domain of the A protein component of the U1 small nuclear ribonucleoprotein analyzed by NMR spectroscopy is structurally similar to ribosomal proteins. Proc Natl Acad Sci U S A 88:2495–2499
Huh SU, Paek KH (2013) Plant RNA binding proteins for control of RNA virus infection. Front Physiol 4:397
Imai Y, Matsuo N, Ogawa S, Tohyama M, Takagi T (1998) Cloning of a gene, YT521, for a novel RNA splicing-related protein induced by hypoxia/reoxygenation. Brain Res Mol Brain Res 53:33–40
Iost I, Dreyfus M, Linder P (1999) Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J Biol Chem 274:17677–17683
Janga SC, Mittal N (2011) Construction, structure and dynamics of post-transcriptional regulatory network directed by RNA-binding proteins. Adv Exp Med Biol 722:103–117
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Johnstone O, Lasko P (2001) Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu Rev Genet 35:365–406
Kang HJ, Jeong SJ, Kim KN, Baek IJ, Chang M, Kang CM, Park YS, Yun CW (2014) A novel protein, Pho92, has a conserved YTH domain and regulates phosphate metabolism by decreasing the mRNA stability of PHO4 in Saccharomyces cerevisiae. Biochem J 457:391–400
Kim TH (2012) Plant stress surveillance monitored by ABA and disease signaling interactions. Mol Cells 33:1–7
Lee SC, Luan S (2012) ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ 35:53–60
Letunic I, Doerks T, Bork P (2012) SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res 40:D302–D305
Liu Y, Zhang D, Wang L, Li D (2013) Genome-wide analysis of mitogen-activated protein kinase gene family in maize. Plant Mol Biol Rep 31:1446–1460
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta DeltaC(T)) method. Methods 25:402–408
Lorkovic ZJ (2009) Role of plant RNA-binding proteins in development, stress response and genome organization. Trends Plant Sci 14:229–236
Lunde BM, Moore C, Varani G (2007) RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol 8:479–490
McPheeters DS, Cremona N, Sunder S, Chen HM, Averbeck N, Leatherwood J, Wise JA (2009) A complex gene regulatory mechanism that operates at the nexus of multiple RNA processing decisions. Nat Struct Mol Biol 16:255–264
Mittal N, Scherrer T, Gerber AP, Janga SC (2011) Interplay between posttranscriptional and posttranslational interactions of RNA-binding proteins. J Mol Biol 409:466–479
Morris AR, Mukherjee N, Keene JD (2010) Systematic analysis of posttranscriptional gene expression. Wiley Interdiscip Rev Syst Biol Med 2:162–180
Nakashima K, Yamaguchi-Shinozaki K (2013) ABA signaling in stress-response and seed development. Plant Cell Rep 32:959–970
Newman MA, Dow JM, Molinaro A, Parrilli M (2007) Priming, induction and modulation of plant defence responses by bacterial lipopolysaccharides. J Endotoxin Res 13:69–84
Ok SH, Jeong HJ, Bae JM, Shin JS, Luan S, Kim KN (2005) Novel CIPK1-associated proteins in Arabidopsis contain an evolutionarily conserved C-terminal region that mediates nuclear localization. Plant Physiol 139:138–150
Pallas V, Gomez G (2013) Phloem RNA-binding proteins as potential components of the long-distance RNA transport system. Front Plant Sci 4:130
Pieterse CM, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SC (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28:489–521
Rafalska I, Zhang Z, Benderska N, Wolff H, Hartmann AM, Brack-Werner R, Stamm S (2004) The intranuclear localization and function of YT521-B is regulated by tyrosine phosphorylation. Hum Mol Genet 13:1535–1549
Rost B (1996) PHD: predicting one dimensional protein structure by profile-based neural networks. Methods Enzymol 266:525–539
Roychoudhury A, Paul S, Basu S (2013) Cross-talk between abscisic acid-dependent and abscisic acid-independent pathways during abiotic stress. Plant Cell Rep 32:985–1006
Schultz J, Milpetz F, Bork P, Ponting CP (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A 95:5857–5864
Stoilov P, Rafalska I, Stamm S (2002) YTH: a new domain in nuclear proteins. Trends Biochem Sci 27:495–497
Stoss O, Novoyatleva T, Gencheva M, Olbrich M, Benderska N, Stamm S (2004) p59fyn-mediated phosphorylation regulates the activity of the tissue-specific splicing factor rSLM-1. Mol Cell Neurosci 27:8–21
Stower H (2014) Gene regulation: translation steps up to a regulatory role. Nat Rev Genet 15:64
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Thomas PE, Wu X, Liu M, Gaffney B, Ji G, Li QQ, Hunt AG (2012) Genome-wide control of polyadenylation site choice by CPSF30 in Arabidopsis. Plant Cell 24:4376–4388
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C (2014) N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505:117–120
Zhang J, Addepalli B, Yun KY, Hunt AG, Xu R, Rao S, Li QQ, Falcone DL (2008) A polyadenylation factor subunit implicated in regulating oxidative signaling in Arabidopsis thaliana. PLoS One 3:e2410
Zhang B, zur Hausen A, Orlowska-Volk M, Jager M, Bettendorf H, Stamm S, Hirschfeld M, Yiqin O, Tong X, Gitsch G, Stickeler E (2010a) Alternative splicing-related factor YT521: an independent prognostic factor in endometrial cancer. Int J Gynecol Cancer 20:492–499
Zhang Z, Theler D, Kaminska KH, Hiller M, de la Grange P, Pudimat R, Rafalska I, Heinrich B, Bujnicki JM, Allain FH, Stamm S (2010b) The YTH domain is a novel RNA binding domain. J Biol Chem 285:14701–14710
Zhong J, Peters AH, Kafer K, Braun RE (2001) A highly conserved sequence essential for translational repression of the protamine 1 messenger RNA in murine spermatids. Biol Reprod 64:1784–1789
Zipfel C (2008) Pattern-recognition receptors in plant innate immunity. Curr Opin Immunol 20:10–16
Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125:749–760
Acknowledgments
This work was supported by the National Key Project for Research on Transgenic Plants (2011ZX08009-003-001), the National Natural Science Foundation of China (Nos. 31101422 and 31272028), the National High-Tech R&D Program (No. 2012AA101504), and the Research Fund for the Doctoral Program of Higher Education of China (20120101110070).
Conflict of Interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Li, D., Zhang, H., Hong, Y. et al. Genome-Wide Identification, Biochemical Characterization, and Expression Analyses of the YTH Domain-Containing RNA-Binding Protein Family in Arabidopsis and Rice. Plant Mol Biol Rep 32, 1169–1186 (2014). https://doi.org/10.1007/s11105-014-0724-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-014-0724-2