Abstract
An Agrobacterium tumefaciens-mediated transformation system along with regeneration of transgenic plants of the halophyte Leymus chinensis (Trin.) was developed. A. tumefaciens strain EHA105 containing the binary vector pCAMBIA2300 was used for transformation, along with an Ipomoea batatas 2-cysteine peroxiredoxin (Ib2-Cys prx) gene under the control of the stress-inducible sweet potato anionic peroxidase 2 (SWPA2) promoter or the cauliflower mosaic virus (CaMV) 35S promoter. Among different pre-culture periods, 7-day pre-culture promoted the highest frequency of transformation. Among the different cocultivation periods, 20 min of cocultivation with bacterial cells (OD600 = 0.4) promoted the highest frequency of transformation. Acetosyringone at 100 μM was added to increases virulence induction. Selection of transgenic shoots was done in the presence of 150 mg l−1 kanamycin. Polymerase chain reaction (PCR) analysis of putative transgenic plants showed the presence of Ib2-Cys prx and nptII genes. The expression of transgene, Ib2-Cys prx was also confirmed in non-stressed and various stressed plants by RT-PCR. The SWPA2::prx transformants showed very low prx gene expression under non-stressed conditions but higher prx gene expression than the CaMV 35S::prx transformants, when exposed to various oxidative stresses. The highest transformation efficiency was found to be 8.97 %. The protocol provides a direct opportunity for improvement of the quality traits of L. chinensis via genetic transformation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The halophyte Leymus chinensis (Trin.), a perennial rhizome grass in the Gramineae family, is widely distributed throughout Northern China, Mongolia, and Siberia (Huang et al. 2004). Due to its strong rhizomes and excellent stress tolerance (Chen 1988), L. chinensis has been used as a soil-binding plant to protect soil from desertification in the arid areas of Northern China. Due to its high vegetative productivity and protein content, good palatability for cattle, and abundant horizontally creeping rhizomes, this species has also been considered as an ideal grassland species for grazing and forage production (Shu et al. 2005). As L. chinensis possesses a great economical and ecological significance, this species has increasingly received considerable attention. However, low sexual reproductivity and low fecundity are the major problems facing the propagation of this species (Huang et al. 2004). Combined with the pressure from environmental stresses and reclamation by humans and animals, the improvement of physiological traits of L. chinensis is urgently needed.
Abiotic stresses, such as drought, salinity, extreme temperatures, and chemical toxicity and oxidative stress, limit plant growth and drastically reduce plant productivity (Boyer 1982). To maintain growth and productivity, plants are equipped with a wide range of antioxidant proteins, including superoxide dismutase, catalase, many types of peroxidases, and molecular chaperones (Storz et al. 1990; Hendrick and Hartl 1993). One of these antioxidant proteins that reduces H2O2, peroxinitrite, and organic hydroperoxide is peroxiredoxin (Prx). Prxs are a group of ubiquitous peroxidase enzymes in which redox-active cysteine residues participate in the reduction of hydrogen peroxide (Chae et al. 1994). Originally, Prxs have been divided into two categories, 1-Cys and 2-Cys Prxs, based on the number of cysteinyl residues (Chae et al. 1994). Due to different structural and mechanistic data, 2-Cys Prxs have been further divided into typical and atypical 2-Cys Prxs. Of these, the typical 2-Cys Prxs are the largest class of Prxs and are identified by the conservation of their two redox-active cysteines, the peroxidatic cysteine (generally near residue 50) and the resolving cysteine (near residue 170, Hofmann et al. 2002). Typical 2-Cys Prxs that can be oxidized directly by hydrogen peroxide have been found to be protective against the toxic effects of hydrogen peroxide (Jang et al. 2004). Other genetic studies have revealed that typical 2-Cys Prxs appear to be involved in a large number of cellular function, such as cell proliferation, differentiation, immune response, growth control, tumor promotion, and apoptotic processes (Hirotsu et al. 1999; Neumann et al. 2003). Moreover, 2-Cys Prx has been transformed into sweet potato by Agrobacterium-mediated transformation, and transgenic plants that overexpress 2-Cys Prx have shown an enhanced tolerance to oxidative stress (Kim et al. 2009).
The use of transgenic techniques has become a common and convenient method for improving the traits of plants (Hiei et al. 1997). In previous reports, we have established an efficient tissue culture and plant regeneration system using wild-type (LcWT07) and new variety (LcJS0107) L. chinensis plants and firstly constructed plant regeneration systems through suspension-derived callus or/and using mature seeds and leaf base and root segments as explants (Sun and Hong 2009, 2010a, b). The highest regeneration frequency of 71.0 % using type 1 callus derived from mature seeds was found to have potentials to produce transgenic plants. Thus, we established a stable Agrobacterium-mediated transformation of L. chinensis by testing explant age, Agrobacterium tumefaciens infection solution concentration, and infection time and successfully obtained kanamycin-resistant plants with a positive transgene insertion. Here, we investigate the effect of expression system with different promoters on transformation efficiency. After, a powerful expression system with an appropriate promoter is a key requisite for expression of foreign genes efficiently in plants (Kim et al. 2003).
Despite a multitude of studies of Agrobacterium-mediated transformation of many plant species including graminaceous plants, L. chinensis transformation study remains lacking and the transformation efficiency is not very high in this grass. For L. chinensis transformation situation, only two reports of successful transformation and plant regeneration have been published; until now. Shu et al. (2005) have transformed the phosphinothricin acetyltransferase gene using microprojectile bombardment transformation and successfully obtained transgenic plants. Wang et al. (2009) have transformed the wheat late embryogenesis abundant gene using Agrobacterium-mediated transformation and obtained transgenic plants with high drought stress tolerance. Thus, a reliable protocol for efficient transformation and plant regeneration in this grass needs to be developed. The results of this investigation would provide rapid and direct opportunities for improvement of the quality traits of L. chinensis via genetic transformation.
Materials and Methods
Plant Materials
Mature seeds of China wild-type L. chinensis plants, LcWT07, were collected from the natural grassland in Siping, Jilin province, China. The grassland located on the Songnen Plain, Jilin Province is the largest natural L. chinensis grassland in the Eurasian continent (Guo 1994). The local climate is semiarid, with windy and dry winters and springs, and warm but comparatively rain-rich summers, followed by short and cool autumns. LcWT07 plants have adapted to these natural conditions as the result of many years of natural evolution. All seeds were de-husked and surface-sterilized with 70% ethanol for 1 min and then with 5 % sodium hypochlorite for 20 min. The sterilized seeds were rinsed five times with sterile water and were then placed on callus induction medium.
Bacterial Strains and Plasmids
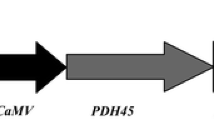
The pCAMBIA2300 vector carries an Ipomoea batatas 2-Cys prx (Ib2-Cys prx) gene and an nptII gene, under the control of the stress-inducible sweet potato anionic peroxidase 2 (SWPA2, Kim et al. 2003) or the cauliflower mosaic virus (CaMV) 35S promoter and CaMV 35S terminator, respectively (Fig. 1). The A. tumefaciens strain EHA105 was used in this study.
Structure of T-DNA regions in plant expression vector pCAMBIA2300-Ib2-Cys prx. The Ib2-Cys prx gene was inserted between the stress-inducible SWPA2 promoter (a) or the CaMV 35S promoter (b) and the CaMV 35S terminator. The nptII gene was inserted between the CaMV 35S promoter and the CaMV 35S terminator. RB right border, LB left border, nptII neomycin phosphotransferase gene, Ib2-Cys prx I. batatas 2-cysteine proxiredoxins
Agrobacterium-Mediated Transformation
The A. tumefaciens strain EHA105 with one binary vector was grown in YEP liquid medium supplemented with 50 mg l−1 kanamycin (Km). The culture was grown at 28°C for 24 h with continuous shaking at 225 rpm until an optical density at a wavelength of 600 nm (OD600) of 0.8 ~ 1.0 was reached. The solution was centrifuged at 5,000 rpm for 10 min. The pellet material was resuspended to OD600 of 0.4 with MMA medium containing Murashige and Skoog (MS) basal salt (Murashige and Skoog 1962), MES 10 mol l−1, pH 5.6, acetosyringone (AS) 100 μM, and sucrose 20 g l−1.
Type 1 embryogenic calli were prepared as described in our previous study (Sun and Hong 2010a), and the 1-month-old type 1 embryogenic calli (from their being derived from explants) were used for transformation. With a pre-culture on most optimum callus induction medium containing MS basic salt, 2.0 mg l−1 2,4-D, and 5.0 mg l−1 l-glutamic acid described in our previous study (Sun and Hong 2010a), for various days (0, 3, 5, 7, 10, and 14 days), the embryogenic calli were immersed in an A. tumefaciens EHA105/pCAMBIA2300 suspension and gently shaken to ensure that all the explants were fully submerged, for various infection times (10, 20, 30, and 60 min). After the incubation, the infected calli were blotted dry on filter paper for removal of excess bacteria and cocultivated on callus induction medium with the addition of 100 μM AS for additional 3, 4, or 5 days at 28°C in the dark. After cocultivation, transgenic calli were washed five times with sterile water containing 250 mg l−1 cefotaxime (CE) and blotted dry on filter paper for removal of excess water. The infected calli were then transferred onto callus induction medium with selection using 250 mg l−1 CE and 150 mg l−1 Km for 20 days of culture at 28°C in the dark.
The regeneration of putative transgenic shoots was carried out on the most optimum plant regeneration medium with MS basic salt, 2.0 mg l−1 kinetin, 0.2 mg l−1 α-naphthalene acetic acid, and 2.0 g l−1 casamino acid described in our previous study (Sun and Hong 2010a) with selection using 250 mg l−1 CE and 150 mg l−1 Km. The cultures were incubated under low light conditions (25 μmol m−2 s−1) with a 16/8-h (light/dark) photoperiod for the first week and then maintained under high light conditions (70 μmol m−2 s−1) with the same photoperiod.
Rooting of regenerated putative transgenic shoots was carried out on rooting medium with half-strength MS basic salt and selection using 150 mg l−1 Km, at 28°C under high light conditions (70 μmol m−2 s−1) with a 16/8-h (light/dark) photoperiod. Well-rooted plants were removed from the culture medium, rinsed with sterile water to remove the media, and transplanted into pots with a mixture of sterilized soil and vermiculite (3:1) under greenhouse conditions (25 ± 2°C, natural light, and 60 % humidity). All solid culture media were supplemented with 30.0 g l−1 sucrose and 4.0 g l−1 Gelrite for solidification, with the pH adjusted to 5.8 prior to the addition of Gelrite.
DNA Extraction and Gene Insertion Assay
Genomic DNAs were extracted from fresh leaf tissues of wild-type and transgenic plants using the sodium dodecyl sulfate method (Dellaporta et al. 1983). Polymerase chain reaction (PCR) analysis for the selection gene (nptII) was carried out following the method above: 0.5 μmol l−1 of each primer, 10 ng DNA template, 200 μmol l−1 each of dATP, dCTP, dGTP, and dTTP mix, 1.0 units of Taq DNA polymerase (Promega, USA), and the corresponding buffer in a total volume of 20 μl. The PCR reaction was carried out as follows: 95°C for 5 min followed by 35 cycles of incubation at 95°C for 1 min, 50°C for 1 min, and 72°C for 1 min, with final extension at 72°C for 7 min. The nptII gene was amplified using the following primer sets: npt2-f1, 5′-GAGGCTATTCGGCTATGACTG-3′ and npt2-r1, 5′-ATCGGGAGCGGCGATACCGTA-3′. To detect the presence of the transgene (Ib2-Cys prx), a gene-specific primer set was designed with the following sequences: prx-f1, 5′-TCTAGAATGGCGTCTGTTGCT-3′ and prx-r1, 5′-GAGCTCCTAAATAGCTGAGAA-3′, using the same PCR program described above. PCR reactions were performed using the ASTEC PC808 PCR detection system (ASTEC, PC808, Japan). PCR products were analyzed by 1 % agarose gel electrophoresis. Photos were taken and analyzed using the MultiDoc-It Digital imaging system (UVP, Cambridge, UK).
Stress Treatment
For stress treatment, 1-month-old plants grown in a greenhouse were used. For saline stress, plants were watered with 200 mM NaCl until the final pH value of the outflow became stable at 25°C for 6 h and then sampling was done. For alkaline stress, plants were watered with 100 mM Na2CO3 until the final pH value of the outflow became stable at 25°C for 6 h and then sampling was also done. For hydrogen peroxide treatment, plants were watered with 2.5 mM H2O2 until the solution filled whole pots at 25°C for 6 h and then sampling was done. All data are mean ± standard error of three replicates.
RNA Extraction and RT-PCR
Total RNAs were isolated from fresh leaf tissues of various treated and nontreated plants using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The NanoPhotometer (IMPLEN, UK) was used to determine RNA concentration and quality. The β-actin gene (accession number: HM623326) was used as control, with the following primers: β-actin-f1: 5′-TGGACTCTGGTGATGGTGTC-3′ and β-actin-r1: 5′-CCTCCAATCCAAACACTGTA-3′. The Ib2-Cys prx gene (accession number AF4532791) was amplified using the following primers: prx-f1, 5′-TCTAGAATGGCGTCTGTTGCT-3′ and prx-r1, 5′-GAGCTCCTAAATAGCTGAGAA-3′. Each PCR reaction contained 0.5 μmol l−1 of each primer and 2-μg RNA template in a total volume of 20 μl using the Maxime RT PreMix Kit (Oligo dT Primer, iNtRON BIOTECHNOLOGY, Korea), according to the manufacturer’s instructions. PCR amplification was performed with an initial denaturation at 95°C for 5 min followed by 35 cycles of incubation at 95°C for 30 s, 50°C for 30 s, and 72°C for 30 s, with a final extension at 72°C for 7 min. PCR reactions were performed using the ASTEC PC808 PCR detection system (ASTEC, PC808, Japan). PCR products were analyzed by 1 % agarose gel electrophoresis. Photos were taken and analyzed using the MultiDoc-It Digital imaging system (UVP, Cambridge, UK).
Statistical Analysis
Survival probability of explants was calculated by the ratio of the number of lived explants after the first selection to all number of explants used for transformation. Daily growth rate of explants was calculated by the average daily increase in frequency of the fresh weight when cultured on selection media. Transformation frequency was calculated by the ratio of the number of putative transgenic plants to all number of explants used for transgenic plant regeneration. Albino transformation frequency was calculated by the ratio of the number of albino plants to all number of explants used for transgenic plant regeneration. Three identical, independent experiments were performed for each experiment. The data presented are the average of three experiments. Statistical analysis was performed by two-way ANOVA using Duncan’s multiple-range test with P < 0.05 (Duncan 1955).
Results and Discussion
This report describes a method for the routine Agrobacterium-mediated transformation of the China wild-type halophyte L. chinensis using nptII as the selectable marker. In this study, we have investigated Agrobacterium-mediated transformation efficiency of L. chinensis LcWT07 variety by evaluating several factors including pre-culture period, infection time, co-culture period, and maintenance stage of infected explants.
Due to no introduction of reporter genes such as green fluorescent protein and β-glucuronidase, the real-time inspection of gene insertion efficiency is difficult to actualize. Thus, putative transgenic plants were selected by kanamycin resistance in this study, and the transformation efficiency was determined by kanamycin-resistant callus induction frequency and kanamycin-resistant plant regeneration frequency. Efficient selection is a necessary prerequisite for successful transgenic plant production mediated by Agrobacterium. Kanamycin is the most widely used selective agent and has been used successfully for selection of A. tumefaciens-mediated transformed tissues (Joyce et al. 2010). In particular, Zhangsun et al. (2007) have reported that the neomycin phosphotransferase (nptII) gene is one of the most effective selectable marker genes and successfully produced kanamycin-resistant transgenic plants in sugarcane. Despite that some studies have reported that not all shoots regenerated from kanamycin-resistant calli were transformed, and untransformed shoots could also be regenerated from kanamycin-resistant calli through the detoxification of the kanamycin by the nptII gene product leading to a reduction in selection pressure in localized areas of the callus (Metz et al. 1995; Sharma and Anjaiah 2000). However, the nptII gene has been declared safe to use in transgenic crops by the US Environmental Protection Agency (1994); Fuchs et al. (1993) have also reported the NPTII protein is nontoxic to humans and rapidly degraded under simulated gastric conditions. Although a dose–response of kanamycin was not performed in this study, the dose of 150 mg l−1 was suggested to be able to meet the need of selecting of transgenic plants. In the only published work of L. chinensis transformation, Wang et al. (2009) have used hygromycin as the selective agent, with the dose of 100 mg l−1 in selection media.
Effect of Pre-Culture Period on Explants Growth Status
Transformation efficiency can be increased by manipulating the explants to be more competent for transformation by pre-culture of explants (Sangwan et al. 1992). The detailed significance of pre-culture period has not been discussed in terms of its influence on transformation, but many scientists have been using a pre-culture treatment to enhance the transformation efficiency (McHughen et al. 1989; Sangwan et al. 1992). Despite that it has been reported not to be required for peduncle explants (Metz et al. 1995), a pre-culture before infection with A. tumefaciens solution could improve the growth status of explants and allowed for maintenance of an optimal callus type. We hypothesized that explants in the good growth status with strong differentiation force might possess active transgene integration and significantly favor in transformation efficiency being good potential targets for transformation, so we first tested the growth status of explants with various pre-culture periods on biomass and daily growth rates (Fig. 2). What we used as explants was the type 1 embryogenic callus derived from mature seeds of L. chinensis LcWT07 variety by our previous study (Sun and Hong 2010a), which has high growth rate (more than 300 % after every 20 days of subculture) and plant regeneration potential (71.0 %). Our findings showed that explants biomass manifold increasingly during pre-culture (Fig. 2a), and the daily growth rates of explants were about 1 % during the first 3 days of pre-culture (Fig. 2b). Until 7 days of pre-culture, the daily growth rate showed a significant increase compared to that after 5 days of pre-culture. With longer pre-culture periods, it maintained relatively high daily growth rates but began to decline (Fig. 2b). We investigated effects of infection time and cocultivation period using various pre-cultured explants in further experiments, and suggested the explants with 7 days pre-culture time were more suitable for transformation than explants with other pre-culture treatments, indicating that the growth rates of explants were related with transformation efficiency.
Effect of Cocultivation Period on Transformation Efficiency
Certain cocultivation period results in high transformation efficiency in a species-, genotypes-, and explants-specific matter (Puddephat et al. 1996). A 4-day period of cocultivation has been reported to increase the efficiency of A. tumefaciens-mediated transformation of peduncle explants but caused necrosis of seedling tissues, ultimately reducing the recovery of transformants and necessitating a reduction to 3 days (Metz et al. 1995). Another study reported that determining of the cocultivation period length depended on effective bacterial eradication; thus, the cocultivation was ended when bacterial growth was evident (Hosoki and Kigo 1994). Similar protocol was used in this work, and our results showed that 3-day cocultivation resulted in evident bacterial growth, while 4- or 5-day cocultivation allowed excess bacterial growth on the face of explants to inhibit the nutrition absorbability (Fig. S1). So 3-day cocultivation was used in further experiments.
Effect of Infection Time on Survival with Selection
To examine the effect of A. tumefaciens infection time on transformation efficiency, the surviving explants number with selection was used as an index in this study. In the exclusive Agrobacterium-mediated transformation report by Wang et al. (2009), the solution concentration of A. tumefaciens strain EHA105 was regulated to OD600 0.4 with sterile distilled water. Moreover, we have performed high OD600 0.8 and low OD600 0.4 to examine the transformation efficiency in terms of A. tumefaciens solution concentration, and the low OD600 showed higher transgene insertion frequency (unpublished data). In subsequent studies, we all use the low OD600 0.4 as the optimal infection solution concentration. In earlier studies, several factors, e.g., virulence (vir) gene-inducing compounds, have been reported as important for improving transformation efficiency in plant species (Opabode 2006). AS, one the best phenolic vir inducer, efficiently increases virulence induction and the transformation rate due to bearing an unsaturated lateral chain (Joubert et al. 2002). Many researchers demonstrated that AS at 100 μM concentration enhances the efficiency of transformation in some plant species, including some grass species (Bettany et al. 2003; Pandey et al. 2010). In this work, AS at 100 μM concentration also was used to improve virulence induction.
The putative transgenic callus induction frequency depends on infection time and transgene insertion. As known infection with A. tumefaciens solution results in an oxidative stress, stress-exposed time increases along with increased infection time, but probability of the transgene insertion either increases along with increased infection time. After 10- or 20-min infection, the explants did not show remarkable changes in putative transgenic callus, or named as kanamycin-resistant callus induction frequency, irrespectively in strain type (Fig. 3). However, after 30-min infection, the kanamycin-resistant callus induction frequency of explants, especially with 0, 3, 5, and 14 days pre-culture, had a remarkable decrease in both strain systems. After 60-min infection, all the explants showed lower kanamycin-resistant callus induction frequency due to long exposed time under stress conditions. In each experiment with different infection time treatments, the explants with 7 days pre-culture showed the highest induction frequencies of kanamycin-resistant calli, irrespectively in strain type (Fig. 3). Despite that the callus induction frequency after 60-min infection significantly decreased to 24.51 and 26.92 % in pSWPA2::prx and pCaMV 35 s::prx, respectively, in this experiment with 60-min infection treatment, the callus induction frequencies using explants with 7 days pre-culture were still significantly higher than those with other explants (Fig. 3).
Putative transgenic callus induction frequency of various explants after selection with various infection times (10, 20, 30, and 60 min) in A. tumefaciens solution. Means followed by the same letter in the same series according to infection time are not significantly different with P < 0.05 according to two-way ANOVA using Duncan’s multiple-range test. Asterisk means the statistical analysis was according to the series of pre-culture period
According to various infection times, the transformation efficiencies (transformants/explants) were also evaluated, shown in Table 1. Of different explants with various infection times, explants with 20-min infection showed the highest frequencies than other explants, irrespectively in vector type. Explants after 10- and 30-min infection with bacterial solutions still produced putative transformants though the transformation frequencies were low; however, explants after 60-min infection fully lost the ability of producing transformants, irrespectively in vector type. Based on these results, 20-min infection was thought to be the most optimal infection time with OD600 0.4 bacterial solutions in further experiments. Generally, the bacterial solution with pCaMV 35S::prx as vector had higher transformation efficiencies than that using pSWPA2::prx as vector, with the highest frequency of 8.97 %. Although this transformation efficiency is lower than those reported in rice (11.1 %; Hoque et al. 2005) and Italian ryegrass (24 ~ 68 %; Bettany et al. 2003), it is comparable to tall fescue (8 %; Dong and Qu 2005), sugarcane (0.8 ~ 4.8 %; Joyce et al. 2010), and wheat (2.2 %; Peters et al. 1999).
Regeneration of Transgenic Plants
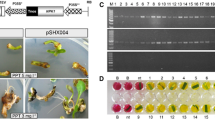
At the early plant regeneration stage, purple spots were produced on the surface of embryogenic calli (Fig. 4a, b). Although the reason had not been understood well until now, it was found that the putative transgenic shoots were generally regenerated from these purple spots, and some regenerated shoots still existed purple (Fig. 4c), and some existed green (Fig. 4d). At the late plant regeneration stage, purple or green leaves of the regenerated shoots continuously grew up until strong enough to be transferred to rooting media with selection (Fig. 4e, f). To induce the roots, the putative transgenic shoots were transferred on rooting medium with selection, and roots could be regenerated within 10 following days, even for the purple regenerated shoots (Fig. 4g, h). During the rooting stage, the purple putative transgenic shoots did not produce roots without producing green leaves but have potentials to produce green leaves first and then roots. Rhizoma might also be induced from the shoot apical meristems of putative transgenic plants (shown in arrow, Fig. 4g). Well-rooted putative transgenic plants were transferred into pots containing a mixture of sterilized soil and vermiculite (3:1) under greenhouse conditions (Fig. 4i, j). In addition, if no green leaves were formed from the purple regenerated shoots during the rooting stage, they needed further culture on solid culture media for that one purple regenerated plant with many leaves and strong root system that had been produced in our study and died after transferring into pots (Fig. 4k). One month culture later, putative transgenic plants regenerated multi-rhizoma were divided into many independent transgenic plants with 5 ~ 8 rhizoma per independent plant to meet the need of vegetative propagation. Due to the poor seed set and low fecundity (Huang et al. 2004) of this grass, a few seeds were obtained from transformants, but viable seeds had not been obtained until now.
Regeneration of transgenic plants. a, b Putative transgenic explants on selection media; c–e transgenic explants with regenerated shoots on selection media; f, g regenerated shoots forming roots on selection media; h, i putative transgenic plants on selection media; j, k transgenic plants grown in pots in greenhouse conditions. Scale bar 2 mm (a, b, g, k), 4 mm (c–f), 10 mm (h), and 20 mm (i, j)
Morphological Characteristics of Plants
All the putative transgenic plants appeared normal in phenotype. To compare the morphological characteristics between wild-type and putative transgenic plants, three space lengths of leaves were measured, including the lengths between root and first leaf, first and second leaf, and second and third leaf. Both the wild-type plants and putative transgenic plants had 1 ~ 2 stalks prior to being transferred from rooting media into pots, and the putative transgenic plants with pSWPA2::prx vector system showed less leaf biomass and slower leaf growth but no remarkable influence in plant height (Table S1). As growing up under a greenhouse condition, the plants regenerated more rhizoma from the shoot apical meristem (Fig. 5a). As L. chinensis is also considered as a pioneer forage species, especially in Northern China, the stalk numbers of plants were also measured after 1 month of culture in the greenhouse condition (Fig. 5b, Table 2). The wild-type plants had about 70 stalks with 3.50 ~ 3.81 leaves per stalk, while the morphological traits of putative transgenic lines showed not only line specific but vector specific: T1-14 line had fewer stalks than T1-11 and T1-17, but not the average leaf number per stalk; T2-5 had more stalks than T2-10 and T2-14, but the average leaf number per stalk of T2-5 was the lowest one among them; and generally, the lines with pSWPA2::prx showed fewer leaf biomass than lines with pCaMV 35S::prx.
Molecular Analysis of Plants
Fresh leaf tissues from independent putative transgenic lines were used for PCR analysis using primers specially amplifying the nptII gene and an internal 800-bp fragment of the Ib2-Cys prx gene. Of 47 putative transgenic lines regenerated from selection medium, 45 (95.74 %) lines were positive for integration of nptII gene (850 bp) when analyzed by PCR, while 36 (76.60 %) lines were positive for integration of Ib2-Cys prx gene (800 bp; Fig. 6). High integration frequency of nptII gene suggested that the kanamycin selection (150 mg l−1) used in this work could satisfy the demand for regeneration inhibition of non-transformants. To investigate the impact of exogenous Ib2-Cys prx gene on endogenous gene expression in the transgenic plants, transcription of Ib2-Cys prx gene was analyzed by RT-PCR analysis. We determined prx gene expression in the leaves of transformants harboring different constructs under various stresses such as 200 mM NaCl, 100 mM Na2CO3, and 2.5 mM H2O2 (Fig. 7). The prx gene expression in CaMV 35S::prx transformants had high levels under non-stressed conditions but was not significantly modulated after either stress. However, the prx gene expression in SWPA2::prx transformants was detected at a very low level under non-stress conditions and significantly induced after 6-h 200 mM NaCl treatment (Fig. 7a). The average gene expression in SWPA2::prx transformants exposed to NaCl stress was comparable to that in transgenic plants containing CaMV 35S::prx. In the case of 100 mM Na2CO3, prx gene expression in SWPA2::prx transformants was also significantly induced by 6-h alkaline stress treatment, with the levels 38.93-fold higher than that before alkaline stress treatment but comparable to that established by the CaMV 35S promoter in T1-11, T1-14, and T1-17 transgenic lines (Fig. 7b). H2O2 treatment also enhanced prx gene expression elevated by the stress inductive SWPA2 promoter in T2-5, T2-10, and T2-14 transgenic lines (Fig. 7c). The prx gene expression in SWPA2::prx transformants had increased 35.70 times after 6-h 2.5 mM H2O2 treatment. Although the average prx gene expression level in SWPA2::prx transformants were 1.27-fold higher than that in CaMV 35S::prx transformants under H2O2 stress treatment, statistical analysis of both levels did not show remarkable differences.
Molecular analysis of transgenic plants. a PCR analysis of DNA isolated from leaves of ten putative transformants and one untransformed control (–) using primers specific for the nptII and Ib2-Cys prx genes, respectively. Lanes 1 ~ 10 putative transgenic plants with pCaMV 35S::prx, +, positive control amplified from pCAMBIA2300-CaMV35S::prx; b. PCR analysis of DNA isolated from leaves of ten putative transformants and one untransformed control (–) using primers specific for the nptII and Ib2-Cys prx genes, respectively. Lanes 1 ~ 10 putative transgenic plants with pSWPA2::prx, +, positive control amplified from pCAMBIA2300-SWPA2::prx
The results indicate that a successful A. tumefaciens-mediated transformation and regeneration of transgenic plants of the halophyte L. chinensis has been established. The protocol can be used for the transfer of genes related to stress tolerance in this grass. The successful gene interaction in CaMV 35S::prx transformants enhances the oxidative stress tolerance with overexpression of prx gene from sweet potato. The SWPA2::prx transformants with exogenous prx gene under the control of an oxidative stress-inducible SWPA2 promoter showed higher expression of prx when exposed to oxidative stress, indicating that it results in the elicitation of stress defense mechanisms by increasing antioxidant enzymes and proteins.
Abbreviations
- 2-Cys Prx :
-
2-Cysteine peroxiredoxin
- nptII :
-
Neomycin phosphotransferase
- MS:
-
Murashige and Skoog
- AS:
-
Acetosyringone
- 2,4-D:
-
2,4-Dichlorophenoxyacetic acid
- CE:
-
Cefotaxime
- Km:
-
Kanamycin
- Kn:
-
Kinetin
- NAA:
-
Naphthalene acetic acid
- PAT:
-
Phosphinothricin acetyltransferase
- LEA:
-
Late embryogenesis abundant
- CaMV:
-
Cauliflower mosaic virus
- OD600 :
-
Optical density at a wavelength of 600 nm
- SDS:
-
Sodium dodecyl sulfate
- PCR:
-
Polymerase chain reaction
References
Bettany AJE, Dalton SJ, Timms E, Manderyck B, Dhanoa MS, Morris P (2003) Agrobacterium tumefaciens-mediated transformation of Festuca arundinacea (Schreb.) and Lolium multiflorum (Lam.). Plant Cell Rep 21:437–444. doi:10.1007/s00299-002-0531-3
Boyer JS (1982) Plant productivity and environment. Science 218:443–448. doi:10.1126/science.218.4571.443
Chae HZ, Robison K, Poole LB, Church G, Storz G, Rhee SG (1994) Cloning and sequencing of thiol-specific antioxidant from mammalian brain: alkyl hydroperoxide reductase and thiol-specific antioxidant define a large family of antioxidant enzymes. Proc Natl Acad Sci USA 91:7017–7021
Chen ZZ (1988) Topography and climate of Xilin River Basin. In: Research on grassland ecosystem, Science, China 3:13-22
Dellaporta SL, Wood J, Hicks HB (1983) A plant DNA minipreparation: version II. Plant Mol Biol Rep 1:19–21. doi:10.1007/BF02712670
Dong SJ, Qu RD (2005) High efficiency transformation of tall fescue with Agrobacterium tumefaciens. Plant Sci 168
Duncan DB (1955) Multiple range and multiple F tests. Biometrics 11:1–42
Fuchs RL, Ream JE, Hammond BG, Naylor MW, Leimgruber RM, Berberich SA (1993) Safety assessment of the neomycin phosphotransferase II (NPTII) protein. Biotechnology 11:1543–1547
Guo JX (1994) Decomposer subsystem of Leymus chinensis grasslands. Jilin Science and Technology, Jilin, China (in Chinese)
Hendrick JP, Hartl FU (1993) Molecular chaperone functions of heat shock proteins. Annu Rev Biochem 62:349–384
Hiei Y, Komari T, Kubo T (1997) Transformation of rice mediated by Agrobacterium tumefaciens. Plant Mol Biol 35:205–218
Hirotsu S, Abe Y, Okada K, Nagahara N, Hori H, Nishino T, Hakoshima T (1999) Crystal structure of a multifunctional 2-Cys peroxiredoxin heme-binding protein 23 kDa/proliferation-associated gene product. Proc Natl Acad Sci USA 96:12333–12338
Hofmann B, Hecht HJ, Flohé P (2002) Peroxiredoxins. Biol Chem 383:347–364. doi:10.1515/BC.2002.040
Hoque ME, Mansfield JW, Bennett MH (2005) Agrobacterium-mediated transformation of Indica rice genotypes: an assessment of factors affecting the transformation efficiency. Plant Cell Tiss Organ Cult 82:45–55
Hosoki T, Kigo T (1994) Transformation of Brussels sprouts (Brassica oleracea var. gemmifera Zenk.) by Agrobacterium rhizogens harbouring a reporter β-glucuronidase gene. J Japan Soc Hort Sci 63:589–592
Huang ZH, Zhu JM, Mu XJ, Lin JX (2004) Pollen dispersion, pollen viability and pistil receptivity in Leymus chinensis. Ann Bot 93:295–301. doi:10.1093/aob/mch044
Jang HH, Lee KO, Chi YH, Jung BG, Park SK, Park JH, Lee JR, Lee SS, Moon JC, Yun JW, Choi YO, Kim WY, Kang JS, Cheong GW, Yun DJ, Rhee SG, Cho MJ, Lee SY (2004) Two enzymes in one: two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell 117:625–635
Joubert P, Beaupère D, Lelièvre P, Wadouachi A, Sangwan RS, Sangwan Norreel BS (2002) Effects of phenolic compounds on Agrobacterium vir genes and gene transfer induction-a plausible molecular mechanism of phenol binding protein activation. Plant Sci 162:733–743
Joyce P, Kuwahata M, Turner N, Lakshmanan P (2010) Selection system and co-cultivation medium are important determinants of Agrobacterium-mediated transformation of sugarcane. Plant Cell Rep 29:173–183
Kim KY, Kwon SY, Lee HS, Hur YK, Bang JW, Kwak SS (2003) A novel oxidative stress-inducible peroxidase promoter from sweet potato: molecular cloning and characterization in transgenic tobacco plants and cultured cells. Plant Mol Biol 51:831–838
Kim MD, Yang KS, Kwon SY, Lee SY, Kwak SS, Lee HS (2009) Selection of transgenic sweetpotato plants expressing 2-Cys peroxiredoxin with enhanced tolerance to oxidative stress. J Plant Biotechnol 36:75–80
McHughen A, Jordan M, Feist G (1989) A preculture period prior to Agrobacterium inoculation increases production of transgenic plants. J Plant Physiol 135:245–248
Metz TD, Dixit R, Earle ED (1995) Agrobacterium tumefaciens-mediated transformation of broccoli (Brassica oleracea var. italica) and cabbage (B. oleracea var. capitata). Plant Cell Rep 15:287–292
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–497
Neumann CA, Krause DS, Carman CV, Das S, Dubey DP, Abraham JL, Bronson RT, Fujiwara Y, Orkin SH, Van Etten RA (2003) Essential role for the peroxiredoxin Prdx1 in erythrocyte antioxidant defence and tumour suppression. Nature 424:561–565. doi:10.1038/nature01819
Opabode JT (2006) Agrobacterium-mediated transformation of plants: emerging factors that influence efficiency. Biotechnology and Molecular Biology Review 1:12–20
Pandey V, Misra P, Chaturvedi P, Mishra MK, Trivedi PK, Tuli R (2010) Agrobacterium tumefaciens-mediated transformation of Withania somnifera (L.) Dunal: an important medicinal plant. Plant Cell Rep 29:133–141
Peters NR, Ackerman S, Davis EA (1999) A modular vector for Agrobacterium mediated transformation of wheat. Plant Mol Biol Rep 17:323–331
Puddephat IJ, Riggs TJ, Fenning TM (1996) Transformation of Brassica oleracea L.: a critical review. Mol Breed 2:185–210
Sangwan RS, Bourgeois Y, Brown S, Vasseur G, Sangwan Norreel B (1992) Characterization of competent cells and early events of Agrobacterium-mediated genetic transformation in Arabidopsis thaliana. Planta 188:439–456
Sharma KK, Anjaiah V (2000) An efficient method for the production of transgenic plants of peanut (Arachis hypogea L.) through Agrobacterium tumefaciens-mediated genetic transformation. Plant Sci 159:7–19
Shu QY, Liu GS, Xu SX, Li XF, Li HJ (2005) Genetic transformation of Leymus chinensis with the PAT gene through microprojectile bombardment to improve resistance to the herbicide Basta. Plant Cell Rep 24:36–44. doi:10.1007/s00299-004-0908-6
Storz G, Tartaglia LA, Farr SB, Ames BN (1990) Bacterial defense against oxidative stress. Trends Genet 6:363–368
Sun YL, Hong SK (2009) Somatic embryogenesis and in vitro plant regeneration from various explants of the halophyte Leymus chinensis (Trin.). J Plant Biotechnol 36:236–243
Sun YL, Hong SK (2010a) Effects of plant growth regulators and l-glutamic acid on shoot organogenesis in the halophyte Leymus chinensis (Trin.). Plant Cell Tiss Organ Cult 100:317–328. doi:10.1007/s11240-009-9653-4
Sun YL, Hong SK (2010b) Establishment of a novel plant regeneration system from suspension-derived callus in the halophytic Leymus chinensis (Trin.). J Plant Biotechnol 37:228–235. doi:10.5010/JPB.2010.37.2.228
U.S. Environmental Protection Agency (1994) Neomycin phosphotransferase: II. Tolerance exemption. 56:4935
Wang LJ, Li XF, Chen SY, Liu GS (2009) Enhanced drought tolerance in transgenic Leymus chinensis plants with constitutively expressed wheat TaLEA 3 . Biotechnol Lett 31:313–319. doi:10.1007/s10529-008-9864-5
Zhangsun D, Luo S, Chen R, Tang K (2007) Improved Agrobacterium-mediated genetic transformation of GNA transgenic sugarcane. Biologia 62:386–393
Acknowledgments
This work was supported by Nutraceutical Bio Brain Korea 21 Project Group.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Sun, YL., Hong, SK. Agrobacterium tumefaciens-Mediated Transformation of the Halophyte Leymus chinensis (Trin.). Plant Mol Biol Rep 30, 1253–1263 (2012). https://doi.org/10.1007/s11105-012-0434-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-012-0434-6