Abstract
Aims
Crops often encounter a soil deficiency of nitrogen (N), the most important macronutrient for plants; however, the molecular mechanism of plant responses to N deficiency remains unclear. In this study, proteome-level changes that occur in bread wheat seedlings suffering from N deficiency were investigated to identify some N deficiency-responsive protein species in bread wheat.
Methods
We utilized isobaric tagging for relative and absolute quantification (iTRAQ) to measure changes in the proteome patterns of N-deficient wheat seedlings and validated the role of abscisic acid (ABA) using the barley stripe mosaic virus-induced gene-silencing (BSMV-VIGS) method.
Results
A total of 1515 N deficiency–responsive protein species were successfully identified in both root and leaf tissues of wheat seedlings suffering from 8-d N deficiency. Of these, abundance of wheat zeaxanthin epoxidase (TaZEP), a key ABA synthesis-related enzyme, was significantly upregulated, and the endogenous ABA contents also markedly increased. After TaZEP gene was further silenced using BSMV-VIGS method, BSMV-VIGS-TaZEP infected wheat seedlings showed enhanced sensitivity to N deficiency, suggesting silencing of TaZEP gene decreased the tolerance to N deficiency remarkably.
Conclusion
Our results identified some N deficiency-responsive protein species and revealed the role of ABA in wheat responses to N deficiency.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nitrogen (N) is one of three most essential macronutrients for plant growth and development. It constitutes approximately 2% of plant dry matter and is also an important constituent of most cellular molecules, including proteins (enzymes), chlorophyll pigments, amino acids, nucleic acids, and phytohormones (Criado et al. 2007; Shi et al. 2010). It plays key roles in many physiological processes, such as protein synthesis, amino acid metabolism, and carbon metabolism (Cai et al. 2012). Plant growth and yield require abundant N, and the element is deficient in most cultivated soils worldwide. Therefore, in many agricultural regions, large amounts of chemical N fertilizers are applied annually to compensate for this deficiency, and the consumption of chemical N fertilizers has increased more than 9-fold worldwide since 1960 (Qin et al. 2019). However, less than 50% of applied nitrogen is effectively absorbed, and the other 50% or more is ineffectively dissipated in the wider environment by leaching into the soil, water, and air, leading to environmental pollution and health hazards (Hakeem et al. 2011; Nazir et al. 2015). Therefore, it is essential to improve the absorption and utilization efficiency of N fertilizer and minimize its input, and implementing these changes requires detailed understanding of the molecular mechanisms of N absorption and utilization.
Plant responses to N deficiency involve morphological, physiological, developmental, and cellular processes with known responses, including the improvement of N absorption and translocation, remobilization of N from source organs to newly growing tissues, and alteration of carbohydrate partitioning (Krapp et al. 2011). These processes are effectively controlled by N deficiency-responsive genes, and thus, identification of N deficiency-responsive genes is a fundamental step to understanding these complex processes (Liang et al. 2013). In addition to identification on the genetic basis of nutrient use efficiency, such as genome-wide association studies, forward genetic tools such as omics are suitable to obtain a comprehensive overview of the changes in response to stress and may help to close the information gap between genotype and phenotype (Marioni et al. 2008). Transcriptomic technology can take into consideration all aspects of nutrient management in plants and is an integrated approach to investigating biotic and abiotic factors, and using this technology combined with other protocols, many N deficiency-responsive genes have been identified in the past few decades (Gelli et al. 2014; Yang et al. 2015; He et al. 2016a; Quan et al. 2016; Wei et al. 2016; Curci et al. 2017; Nawaz et al. 2018). These studies have transcriptionally provided a basis for elucidating the underlying molecular mechanism of physiological changes associated with N deficiency and have improved our understanding of plant responses to N deficiency. Phytohormones regulate many developmental processes and adaptive stress processes and are considered to be the foremost signaling molecules in plants (Lee and Luan 2012), and some studies have shown that the genes involved in the biosynthetic pathway of some plant hormones (e.g., abscisic acid, jasmonic acid, and gibberellin) are significantly regulated (Ristova et al. 2016; Kuang et al. 2017). For example, ABA contents significantly increase under low-N conditions and exogenous ABA application promotes the nitrogen use efficiency in plants (Oka et al. 2012; Han et al. 2017). ABA is synthesised via the carotenoid biosynthetic pathway, zeaxanthin epoxidase (ZEP) enzyme catalyses the conversion of zeaxanthin to alltrans-violaxanthin in a two-step oxidation, and is considered as one of the key enzymes in this biosynthetic pathway because its mutant displays the remarkably decreased levels of ABA (Agrawal et al. 2001; Cuming and Stevenson 2015).
However, many questions need to be further answered because gene expression can also be regulated at the posttranscriptional, translational, and posttranslational levels (Hu et al. 2015). Cellular proteins directly perform most of the catalytic work required by biological processes, and proteomics is an alternative approach to complement transcriptomic analyses because changes at the transcriptional level are not always reflected at the translational level (Findlay et al. 2009; Grobei et al. 2009; Kosová et al. 2011). Some N deficiency-responsive protein species in higher plants have been identified using a proteomic approach in some plant species, with most of the focus on two model plant species (Arabidopsis and rice) (Møller et al. 2011; Deng et al. 2014; Jin et al. 2015; Qin et al. 2019). To some extent, different plant species have diverse responses to the same abiotic or biotic stresses (Kang et al. 2015), and it is important to identify N deficiency-responsive protein species in important food crops because we are facing challenges to the worldwide food supply. Bread wheat is one of the three most important cereal crops globally and is widely cultivated worldwide. Its production accounts for over 20% of the world’s food supply (He et al. 2015). To our knowledge, however, quantitative proteomics studies on N deficiency have not been reported in this species so far because this species is characterized by a very large and rather complex, 17-gigabase-pair (Gb) allohexaploid genome and more than 85% of its genome consists of highly repetitive sequences (Brenchley et al. 2012), although the draft genome sequences of bread wheat and its diploid ancestors have recently been completed (Jia et al. 2013; Ling et al. 2013; IWGSC 2014 and 2018).
Short-term nutrient deficiency is a result of a significant decrease in the level of the corresponding nutrients, whereas long-term nutrient deficiency is characterized by the major alteration of metabolic activities and the presence of a distinct phenotypic symptom (Liang et al. 2013). More nutrient-responsive proteins (genes) have been identified under long-term nutrient deficiency than under short-term deficiency (Ma et al. 2014). Identifying differentially abundant protein species in plants suffering from long-term nutrient deficiency could reveal the in-depth mechanisms of nutrient deficiencies (Liang et al. 2013). When wheat seedlings suffered from 8 d of N deficiency in our previous study, the phenotype, morphological and physiological parameters (e.g., plant height, shoot fresh and dry weights, and nitrate and protein contents) of wheat seedlings were significantly inhibited (Guo et al. 2014). These results suggested that wheat seedlings at this time point suffered from long-term N deficiency. At this time point in the present study, an iTRAQ-based large-scale quantitative proteomics analysis was used to identify a large number of N deficiency-responsive protein species in the root and leaf tissues of wheat seedlings responding to long-term N deficiency. This technique was sensitive enough to delineate 1515 N deficiency-responsive protein species. Moreover, an important role of phytohormone ABA in wheat seedlings responding to long-term N deficiency was verified using both physiological protocols and BSMV-VIGS. This study identifies some N deficiency-responsive protein species in important crops in response to N deficiency and reveals ABA roles.
Methods
Plant growth conditions and N deficiency treatment
A spring bread wheat cultivar Yumai 34, characterized with good quality, and high biotic and abiotic tolerance, and released in 1994, was developed by the Agricultural and Forestry Science Institute of Zhengzhou and its seeds were kindly provided by Dr. Xiangzheng Liao in this institute. Seeds (sterilized in 0.01% HgCl2, washed in distilled water) were grown hydroponically in glass dishes (15 cm diameter) in full-strength Hoagland solution (Hoagland and Arnon 1950; Elberse et al. 2003). These glass dishes were placed in an FPG-300C-30D illumination incubator (Ningbo Laifu Technology Co., Ltd., China) with a 14-h photoperiod, 25/15 °C day/night temperatures, relative humidity of 60/75% (day/night), and light intensity of 250 μmol m−2 s−1. Each dish contained approximately 50 seedlings. After two weeks, the wheat seedlings were divided into two groups: one group remained in Hoagland medium in full N conditions (6 mM) as a control, and the other group was transferred to N-free (DN) Hoagland medium for N deficiency. KNO3 and CaNO3 were replaced by KCl and CaCl2, respectively (Guo et al. 2014). The nutrient solutions were renewed every 2 days, and the pH was adjusted to 5.5~6.0 with HCl or NaOH each time. Root and leaf tissues of the wheat seedlings were collected at 0, 2, 4, 6, and 8 days after incubation in two nutrient media, immediately frozen in liquid nitrogen, and stored at −80 °C for extracting the total proteins.
iTRAQ labeling, LC-MS/MS analysis, and protein identification
For iTRAQ, three biological replicates of protein samples were extracted from root and leaf tissues. Total proteins were extracted using the trichloroacetic acid/acetone method (Li et al. 2013) and then reduced, alkylated, and digested using the filter–aided sample preparation procedure (Wisniewski et al. 2009). The digested peptide mixture was labeled using the iTRAQ reagents 8-plex Kit according to the manufacturer’s instructions (AB Sciex, Inc., Foster City, CA, USA). Extracted protein samples were labelled with 113, 114, 115, 116, 117, 118, 119, and 121 labels in each independent 8-plex iTRAQ reagent, each label represented one sample, and three 8-plex iTRAQ tags containing three biological replicates of each sample were performed. In three 8-plex iTRAQ tags, 113-control root, 114-N-deficient root, 117-control leaf, and 118-N-deficient leaf have been labelled and used in this study, whereas the other four labels (115, 116, 119, and 121) were used in other experiments (Li et al. 2017).
The MS/MS data were analyzed by the cHiPLC Nanoflex microchip system (Dublin, CA, USA) with nanoLC–MS/MS (Nano Ultra 2D Plus, Eksigent) equipped with an AB SCIEX TripleTOF 5600 MS (Toronto, Concord, Canada) (Kang et al. 2015). MS/MS spectra were identified using the MASCOT engine software (Matrix Science, London, UK; version 2.2) embedded in the Proteome Discoverer 1.4 software (Thermo Electron, San Jose, CA). The protein species were filtered using the cRAP database (ftp://ftp.thegpm.org/fasta/cRAP) (Torabi et al. 2009). Moreover, the functional/biological processes were searched by using NCBInr databases (http://blast.ncbi.nlm.nih.gov) (version 10/06/2015, 4,075,097 entries), and then grouped into biological functions using Gene Ontology terms (Wang et al. 2012). For protein identification, the following parameters were used: Viridiplantae as the taxonomy, MS/MS as the ion search type, trypsin as the digestion enzyme, carbamidomethyl (C), iTRAQ8plex (N-term), and iTRAQ8plex (K) as the fixed modfication, oxidation (M) as the variable modification, the matched unique peptides at ≥1, monoisotopic as the mass values, the max missed cleavage at 2.0, the fragment mass tolerance at 0.1 Da, peptide mass tolerance at ±20 ppm, and the peptide false discovery rate (FDR) at ≤0.01 as described in our previous study (Li et al. 2017).
For protein quantification, the MS/MS spectra revealed unique proteins that were identified by extract peak intensities of the iTRAQ labeling tags. Then, they were normalized among samples with the built-in bias correction function of ProteinPilot (AB Sciex, Inc). Using the software’s standard procedures, we generated relative protein quantification for comparisons of interest. In one 8-plex iTRAQ reagent, only one replicate of each sample was labelled and ratios of the identified protein species between DN and CK (e.g., replicate 1 of N-deficient root vs replicate 1 of N-sufficient root) were calculated and compared according to the protein abundance. According to methods of Ma et al. (2012), we used the 1.2-fold cutoff for further analysis in order to contain more candidate genes involving in wheat response to N deficiency because there were fewer up- or down-regulated protein species with the abundance change more than 1.5 or 2.0-fold cutoff.
Determination of the contents of selected phytohormones
Root and leaf tissues of wheat seedlings were harvested separately at 8 days after incubation in two nutrient media. Sampled tissues (2.5 g) were homogenized with 3 cm3 10% trichloroacetic acid, then incubated overnight at 20 °C, and centrifuged at 8000 g for 1 h at 4 °C. Precipitates were dried at 40 °C in vacuum and homogenized with lysate solution (1.35 g of urea, 0.1 g of CHAPS, and 2.5 mm3 of double-distilled water). The contents of ABA, auxin (IAA), salicylic acid (SA), jasmonic acid (JA), cytokinin (CTK), and ethylene (ETH) were determined using an enzyme–linked immunosorbent assay kit (R&D, USA) according to the manufacturer’s instructions.
In vitro transcription of virus-induced genes and plant inoculation
ABA role in response to N deficiency was identified using the BSMV-VIGS method by identifying the function of wheat TaZEP gene. Because of asymmetric expression patterns of two or more homoeoalleles in allopolyploid plants (Møller et al. 2011), only one copy (gi|474,244,839) of TaZEP was identified in our study. In this study, cDNA sequences of three copies were identified in the genome of bread wheat, and they had high similarities (98.2%) (Supplementary Fig. S1). To prevent functional complementation and allow commonly silencing of three copies (homologues) of TaZEP gene in genome of bread wheat, a conserved cDNA fragment (254 bp), which shares high similarities (99.5%) among three homologues (Supplementary Fig. S1), was used to construct silencing vector (BSMV-TaZEP) to possibly result in the simultaneous silencing of three homologues in the wheat genome. In BSMV-TaZEP-inoculated wheat plants, transcription levels of TaZEP gene were to identify the total expression profile of all three homologues. PCR primers specific to the family member but shared by the three homologues of the same gene were designed and used for quantitative real-time PCR (qRT-PCR) (Song et al. 2012; Grün et al. 2018), and primers and the amplified fragment are indicated in Supplementary Fig. S1.
In this study, the BSMV-VIGS method was performed as previously described (Zhang et al. 2016). The α, β, and γ and α-, β-, and γ-ZEP transcripts of the BSMV genome were synthesized using RiboMAX™ Large Scale RNA Production System-T7 (Promega, USA). The RNA-α, RNA-β, and RNA-γ and RNA-α-, −β-, and -γ-ZEP transcripts were separately mixed in a 1:1:1 ratio, added to diethylpyrocarbonate-treated distilled water (1:1), and subsequently added with an abrasive FES buffer (Scofield et al. 2005). BSMV-γ:00 and BSMV-TaZEP were composed of α, β, and γ transcripts and RNA-α-, −β-, and -γ-ZEP transcripts, respectively. Two-week-old wheat seedlings were separately inoculated with BSMV-γ:00 and BSMV-TaZEP, and 10 mm3 viral RNA was inoculated on the downmost second leaf of each wheat seedling (Supplementary Fig. S2). Inoculated leaves were sprayed with nuclease-free water and then covered with plastic film to maintain high relative humidity (85.0%) and darkness for 24 h in an illumination incubator. After inoculation, the wheat seedlings were immediately transferred to N-free Hoagland solution for the N deficiency stress treatment. At 8 days after inoculation, wheat seedlings with chlorosis were hypothesized to be BSMV-infected plants (Liu et al. 2016). In this study, 40 (80.0%) and 43 (86.0%), and 38 (76.0%) and 44 (88.0%) of the 50 BSMV-γ:00-infected or BSMV-TaZEP-inoculated wheat seedlings showed chlorosis under full-length N supply and nitrogen deficiency conditions, respectively (Supplementary Table S1). At the same time, the inoculated and most adjacent and higher leaves of BSMV-inoculated wheat seedlings were sampled, and qRT-PCR was performed to determine the transcription levels of TaZEP gene for further identification of the BSMV-TaZEP-inoculated wheat seedlings. The primers are indicated in Supplementary Fig. S1. At 8 days after inoculation, dry weight and ABA contents of the BSMV-TaZEP-inoculated wheat seedlings identified above were examined as described. N contents were measured as described by He and his colleagues (He et al. 2016b).
Statistical analysis
In this study, all experiments were repeated in triplicate, and each sampling was analyzed separately. The results were analyzed using SPSS 17.0 statistical software (SPSS Inc., Chicago, IL., USA), and Duncan’s multiple range test (DMRT) was used to identify significant (P < 0.05) differences among the group means.
Results
A large number of N deficiency-responsive protein species identified using the iTRAQ method in wheat seedlings suffering from N deficiency
To identify the N deficiency-responsive protein species, proteome profiles were generated using the iTRAQ method in the root and leaf tissues of wheat seedlings subjected to N deficiency for 8 days. Three independent biological replicates were used for each sample for iTRAQ labeling, and then the spectra were analyzed using the NCBInr database. A total of 10,001 peptides from 151,654 spectra (Supplementary Table S2), and 7336 protein species were identified (Supplementary Table S3), of which 4571 occurred in all three biological replicates and were included in subsequent analyses (Supplementary Table S4). The mass spectrometry proteomics data for the present study are available via ProteomeXchange with the dataset identifier PXD71498.
In this study, the ratios (≥ 1.20- and ≤ 0.83-fold) in N deficient vs. control plants were considered to be changes in the altered abundance of N deficiency-responsive protein species, and Duncan’s multiple-range tests were performed to determine the differences (P < 0.05). The protein species identified were further filtered using the cRAP database. Terminology for the identified protein species was based on the method of Schlüter and his colleagues (Schlüter et al. 2009). Based on this method, some protein species were identified with the same names and accession numbers but had different isoelectric points (pIs) or molecular weights (MWs) (Supplementary Table S5). These proteins should be classified into different protein species because one gene could have different protein products and diverse cellular tasks (Kosová et al. 2018). These could be due to nucleotide polymorphisms, proteolytic cleavage, alternative splicing, post-translational modifications, etc. (Schlüter et al. 2009). For example, two phenylalanine ammonia-lyase enzymes (gi|357,150,254) were identified in root proteomic data, but they should be treated as two different protein species because they had different pIs and MWs.
Based on the above criteria, in the present study, 1515 N deficiency-responsive protein species were identified in both root and leaf tissues of wheat seedlings suffering from 8-d N deficiency. Of these protein species, 866 (427 ≥ 1.20- and 439 ≤ 0.83-fold) and 649 (348 ≥ 1.20- and 301 ≤ 0.83-fold) were from the root and leaf tissues, respectively (Table 1, Supplementary Table S5). Our findings indicated that most of the N deficiency-responsive protein species were expressed at low abundance, and the abundance of most N deficiency-responsive protein species in the root (720/866, 83.1%) and leaf (543/649, 83.7%) tissues of N-deficient wheat seedlings was altered by 1.20 ~ 1.49- or 0.67 ~ 0.83-fold, respectively (Fig. 1).
Proportions of the identified protein species with decreased (blue) and increased (yellow) abundances in root (a) and leaf (b) tissues of N-deficient wheat seedlings with respect to control plants. Two weeks-old wheat seedlings were transferred to N-deficient full-length Hoagland medium for 8 d. At this timepoint, some growth and physiological parameters (plant height, root length, and root or leaf dry weights) showed significant changes between N-deficient wheat seedlings and N-sufficient wheat seedlings, and thus, iTRAQ proteomics was used to measure the global protein expression patterns in the root (a) and leaf (b) tissues of N-deficient wheat seedlings. Venn diagrams show the proportions of the identified protein species with low (blue) and high (yellow) abundances. Only protein species commonly identified in three biological replicates for each sample were considered. Names and abundance of the identified representative protein species related to ABA biosynthesis (e.g. zeaxanthin epoxidase, ZEP; short-chain dehydrogenase, SDR; farnesyl pyrophosphate synthase, FPS) are indicated in brackets
Functional classification of N deficiency-responsive protein species and differences in the root and leaf tissues
According to the biological functions listed on the UniProt and Gene Ontology website (Li et al. 2014), the 1515 N deficiency-responsive protein species identified were classified into ten and eleven functional categories in the root and leaf tissues, respectively (Fig. 2; Supplementary Table S5). The functional categories in the root tissue were involved in hormone synthesis (2.5%), signal transduction (5.0%), nucleotide metabolism (5.3%), lipid and phosphate metabolism (1.7%), protein metabolism (22.1%), carbohydrate metabolism (12.6%), stress and defense (14.1%), transportation (9.6%), other metabolisms (3.3%), and unknown function (23.8%) (Fig. 2a). Analogous functional categories in the leaf tissue of wheat seedlings were related to hormone synthesis (2.0%), signal transduction (4.6%), nucleotide metabolism (5.7%), lipid and phosphate metabolism (2.2%), protein metabolism (18.2%), carbohydrate metabolism (9.9%), photosynthesis (5.7%), stress and defense (14.2%), transportation (11.4%), other metabolisms (2.6%), and unknown function (23.6%) (Fig. 2b). Predicted, hypothetical, uncharacterized, and unnamed N deficiency-responsive protein species were classified into the unknown function category. A large proportion (23.8% and 23.6% in root and leaf tissues, respectively) of unknown function protein species were not annotated or identified. A high proportion of unknown function protein species were not annotated (Fig. 2; Supplementary Table S5).
Functional categorization of N-responsive protein species in root and leaf tissues of bread wheat seedlings suffering from N deficiency for 8 days. Two weeks-old wheat seedlings were transferred to N-deficient full-length Hoagland medium for 8 days, and an iTRAQ proteomics was performed in the root and leaf tissues of N-deficient wheat seedlings. The N deficiency-responsive protein species identified in root (a) and leaf (b) tissues were functionally categorized according to the biological functions listed on the UniProt and Gene Ontology website (Li et al. 2014). Names of the identified and representative protein species related to ABA synthesis (ZEP, SDR, and FPS) are indicated in brackets, and ZEP, SDR, and FPS have been annotated in Fig. 1
The amount and percentage of the carbohydrate metabolism-related protein species identified in root tissue (109/866, 12.6%) were greater than those in leaf tissue (64/649, 9.9%). The amount and percentage (66/109, 60.6%) of downregulated carbohydrate metabolism-related protein species in root tissue were greater than those (36/64, 56.2%) in leaf tissue. Some (155/1515, 10.2%) of the identified N deficiency-responsive proteins were found in both the root and leaf tissues of N-deficient wheat seedlings (Table 1, Supplementary Table S6). However, most (1360/1515, 89.8%) of the protein species identified appeared specifically between the root and leaf tissues.
Some phytohormones could be involved in N deficiency in wheat seedlings
Our study identified a few protein species related to the synthesis of important plant hormones, such as ABA, jasmonic acid (JA), ethylene (ETH), auxin (IAA), salicylic acid (SA), and cytokinin (CTK). In the root tissue, 2, 4, 9, 2 and 5 protein species were involved in the synthesis of ABA, SA, JA, CTK, and ETH, respectively. In the leaf tissue, 3, 3, 1, 3, 2 and 1 were separately related to the synthesis of ABA, JA, IAA, SA, CTK, and ETH (Supplementary Table S5). However, there were some differences in the changes in the contents of these plant hormones between the root and leaf tissues (Fig. 3).
Contents of the phytohormones in N-deficient wheat seedlings. Two weeks-old wheat seedlings were transferred to full N conditions (6 mM, CK) or N-free Hoagland solution for the N-deficient (DN) full-length Hoagland medium for 8 d, and contents of phytohormones ABA (a), JA (b), ETH (c), IAA (d), CTK (e), SA (f) both in root and leaf tissues of N-sufficient and -deficient wheat seedlings were measured. Each value is mean ± standard deviation (SD) of three biological replicates, and different letters indicate significant differences (P < 0.05) relative to control by using two-way ANOVA followed by Duncan’s multiple range test
Phytohormone contents (IAA, ABA, SA, JA, CTK and ETH) were measured next in both root and leaf tissues of wheat seedlings at 8 days after N deficiency. Our data indicated that ABA, JA, and ETH contents markedly increased by 37.3% and 43.6%, 17.5% and 23.6%, and 17.2% and 14.0% in both root and leaf tissues, respectively (Fig. 3a-c). However, the contents of IAA, CTK, and SA significantly decreased by 60.5% and 58.0%, 69.4% and 70.0%, and 46.8% and 37.2% in both root and leaf tissues, respectively (Fig. 3d-e). At 8 d after N deficiency, the contents of ABA in both root and leaf tissues were increased by 4.50- and 5.20-fold, respectively, far higher than those of other phytohormones (Fig. 3a-c).
Roles of ABA hormone in N-deficient wheat seedlings
A conserved fragment (254 bp) of TaZEP gene was used to construct a BSMV-derived TaZEP silencing vector, which was inoculated into the leaves of wheat seedlings. BSMV-γ:00-inoculated wheat plants were used as control. Our results indicated that BSMV-VIGS-induced chlorosis appeared in the middle of seedling leaf tissue at 8 days after inoculation (Fig. 4), and the efficiency of the BSMV-VIGS was over 76% (Supplementary Table S1). Total transcription levels of three TaZEP homologues and contents of ABA in the leaf tissue of BSMV-TaZEP-inoculated wheat seedlings were markedly reduced by 56.7% and 61.6%, respectively (Fig. 5), suggesting that TaZEP was successfully silenced and ABA synthesis was greatly inhibited in wheat seedlings. Our data indicated that, compared to BSMV-γ:00-inoculated wheat seedlings, the growth of the BSMV-TaZEP-inoculated wheat seedlings was substantially inhibited (Fig. 4) and the total dry weights and N contents of the BSMV-TaZEP-silenced wheat seedlings were significantly reduced (Fig. 5). These results suggested that silencing the gene encoding TaZEP enzyme resulted in a decrease in the tolerance to N deficiency in wheat seedlings, suggesting a role of ABA in response to N deficiency.
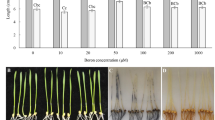
Phenotypes of the BSMV-TaZEP vector-inoculated wheat seedlings at 8 days after nitrogen deficiency. Two-week-old wheat seedlings were sprayed with nuclease-free water (WT), or separately inoculated with 10 mm3 BSMV-γ:00 (γ:00) and BSMV-TaZEP (TaZEP) viral RNA on the downmost second leaf of the wheat seedlings. After inoculation, wheat seedlings were immediately transferred to full N conditions (6 mM, CK) or N-free Hoagland solution for the N deficiency (DN) for 8 days to observe their phenotypes. The efficiency of the BSMV-VIGS was over 76%, and the number is indicated in Supplementary Fig. S1
Transcription levels of TaZEP gene, and some physiological parameters on BSMV-TaZEP vector-inoculated wheat seedlings suffering from nitrogen deficiency. Two-week-old wheat seedlings were separately inoculated with 10 mm3 BSMV-γ:00 (γ:00) and BSMV-TaZEP (TaZEP) viral RNA on the downmost second leaf of the wheat seedlings. After inoculation, the wheat seedlings were immediately transferred to full N conditions (6 mM, CK) or N-free Hoagland solution for the N deficiency (DN) for 8 days to measure the transcription levels of TaZEP gene (a), dry weight (b), ABA contents in both root (c) and leaf (d) tissues, total nitrogen contents in both root (e) and leaf (f) tissues. The transcription levels of TaZEP gene were to identify the total expression profile of all three homologues. BSMV-γ:00 vector-inoculated wheat seedlings were used as positive control. Each value is mean ± standard deviation (SD) of three biological replicates, and different letters indicate significant differences (P < 0.05) relative to control by using two-way ANOVA followed by Duncan’s multiple range test
Discussion
A large number of identified N deficiency-responsive protein species helped further reveal the adaptive mechanism of N deficiency in both root and leaf tissues of bread wheat
In this study, we identified 866 and 649 N deficiency-responsive protein species in the root and leaf tissues, respectively (Supplementary Table S5). Numbers of the identified N deficiency-responsive protein species in this study were far more than those (≤ 65) in the above previous studies (Møller et al. 2011; Deng et al. 2014; Meise et al. 2017). The differences in the number of N deficiency-responsive protein species identified from our study and those of previous studies could be attributed to proteomic technologies, experimental designs, the divergences of genome sequences between such distantly related species, and the different statistical cut-off and fold-change thresholds. Thus, our work provided more molecular information on important crops responses to N deficiency. In the present study, however, a large proportion (359/1515, 23.7%) of unknown function protein species were not annotated (Fig. 2, Supplementary Table S5), possibly because genome of bread wheat is huge and complex, and this has resulted in slower research progress compared with homologous species (rice, maize, etc) of relatively simple genomes (Brenchley et al. 2012).
There were some differences in the categories of N deficiency-responsive protein species identified between the root and leaf tissues of N-deficient wheat seedlings (Supplementary Table S6), suggesting that they may have similar biological functions in the response to N deficiency in the two tissues. However, most (1360/1515, 89.8%) of the protein species identified appeared specifically between the root and leaf tissues (Supplementary Table S6), implying that root and leaf tissues have differential N deficiency-responsive mechanisms. The differences in the protein species identified between the root and leaf tissues might be related to their different functions, growth environments, and sensitivities to long-term N deficiency. The different proteome profiles for root and leaf tissues further suggest that long-term N deficiency may have profound and varying effects on growth in the root and leaf tissues of wheat seedlings.
Comparison of proteome profiles in this study with transcriptome profiles in previous studies in plant responses to N deficiency
Our leaf proteome profile of N-deficient bread wheat seedlings was compared with the barley leaf transcriptome profile of N-deficient barley seedlings (Wei et al. 2016), because the plant materials (bread wheat and barley) used in these two studies were close relatives. Moreover, there were many similarities between the two studies, such as similar sampling tissues (leaves) and similar environment conditions (N-deficient solutions). We found that the 176 (176/528, 33.3%) function-known leaf protein species in our study were commonly identified in the leaf transcriptome analysis of N-deficient barley seedlings (Wei et al. 2016). These common N deficiency-responsive protein species (genes) were involved in carbohydrate metabolism, protein metabolism, stress and defense, and photosynthesis (Supplementary Table S7). However, most (352/528, 66.7%) of the N deficiency-responsive protein species in our results were not identified in the leaf transcriptome profiles of N-deficient barley seedlings (Supplementary Table S7) (Wei et al. 2016), further suggesting that these protein levels rarely correlated with mRNA expression levels because of posttranscriptional, translational, posttranslational modifications, as well as differential mRNA and protein degradation rates (Torabi et al. 2009; Vogel and Marcotte 2012; Hu et al. 2013). Therefore, our proteome identified some N deficiency-responsive protein species and enhanced our further understanding of plant responses to N deficiency.
Roles of phytohormone ABA in the N-deficient wheat seedlings
Most stress and defense-, signal transduction-, photosynthesis-, carbohydrate metabolism-, protein metabolism-, nucleotide metabolism-, and other metabolism-related genes have been identified, and their functions have also been discussed in previous N-transcriptomic and proteomics results (Gelli et al. 2014; Yang et al. 2015; He et al. 2016a; Quan et al. 2016; Wei et al. 2016; Curci et al. 2017; Nawaz et al. 2018). Thus, the protein species involved in the functional categories above and identified in this study were not discussed further. However, phytohormones are very important elements of signaling pathways in plants, and they regulate most developmental and physiological processes (Lee and Luan 2012; Chao et al. 2016). In addition, data on the protein species that regulate phytohormones are scarce (Møller et al. 2011; Deng et al. 2014; Meise et al. 2017), possibly because of the low abundance of hormone-related enzymes.
In the present study, a large number (35) of protein species involved in synthesis of phytohormones ABA, JA, ETH, IAA, CTK and SA were first identified in both root and leaf tissues of wheat seedlings (Supplementary Table S5), and their contents were greatly changed in these two tissues (Fig. 3). These implied that they could be involved in the response to N deficiency in wheat seedlings. However, there were some differences in the changes in the contents of these plant hormones between the root and leaf tissues (Fig. 3), partly accounting for their differential roles or the caused differences in growth parameters (e.g., root and shoot fresh and dry weights) in these two tissues of N-deficient wheat seedlings (Zhang et al. 2019). At 8 d after N deficiency, the contents of ABA in both root and leaf tissues were far higher than those of other phytohormones (Fig. 3a-c), similar to the altered abundance of the hormone synthesis-related protein species identified (Supplementary Table S5). Thus, we speculated that ABA could play an important role in wheat seedlings in response to N deficiency.
Bread wheat (Triticum aestivum L.) has a very large, 17-gigabase-pair (Gb), allohexaploid genome (2n = 6x = 42, AABBDD; 2n is the number of chromosomes in each somatic cell, and 6x is the basic chromosome); therefore, each wheat gene potentially exists as a trio of A, B, and D homoeologs (Feldman et al. 2012). This species may be the last cereal to be genetically transformed because the efficiency of the two main transformation methods (particle bombardment and cocultivation with Agrobacterium tumefaciens) in this species is very low due to cultivar specificity, multiple-copy insertions, time consumption, low transformation efficiency, etc. (Wu et al. 2003; Zhao et al. 2006). BSMV-VIGS mainly exploits the BSMV-mediated antiviral defense mechanism of plants to explore the function of endogenous genes based on homology-dependent gene silencing because it is not required for plant transformation, facilitates rapid generation of gene knockdown phenotypes, offers a fast and rapid transient assay for silencing of gene expression, and has been developed as more successful genetics approach to assess gene functions than simple constitutive transgene in polyploid plant species (Cao et al. 2011; Tufan et al. 2011). Although results from BSMV-VIGS are not comparable to results from stable transformation experiments (e.g., RNAi or CRISPR/Cas9), BSMV-VIGS findings are useful for increasing our understanding of gene function and may lead to the development of a strategy that is more successful than simple constitutive transgene overexpression (Borisjuk et al. 2019).
ZEP is one key enzyme of ABA synthesis in higher plants (Cuming and Stevenson 2015), and its abundance was greatly regulated in leaf tissue of wheat seedlings suffering from N deficiency (Supplementary Table S5). To verify the above speculation, in this study, the function of ABA in wheat seedlings response to N deficiency was determined by using BSMV-VIGS-mediated ZEP silence (Supplementary Fig. S1). After 8 d of N deficiency, BSMV-TaZEP-inoculated wheat seedlings displayed the inhibited growth (Fig. 4), and the dry weights, ABA and total nitrogen contents in both root and leaf tissues of BSMV-VIGS-TaZEP silenced wheat seedlings were markedly reduced (Fig. 5). These results suggested that silencing the gene encoding TaZEP enzyme resulted in a decrease in the tolerance to N deficiency in wheat seedlings, suggesting a role of ABA in response to N deficiency.
Based on our experimental data, we proposed a putative ABA-dependent schematic model in higher plants under N deficiency (Fig. 6). In this pathway, plant cells could perceive external N deficiency in cell membrane, and N deficiency signaling is subsequently transduced by Ca2+ sensors, kinases, or other signaling components, which then induce translational changes in many functional protein species related to phytohormones (e.g., ABA). ABA and other phytohormones then regulate the downstream transcriptional, translational, and posttranslational responses, and, finally, plants exert many morphological and physiological adaptive changes that assist survival under N-deficiency stress.
Conclusions
Based on large-scale proteomics, in this study, we identified 1515 N deficiency-responsive protein species with significantly altered abundance in wheat seedlings, and these species were functionally related to diverse biological processes. Of the identified protein species, the abundance of an important ABA synthesis-related enzyme (TaZEP) was significantly enhanced, and ABA contents were also increased, suggesting the possible role of ABA in response to N deficiency. Silencing of TaZEP gene markedly decreased the tolerance of wheat seedlings to N deficiency. These results suggest a role for ABA in wheat response to N deficiency. Extensive further investigations are merited to integrate ABA and other signaling regulatory components into a comprehensive signaling network, which would greatly increase our understanding of plant responses to N-deficiency stress.
Abbreviations
- ABA:
-
abscisic acid
- BSMV-VIGS:
-
barley stripe mosaic virus-virus induced gene-silencing
- CTK:
-
cytokinin
- DW:
-
dry weight
- ETH:
-
ethylene
- FW:
-
fresh weight
- IAA:
-
auxin
- iTRAQ:
-
isobaric tagging for relative and absolute quantification
- JA:
-
jasmonic acid
- MW:
-
molecular weights
- pI:
-
isoelectric points
- qRT-PCR:
-
quantitative real-time PCR
- SA:
-
salicylic acid
- ZEP:
-
zeaxanthin epoxidase
References
Agrawal GK, Yamazaki M, Kobayashi M, Hirochika R, Miyao A, Hirochika H (2001) Screening of the rice viviparous mutants generated by endogenous retrotransposon Tos17 insertion. Tagging of a zeaxanthin epoxidase gene and a novel OsTATC gene. Plant Physiol 125:1248–1257
Borisjuk N, Kishchenko O, Eliby S, Schramm C, Anderson P, Jatayev S, Kurishbayev A, Shavrukov Y (2019) Genetic modification for wheat improvement: from transgenesis to genome editing. Biomed Res Int 2019:6216304
Brenchley R, Spannagl M, Pfeifer M, Barker GL, D'Amore R, Allen AM et al (2012) Analysis of the bread wheat genome using whole–genome shotgun sequencing. Nature 491:705–710
Cai H, Lu Y, Xie W, Zhu T, Lian X (2012) Transcriptome response to nitrogen starvation in rice. J Biosci 37:731–747
Cao A, Xing L, Wang X, Yang X, Wang W, Sun Y, Qian C, Ni J, Chen Y, Liu D, Wang X, Chen P (2011) Serine/threonine kinase gene Stpk-V, a key member of powdery mildew resistance gene Pm21, confers powdery mildew resistance in wheat. Proc Natl Acad Sci U S A 108:7727–7732
Chao WS, Doğramaci M, Horvath DP, Anderson JV, Foley ME (2016) Phytohormone balance and stress-related cellular responses are involved in the transition from bud to shoot growth in leafy spurge. BMC Plant Biol 16:47
Criado MV, Roberts IN, Echeverria M, Barneix AJ (2007) Plant growth regulators and induction of leaf senescence in nitrogen–deprived wheat plants. J Plant Growth Regul 26:301–307
Cuming AC, Stevenson SR (2015) From pond slime to rain forest: the evolution of ABA signalling and the acquisition of dehydration tolerance. New Phytol 206:5–7
Curci PL, Cigliano RA, Zuluaga DL, Janni M, Sanseverino W, Sonnante G (2017) Transcriptomic response of durum wheat to nitrogen starvation. Sci Rep-UK 7:1176
Deng G, Liu L, Zhong X, Lao C, Wang H, Wang B, Zhu C, Shah F, Peng D (2014) Comparative proteome analysis of the response of ramie under N, P and K deficiency. Planta 239:1175–1186
Elberse IAM, van Damme JMM, Tienderen PHV (2003) Plasticity of growth characteristics in wild barley (Hordeum spontaneum) in response to nutrient limitation. J Ecol 91:371–382
Feldman M, Levy AA, Fahima T, Korol A (2012) Genomic asymmetry in allopolyploid plants: wheat as a model. J Exp Bot 63:5045–5059
Findlay GD, MacCoss MJ, Swanson WJ (2009) Proteomic discovery of previously unannotated, rapidly evolving seminal fluid genes in Drosophila. Genome Res 19:886–896
Gelli M, Duo Y, Konda AR, Zhang C, Holding D, Dweikat I (2014) Identification of differentially expressed genes between sorghum genotypes with contrasting nitrogen stress tolerance by genome-wide transcriptional profiling. BMC Genomics 15:60–66
Grobei MA, Qeli E, Brunner E, Rehrauer H, Zhang R, Roschitzki B, Basler K, Ahrens CH, Grossniklaus U (2009) Deterministic protein inference for shotgun proteomics data provides new insights into Arabidopsis pollen development and function. Genome Res 19:1786–1800
Grün A, Buchner P, Broadley MR, Hawkesford MJ (2018) Identification and expression profiling of Pht1 phosphate transporters in wheat in controlled environments and in the field. Plant Biol 20:374–389
Guo T, Xuan H, Yang Y, Wang L, Wei L, Wang Y, Kang G (2014) Transcription analysis of genes encoding the wheat root transporter NRT1 and NRT2 families during nitrogen starvation. J Plant Growth Regul 33:837–848
Hakeem KR, Ahmad A, Iqbal M, Gucel S, Ozturk M (2011) Nitrogen-efficient rice cultivars can reduce nitrate pollution. Environ Sci Pollut Res 18:1184–1193
Han YL, Liao JY, Yu Y, Song HX, Rong N, Guan CY, Lepo JE, Ismail AM, Zhang ZH (2017) Exogenous abscisic acid promotes the nitrogen use efficiency of Brassica napus by increasing nitrogen remobilization in the leaves. J Plant Nutr 40:2540–2549
He M, Zhu C, Dong K, Zhang T, Cheng Z, Li J, Yan Y (2015) Comparative proteome analysis of embryo and endosperm reveals central differential expression proteins involved in wheat seed germination. BMC Plant Biol 15:1–17
He X, Ma H, Zhao X, Nie S, Li Y, Zhang Z et al (2016a) Comparative RNA-seq analysis reveals that regulatory network of maize root development controls the expression of genes in response to N stress. PLoS One 11:3
He L, Zhang H, Zhang Y, Song X, Feng W, Kang G, Wang C, Guo T (2016b) Estimating canopy leaf nitrogen concentration in winter wheat based on multi-angular hyperspectral remote sensing. Eur J Agron 73:170–185
Hoagland DR, Arnon DI (1950) The water-culture method for growing plants without soil. Calif Agric Exp Stn Circ 347:1–32
Hu G, Jin K, Yoo MJ, Grupp K, Chen S, Wendel JF (2013) Proteomic profiling of developing cotton fibers from wild and domesticated Gossypium barbadense. New Phytol 200:570–582
Hu J, Rampitsch C, Bykova NV (2015) Advances in plant proteomics toward improvement of crop productivity and stress resistance. Front Plant Sci 6:209
International Wheat Genome Sequencing Consortium (IWGSC) (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345:1251788
International Wheat Genome Sequencing Consortium (IWGSC) (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:661
Jia J, Zhao S, Kong X, Li Y, Zhao G, He W et al (2013) Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 496:91–95
Jin X, Li W, Hu D, Shi X, Zhang X, Zhang F, Fu Z, Ding D, Liu Z, Tang J (2015) Biological responses and proteomic changes in maize seedlings under nitrogen deficiency. Plant Mol Biol Rep 33:490–504
Kang G, Li G, Wang L, Wei L, Yang Y, Wang P, Yang Y, Wang Y, Feng W, Wang C, Guo T (2015) Hg–responsive proteins identified in wheat seedlings using iTRAQ analysis and the role of ABA in Hg stress. J Proteome Res 14:249–267
Kosová K, Vítámvás P, Prášil IT, Renaut J (2011) Plant proteome changes under abiotic stress–contribution of proteomics studies to understanding plant stress response. J Proteome 74:1301–1322
Kosová K, Vítámvás P, Urban MO, Prášil IT, Renaut J (2018) Plant abiotic stress proteomics: the major factors determining alterations in cellular proteome. Front Plant Sci 9:122
Krapp A, Berthomé R, Orsel M, Mercey-Boutet S, Yu A, Castaings L, Elftieh S, Major H, Renou JP, Daniel-Vedele F (2011) Arabidopsis roots and shoots show distinct temporal adaptation patterns toward nitrogen starvation. Plant Physiol 157:1255–1282
Kuang Q, Zhang S, Wu P, Chen Y, Li M, Jiang H, Wu G (2017) Global gene expression analysis of the response of physic nut (Jatropha curcas L.) to medium- and long-term nitrogen deficiency. PLoS One 12:e0182700
Lee SC, Luan S (2012) ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ 35:53–60
Li G, Peng X, Xuan H, Wei L, Yang Y, Guo T, Kang G (2013) Proteomic analysis of leaves and roots of common wheat (Triticum aestivum L.) under copper-stress conditions. J Proteome Res 12:4846–4861
Li A, Liu D, Wu J, Zhao X, Hao M, Geng S, Yan J, Jiang X, Zhang L, Wu J, Yin L, Zhang R, Wu L, Zheng Y, Mao L (2014) mRNA and small RNA transcriptomes reveal insights into dynamic homoeolog regulation of allopolyploid heterosis in nascent hexaploid wheat. Plant Cell 26:1878–1900
Li G, Wu Y, Liu G, Xiao X, Wang P, Gao T, Xu M, Han Q, Wang Y, Guo T, Kang G (2017) Large-scale proteomics combined with transgenic experiments demonstrates an important role of jasmonic acid in potassium deficiency response in wheat and rice. Mol Cell Proteomics 16:1889–1905
Liang C, Tian J, Liao H (2013) Proteomics dissection of plant responses to mineral nutrient deficiency. Proteomics 13:624–636
Ling HQ, Zhao S, Liu D, Wan J, Sun H, Zhang C et al (2013) Draft genome of the wheat A-genome progenitor Triticum urartu. Nature 496:87–90
Liu G, Wu Y, Xu M, Gao T, Wang P, Wang L, Guo T, Kang G (2016) Virus-induced gene silencing identifies an important role of the TaRSR1 transcription factor in starch synthesis in bread wheat. Int J Mol Sci 17:1557
Ma TL, Wu WH, Wang Y (2012) Transcriptome analysis of rice root responses to potassium deficiency. BMC Plant Biol 12:161
Ma C, Zhou J, Chen G, Bian Y, Lv D, Li X, Wang Z, Yan Y (2014) iTRAQ-based quantitative proteome and phosphoprotein characterization reveals the central metabolism changes involved in wheat grain development. BMC Genomics 15:1–20
Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y (2008) RNA-Seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18:1509–1517
Meise P, Jozefowicz AM, Uptmoor R, Mock HP, Ordon F, Schum A (2017) Comparative shoot proteome analysis of two potato (Solanum tuberosum L.) genotypes contrasting in nitrogen deficiency responses in vitro. J Proteome 166:68–82
Møller ALB, Pedas P, Andersen B, Svensson B, Schjoerring JK, Finnie C (2011) Responses of barley root and shoot proteomes to long-term nitrogen deficiency, short-term nitrogen starvation and ammonium. Plant Cell Environ 34:2024–2037
Nawaz MA, Chen C, Shireen F, Zheng Z, Sohail H, Afzal M, Ali MA, Bie Z, Huang Y (2018) Genome-wide expression profiling of leaves and roots of watermelon in response to low nitrogen. BMC Genomics 19:456
Nazir M, Pandey R, Siddiqi TO, Ibrahim MM, Qureshi MI, Abraham G, Vengavasi K, Ahamad A (2015) Nitrogen-deficiency stress induces protein expression differentially in low-N tolerant and low-N sensitive maize genotypes. Front Plant Sci 7:298
Oka M, Shimoda Y, Sato N, Inoue J, Yamazaki T, Shimomura N, Fujiyama H (2012) Abscisic acid substantially inhibits senescence of cucumber plants (Cucumis sativus) grown under low nitrogen conditions. J Plant Physiol 169:789–796
Qin L, Walk TC, Han P, Chen L, Zhang S, Li Y, Hu X, Xie L, Yang Y, Liu J, Lu X, Yu C, Tian J, Shaff JE, Kochian LV, Liao X, Liao H (2019) Adaption of roots to nitrogen deficiency revealed by 3D quantification and proteomic analysis. Plant Physiol 179:329–347
Quan X, Zeng J, Ye L, Chen G, Han Z, Munawar J, Zhang G (2016) Transcriptome profiling analysis for two Tibetan wild barley genotypes in responses to low nitrogen. BMC Plant Biol 16:30
Ristova D, Carré C, Pervent M, Medici A, Kim GJ, Scalia D et al (2016) Combinatorial interaction network of transcriptomic and phenotypic responses to nitrogen and hormones in the Arabidopsis thaliana root. Sci Signal 9:451
Schlüter H, Apweiler R, Holzhütter HG, Jungblut PR (2009) Finding one's way in proteomics: a protein species nomenclature. Chem Cent J 3:11
Scofield SR, Huang L, Brandt AS, Gill BS (2005) Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiol 138:2165–2173
Shi W, Xu W, Li S, Zhao X, Dong G (2010) Responses of two rice cultivars differing in seedling–stage nitrogen use efficiency to growth under low–nitrogen conditions. Plant Soil 326:291–302
Song J, Jiang L, Jameso PE (2012) Co-ordinate regulation of cytokinin gene family members during flag leaf and reproductive development in wheat. BMC Plant Biol 12:78
Torabi S, Wissuwa M, Heidari M, Naghavi MR, Gilany K, Hajirezaei MR, Omidi M, Yazdi-Samadi B, Ismail AM, Salekdeh GH (2009) A comparative proteome approach to decipher the mechanism of rice adaptation to phosphorus deficiency. Proteomics 9:159–170
Tufan HA, Stefanato FL, McGrann GRD, MacCormack R, Boyd LA (2011) The barley stripe mosaic virus system used for virus-induced gene silencing in cereals differentially affects susceptibility to fungal pathogens in wheat. J Plant Physiol 168:990–994
Vogel C, Marcotte EM (2012) Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet 13:227–232
Wang X, Bian Y, Cheng K, Zou H, Sun S, He J (2012) A comprehensive differential proteomic study of nitrate deprivation in Arabidopsis reveals complex regulatory networks of plant nitrogen responses. J Proteome Res 11:2301–2315
Wei Z, Zeng X, Qin C, Wang Y, Bai L, Xu Q et al (2016) Comparative transcriptome analysis revealed genes commonly responsive to varied nitrate stress in leaves of tibetan hulless barley. Front Plant Sci 7:298
Wisniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6:359–362
Wu H, Sparks C, Amoah B, Jones HD (2003) Factors influencing successful Agrobacterium-mediated genetic transformation of wheat. Plant Cell Rep 21:659–668
Yang W, Yoon J, Choi H, Fan Y, Chen R, An G (2015) Transcriptome analysis of nitrogen-starvation responsive genes in rice. BMC Plant Biol 15:31
Zhang N, Huo W, Zhang L, Chen F, Cui D (2016) Identification of winter-responsive proteins in bread wheat using proteomics analysis and virus-induced gene silencing. Mol Cell Proteomics 15:2954–2969
Zhang Y, Zhou Y, Chen S, Liu J, Fan K, Li Z, Liu Z, Lin W (2019) Gibberellins play dual roles in response to phosphate starvation of tomato seedlings, negatively in shoots but positively in roots. J Plant Physiol 234–245:145–153
Zhao T, Zhao S, Chen H, Zhao Q, Hu Z, Hou B, Xia G (2006) Transgenic wheat frogeny resistant to powdery mildew generated by Agrobacterium inoculumto the basal portion of wheat seedling. Plant Cell Rep 25:1199–1204
Acknowledgements
This study was financially supported by the Science and Technology Innovation Program for Increase in Yield and Efficiency of Food Crop (2016YFD0300105) and the National Key Technology Support Program (2015BAD26B01).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Ad C. Borstlap.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Table S1
(DOC 33 kb)
Supplementary Table S2
(XLS 3280 kb)
Supplementary Table S3
(XLS 3016 kb)
Supplementary Table S4
(XLS 1956 kb)
Supplementary Table S5
(XLS 535 kb)
Supplementary Table S6
(XLS 90 kb)
Supplementary Table S7
(DOC 32 kb)
Supplementary Fig. S1
(DOC 41 kb)
Supplementary Fig. S2
(DOC 409 kb)
Rights and permissions
About this article
Cite this article
Kang, G., Wu, Y., Li, G. et al. Proteomics combined with BSMV-VIGS methods identified some N deficiency-responsive protein species and ABA role in wheat seedling. Plant Soil 444, 177–191 (2019). https://doi.org/10.1007/s11104-019-04260-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-019-04260-1