Abstract
Background and Aims
Modelling organic matter decomposition is fundamental to understand biogeochemical cycling in terrestrial ecosystems. Current models use C/N or Lignin/N ratios to describe susceptibility to decomposition, or implement separate C pools decaying with different rates, disregarding biomolecular transformations and interactions and their effect on decomposition dynamics. We present a new process-based model of decomposition including a description of biomolecular dynamics obtained by 13C-CPMAS NMR spectroscopy.
Methods
Baseline decay rates for relevant molecular classes and intermolecular protection were calibrated by best fitting of experimental data from leaves of 20 plant species decomposing for 180 days in controlled optimal conditions. The model was validated against field data from leaves of 32 plant species decomposing for 1-year at four sites in Mediterranean ecosystems.
Results
Simulations correctly reproduced mass loss data and variations of selected molecular classes both in controlled conditions and in the field, for a wide range of plant molecular composition and environmental conditions.
Conclusions
Our innovative approach accurately predicted decomposition of a wide range of litters across different climates. Prediction accuracy emerged from the species-specific partitioning of molecular types and from the representation of intermolecular interactions. Further application should be planned in other ecosystems based on long-term decomposition datasets.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Plant litter decomposition is a main source of nutrients and organic compounds that sustain plant productivity, contribute to soil organic matter formation, and affect most biogeochemical cycles (Kögel-Knabner 2002; De Deyn et al. 2008; Cotrufo et al. 2013). The accumulation of soil organic carbon and nutrients stocks depends on decomposition rates and progressive preservation of hydrophobic biomolecules (Attiwill and Adams 1993; Piccolo 1996, 2002). The major determinants of decomposition rates include climatic variables and plant molecular properties, which interact and selectively prevail across different spatial scales (Liski et al. 2003; Bradford et al. 2016). At global and regional scales litter decay rates are most controlled by temperature and water availability (Aerts 1997). At the local scale, where climate is relatively uniform, the molecular composition of plant residues determines its susceptibility to microbial attack (Meentemeyer 1978; Rovira and Vallejo 2007), which therefore controls its decomposition dynamics. The molecular quality of decaying materials is a function of the availability of organic substrates to soil biota, and usually corresponds to a prevalence of polar biomolecules (Piccolo 2002; Rovira and Vallejo 2007).
However, a definition of litter quality based on molecular composition (Swift et al. 1979) is operationally difficult. Indeed, plant residues contain multiple types of biomolecules with different aqueous solubility, and, hence, different susceptibility to microbial decomposition (e.g. peptides, carbohydrates, lipids, lignin, organic acids, and polyphenols), whose relative fractions vary with decay stage (Berg and McClaugherty 2013). Differential plant residue degradation (a function of molecular composition) ultimately determines the nature of organic matter in soil, which is a heterogeneous association of both plant biomolecules that remain unaltered or only partially transformed and metabolites produced by microbial activity (Piccolo 2002; Nebbioso and Piccolo 2011; Nebbioso et al. 2015). Nevertheless, a substantial effort has been made during the last decades to find effective indicators of plant molecular quality that provide reliable predictions of decomposition rates (Tian et al. 1995; Cornelissen and Thompson 1997; Cornwell et al. 2008). Traditional approaches have been based on correlating simple litter characteristics with decay rates for predictive purposes (Meentemeyer 1978; Melillo et al. 1982; Coq et al. 2010). For example, lignin content, the most abundant plant biopolymer after cellulose, is often negatively correlated with decay rate, especially when root and woody plant tissues are included in analysed samples (Taylor et al. 1989), but not in the case of litter photodegradation in dryland systems (Austin and Vivanco 2006; Baker and Allison 2015). Consistent negative correlations with litter decay rates have been also reported for lignin to nitrogen (Lignin/N) (Melillo et al. 1982) and carbon to nitrogen (C/N) ratios (Taylor et al. 1989), with the latter being more predictive of decay rates in case of substrates with limited lignin content, such as understory herbs (Taylor et al. 1989).
Lignin/N and C/N ratios are extensively used in C-cycle models, as descriptors of organic matter quality to control mass loss rate (Smith et al. 1997; Del Grosso et al. 2001; Burke et al. 2003; Shibu et al. 2006). The main examples are biogeochemical models simulating carbon dynamics and other ecosystem processes, such as CENTURY (Parton et al. 1994), ROTHAMSTED (RothC) (Coleman and Jenkinson 1996), LPJ (Sitch et al. 2003), and the Biome- (Hunt et al. 1996) and Forest- (Running and Gower 1991) BGC (Bio Geochemical Cycles) models. Other models, specifically designed for application to agro-ecosystems, include DNDC (Li et al. 2003), DayCent (Del Grosso et al. 2001) and CANDY (Franko et al. 1995).
Although widely used, research has found that simple indicators such as Lignin/N or C/N are not always reliable indicators of decomposition rates. For instance, Berg and McClaugherty (2013) suggested that the use of C/N ratio to predict decay rate throughout the decomposition process should be avoided, because, irrespectively of its initial value, it progressively decreases as C is lost trough respiration, while N is immobilized in the microbial biomass (Bonanomi et al. 2010). More recently, Hättenschwiler et al. (2011) revisited the commonly held view that N and lignin control the rate of plant litter decomposition, and indicated that, at least in tropical ecosystems, non-lignin plant carbon molecules at low concentration play the major role. In line with this, a recent study based on a controlled experiment of the decomposition of 64 litter types, showed a weak association between C/N ratio and decay rates (Bonanomi et al. 2013). These results showed a negative and positive relation for undecomposed and half-way decomposed materials, respectively, and a not significant association for more decomposed organic matter. Moreover, the litter molecular composition is known to progressively change as a function of the relative susceptibility of biomolecules to breakdown, with rapid mineralization of labile sugars, and selective preservation of less degradable lipids, lignin, and polyphenols (Davidson and Janssens 2006; Rovira and Vallejo 2007). Berg and Matzner (1997) attempted to capture such compositional changes in a three-phase model, whereby labile biomolecules and mineralized nutrients (i.e. N and P) controlled decay rate up to 30–40 % of mass loss, while lignin became progressively more important from then onwards. The model has been successfully tested on a range of biomasses including hardwoods and coniferous plants (Berg and McClaugherty 2013).
The mass partitioning of plant cellular material into different pools broadly corresponding to structural classes of biomolecules was attempted in further modelling descriptions (Gillon et al. 1994; Liski et al. 2005; Adair et al. 2008; Incerti et al. 2011). These models corresponded to an exponential decay following a first-order kinetics (Olson 1963) and were proved to largely fit experimental decomposition datasets. However, they were criticized as implicitly based on wrong assumptions, including the independent decomposition of the biomolecular pools without considering the interactions among them and their progressive structural transformation with time (Rovira and Rovira 2010). Indeed, in some models the C pools do interact, as in the case of the model by Adair et al. (2008), where the lignin/(cellulose + lignin) ratio alters the decomposition rate of the cellulosic pool, but such effect does not change over time. Hence, to amend these shortcomings, single-pool models based on decay rates that change over time were proposed (Yang and Janssen 2000; Rovira and Rovira 2010). However, these models did not yet explicitly represented the structural changes of organic matter during decomposition. In fact, the net balance of all biological and chemical degradation processes are simply implemented as a variation of decay rate with time, on the basis of different dataset-specific curves, whose parameters only allow an a posteriori interpretation of the involved processes. In this framework, a more complex model, that explicitly represents the transformation of plant biomolecules, their mutual interactions and their effect on organic matter decay rate, has not yet been proposed. This is particularly relevant, considering that, as the altered organic compounds enter the soil, a more complex system develops due to interactions with the soil mineral fraction (Piccolo 1996, 2002). Hence, a reliable implementation of a model of litter decomposition dynamics should be based not only on explicit and reliable descriptors of organic matter molecular composition, but should also include its interactions with the soil mineral constituents.
In the last decade chemical throughput methods as pyrolysis-gas chromatography/mass spectrometry (Huang et al. 1998; Spaccini and Piccolo 2007), near infrared reflectance spectroscopy (Gillon et al. 1999) and nuclear magnetic resonance (NMR) spectroscopy, either in the liquid- (Piccolo et al. 1990) or the solid-state (Conte et al. 2004) or in combination with chemometry (Smejkalova et al. 2008) have been applied to characterize organic matter at molecular level. In particular, solid-state 13C-CPMAS NMR spectra were found useful to provide an overview of the total organic chemical composition of complex matrices in soil organic matter (Conte et al. 2004; Piccolo et al. 2005). 13C-CPMAS NMR spectra were found suitable to highlight plant molecular changes related to long-term decomposition (Preston et al. 2009), phytotoxicity (Bonanomi et al. 2011) and autotoxicity (Mazzoleni et al. 2015), as well as organic matter properties in litter mixtures (Bonanomi et al. 2014). In particular, Bonanomi et al. (2013) reported a negative association between litter decay rate and 13C-CPMAS-NMR spectral regions corresponding to alkyl, methoxyl and N-alkyl C, and a positive correlation for O-alkyl and di-O-alkyl C regions. Moreover, restricted 13C NMR regions were significantly associated with decay rate, and used to implement a multiregressive equation that significantly fitted experimental decomposition data (Bonanomi et al. 2013).
Following the conceptual frame introduced by Mazzoleni et al. (2013), the aim of this work was thus to overcome the existing lack of prediction models that include molecular changes during decomposition processes. The underlying hypothesis is that a correct and reliable prediction of decomposition dynamics requires detailed information on organic matter biomolecular content obtained by 13C-CPMAS NMR spectroscopy. Hence, we hereby present OMDY (Organic Matter DYnamics), a new process-based model of decomposition dynamics, developed with a system dynamic approach, calibrated by experimental data in controlled conditions, and validated by assessing its predictivity against original field decomposition data in four different Mediterranean sites.
Materials and methods
Plant litter decomposition experiments in controlled condition
The experiment was planned to estimate the value of maximum potential decomposition rates of leaves of Mediterranean plant species, as a function of molecular content (i.e. the maximum potential decay rate for each initial model layer), as well as to calibrate the model parameters describing the process of intermolecular protection. Experimental design and set-up followed a previous litterbag decomposition experiment focused on the effects of molecular composition, as assessed by 13C CPMAS NMR, on plant decay rate (Bonanomi et al. 2013). The experiment was conducted under controlled conditions, in order not to limit decay rates by temperature and water availability (Gholz et al. 2000).
A set of 20 plant species widespread in the Mediterranean environments and representing a wide range of molecular composition (Lecerf et al. 2007) were selected from natural plant communities of different vegetation types located in the Campania Region, Southern Italy. The species pool included four perennial (Acanthus mollis L., Ampelodesmos mauritanicus (Poir.) Dur. & Schinz, Festuca drymeia M. et K., Medicago sativa L.) and one annual (Lepidium sativum L.) herbaceous plant, two evergreen shrubs (Arbutus unedo L., Coronilla emerus L.), one vine (Hedera helix L.), four evergreen trees (Cupressus sempervirens L., Picea abies (L.) H. Karst., Pinus halepensis Mill., Quercus ilex L.), and eight deciduous trees (Alnus cordata (Loisel.) Desf., Castanea sativa Mill., Fagus sylvatica L., Fraxinus ornus L., Populus nigra L., Quercus pubescens Willd., Robinia pseudoacacia L., Salix alba L.). For each species, more than 20 individuals were randomly selected at the sampling sites, and freshly abscised leaves were collected by placing nets under the plants, dried (30 °C in a ventilated chamber until constant weight was reached) and then stored at room temperature.
Litterbag experiments (Berg and McClaugherty 2013) were carried out in microcosms placed in a growth chamber under controlled constant temperature (25 ± 2 °C) and water (watered every two days to field capacity with distilled water). Terylene litterbags (20 × 20 cm2, mesh size 2 mm) were filled with 6 g of dry leaf litter and placed inside trays (30 cm deep, 100 cm for each side). A microbial inoculum was obtained by mixing 10 g of topsoil (10 cm), collected from the sampling site, with 90 g of distilled water, sprayed over the litterbags in order to improve the start up of the decomposition process. A total of 480 litterbags (20 species × 3 sampling dates × 8 replicates) were harvested after 30, 90 and 180 days of decomposition. Bags were dried at the laboratory (40 °C until constant weight was reached) and the remaining material weighed afterwards.
Field experiments of litter decomposition
A 1-year long multi-site plant leaf litter decomposition experiment was carried out in Campania Region (Southern Italy) in order to validate the model. The field experiments were conducted on soils at four sites representing some of the major ecosystem types within the Mediterranean biome (Table 1 and Supplementary Table S1).
The two Portici sites are located on the south-eastern slope of Monte Vesuvius. The first site is a mixed forest dominated by the evergreen oak Quercus ilex with presence of deciduous trees as Fraxinus ornus L., Celtis australis L., Alnus cordata (Loisel.) Desf., Robinia pseudoacacia L., and the vines Hedera helix L. and Rubus ulmifolius Schott in the understory. The second Portici site located is an early-successional stage of an old field site dominated by the grass Dactylis glomerata L. Other common species included the annual grass Avena spp., the forbs Plantago lanceolata L., Taraxacum officinale Weber ex F.H.Wigg., Oxalis pes-caprae L., and scattered trees including the native Quercus pubescens Willd. and Ulmus minor Mill. and the invasive Ailanthus altissima (Mill.) Swingle and Broussonetia papyrifera (L.) L’Hér. ex Vent.
The Paestum site is located in a Nature Reserve on a flat coastal area with alluvial deposit sand dunes composed of loose siliceous-calcareous marine sand. Vegetation is a mosaic of different plant patches of low Mediterranean maquis, i.e. the vegetation dominated by sclerophyll evergreen shrubs typical of the Mediterranean basin. The dominant species are the shrubs Myrtus communis L., Pistacia lentiscus L., Juniperus phoenicea L., Phylliera latifolia L., and the trees Pinus halepensis Mill. and the invasive Eucalyptus camaldulensis Dehnh. and Acacia longifolia (Andr.) Willd.
Finally, the Cicerale site is a riparian forest along the Alento river dominated by the native trees Populus nigra L. and Salix alba L., where the invasive, nitrogen-fixing tree Amorpha fruticosa L. also occurs. All sites have a Mediterranean climate with a relatively high mean annual rainfall well distributed in winter, spring and fall and a moderate dry summer (Table 1, Supplementary Fig. S1), and are characterized by soil properties reported in Supplementary Table S2.
At each site plant leaves were collected by net traps during the period of maximum leaf fall, from more than 20 randomly selected individuals for each species. Freshly abscissed leaves were first air dried (30 °C in a ventilated chamber until constant weight was reached) and then stored at room temperature. In September 2012, terylene litterbags (20 × 20 cm2, mesh size 2 mm) were filled with 6 g of dry leaf organic matter and incubated at different randomly-selected locations on the soil surface by using metal pegs.
Litterbags were harvested after 30, 90, 180 and 360 days of decomposition. A total of 1024 litterbags were harvested (32 species × 4 sampling dates × 8 replicates), distributed among the sampling sites according to litter species (Table 1, Supplementary Table S1). Bags were dried (30 °C until constant weight was reached) and the remaining material weighed.
Assessment of litter chemical quality by 13C-CPMAS NMR
All organic materials were characterised by solid-state 13C-CPMAS NMR spectra. Plant samples from experiments in controlled conditions were analysed at different times, corresponding to leaves decomposition for 0, 30, 90 and 180 days (total of 80 litter types), whereas only undecomposed materials (32 litter types) were analysed for the field experiment.
A 300 MHz wide-bore magnet (AV-300 Bruker) equipped with a 4 mm wide-bore magic angle spinning (MAS) probe was used. NMR spectra were obtained with MAS as follows: rotor spin, 13,000 Hz; recycle time, 1 s; contact time, 1 ms; acquisition time, 20 ms and 2000 scans (Smejkalova et al. 2008). Spectral regions significantly associated with plant litter decay rate (Mathers et al. 2007; Pane et al. 2011; Bonanomi et al. 2013) were integrated for semiquantitative carbon molecular distribution as shown earlier (Piccolo et al. 2005): 0–45 ppm = alkyl C; 46–60 ppm = methoxyl and N-alkyl C; 61–90 ppm = O-alkyl C; 91–110 ppm = di-O-alkyl C.
Model description and implementation
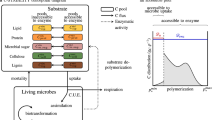
The OMDY model (Fig. 1) is a set of four ordinary differential equations (ODEs) describing the mass dynamics of plant litter organic carbon (C). Total organic C (Ctot, Table 2) is partitioned into four state variables, representing different C molecular classes (C i , with i = I,. .., IV, Table 2) according to plants specific composition. The carbon molecular distribution corresponds to selected resonance regions of 13C-CPMAS NMR spectra, in order to represent lipid alkyl C (δ 0–45), lignin methoxyl and peptidic N-alkyl C (δ 46–60), hydroxylated lipid O-alkyl C (δ 61–90) and carbohydrate di-O-alkyl C (δ 91–110), as previously associated to decay rates (Bonanomi et al. 2013). In a preliminary data analysis, we tested the Pearson’s correlation between the peak of each 1 ppm-wide 13C NMR signal along the spectrum (N = 200) recorded for the litter materials decomposing in controlled conditions, and the simple exponential decay rates of the same materials. Then, the molecular carbons resonating at δ 10–20, δ 50–60, δ 68–78, and δ 95–105 were selected, due to their significant correlation to litter decomposition processes (Fig. 2).
Schematic representation of the OMDY model. Organic carbon in decomposing materials is described in terms of chemical composition by four different spectral regions (gray shaded) of solid-state 13C NMR spectrum, corresponding to model layers. Decomposition proceeds with rates depending on intermolecular interactions (see text for details). Microbial turnover is accounted by the change in molecular composition during decomposition processes. The limiting effects of temperature and moisture on decay rates (not shown in the figure) are also implemented
Relationships between organic matter biomolecular quality and decomposition rates. Correlation profile (Pearson’s r) between decay rate of 60 plant leaf types (20 plant species at 3 decomposition stages) decomposing in controlled optimal conditions and 13C-CPMAS NMR spectral signals of the same organic materials. Dashed lines indicate threshold values of statistical significance for r (α = 0.05/N = 0.000263, after applying the Bonferroni’s correction for multiple comparisons, where N = 190 is the number of tested spectral signals). Black and grey bars indicate spectral signals significantly associated and regions unrelated to decay rate, respectively
Decomposition of each C molecular group proceeded by following a simple exponential decay (Olson 1963) as controlled by a baseline rate, which may be limited by intermolecular interactions, climate related factors, and specific temperature and plant leaf moisture. Microbial turnover contributes to C molecular dynamics by re-entering microbial biomass into the system as decomposition occurs. Then, the decomposition process is described by the following ODEs:
where t is time unit (day), k i are baseline constants related to specific molecular decomposition rates, P is the total intermolecular protection as in Eq. 2, k T and k W are scalars values accounting for the limiting effects of temperature and moisture, respectively (Incerti et al. 2011), and m i and M are microbial specific molecular turnover rate and microbial mass calculated as in Eqs. 4 and 5, respectively.
The intermolecular protection parameter represents the different resistance to decomposition of the diverse biomolecules. This is reflected by the variable decay rates observed for the different regions of the 13C NMR spectra (Bonanomi et al. 2011). Such differences can be ascribed not only to different baseline decay rates for different C molecular classes, but also to interactions among the decaying biomolecules (Rovira and Rovira 2010). For instance, the combined presence of molecular classes of different solubility may reduce the decomposition rates of most labile components, since they can become incorporated, and thus protected, into hydrophobic domains composed by more apolar biomolecules (Piccolo et al. 2004). This phenomenon is implicitly represented in the model by a weighing score (p i ), attributed to each biomolecular class, that reflects its relative contribution to intermolecular interactions. Positive scores of p i indicate a protective action during decomposition, negative scores mean an enhancement of decomposition rate due to easily decomposable compounds, and zero is given to neutral (i.e. non-interacting) compounds. Such representation is seemingly rough, because the physical location of different molecular classes was not explicitly represented in the model, while molecular protection can occur only by direct physical contact between different molecular types. However, in our representation, the chance of occurrence of a molecular interaction, eventually affecting decomposition rate, is implicitly represented by the relative abundance of a given molecular class, with larger abundance corresponding to a greater probability of mutual interaction. Then, the total intermolecular protection (P, Table 2) is calculated by summing up the relative contributions of all chemical classes, weighted by their respective score, as follows:
In the model, temperature and moisture content are considered as the main climate-dependent factors affecting organic matter decomposition. Daily temperature (T, °C) is an input parameter, while volumetric moisture content (θ, %) is calculated by applying a NASA-CASA-like equation, as proposed by Potter et al. (1993) for soil and modified by Incerti et al. (2011) for litter. Specifically, moisture content of litter material is a function of daily rainfall (R, mm) and potential evapo-transpiration (ETP, mm), the latter calculated according to Hargreaves and Samani (1985), obtaining the equation:
where Lth (mm) and Lpo (mm3 air mm−3 bulk) are thickness and porosity of the litter layer, and a and b texture-dependent empirical coefficients.
The effects of T and θ on decomposition rates are implemented as two scalar limiting factors (k T and k W , respectively, Table 2), both ranging between 0 and 1, with the minimum having a multiplying reducing effect on the baseline decomposition rates (Eq. 1). In this way, equal weight is given to the relative importance of temperature and organic matter moisture, as it is considered the reducing effect on decay rates produced by the most limiting of the two independent factors. The equations for temperature and moisture scalars were derived from literature. Besides the number of curves proposed earlier (for temperature see reviews in Lloyd and Taylor 1994; Kirschbaum 1995; Rodrigo et al. 1997; Burke et al. 2003; Del Grosso et al. 2005), we followed an approach previously validated in different sites from our survey area (Incerti et al. 2011). In particular, a logistic increase of decomposition rates with temperature was used, reaching its maximum value at 30 °C, and decreasing at values larger than 35 °C. In the case of moisture, a sigmoid curve in the θ interval was used, where no decomposition occurs at a low level of water content. Conversely, the increase of water content is proportionally related to an increase in decomposition rate up to θ = 90 %. A negative effect comes into play at larger water content, taking into account anoxic conditions.
The model structure, based on molecular differences among model layers, also provides a conceptual frame for implementing a sub-model on microbial turnover. During decomposition processes, a percent of plant organic matter becomes part of microbial biomass. Although the model does not explicitly describe the processes of microbial feeding, growth and reproduction, it simply calculates the newly formed microbial biomass (M, Table 2) according to a “metabolic ratio” (m e , Table 2) of the mineralisation flows.
Then, microbial death is implicitly modelled by re-entering the microbial mass into the 4 molecular classes through a partitioning vector (m i , Table 2), related to NMR spectral intervals highlighting organic matter molecular distribution (Piccolo et al. 2005) and microbial community composition:
where B and F are the relative fractions of bacterial and fungal mass, respectively, and b i and f i are the relative abundance of the i-th NMR spectral interval attributed to microbial mass, respectively (Kögel-Knabner 2002).
The OMDY model, developed according to a system dynamic approach using the software package SIMILE®(Muetzelfeldt and Massheder 2003), was calibrated using experimental data in controlled conditions. In particular, optimal parameter values were obtained from simulation runs with daily time steps, fitting the amount of carbon in each of the four 13C-CPMAS-NMR-defined fractions (C i ) at 30, 90 and 180 days, as described below. Finally, the model was validated against original field experiments on 32 leaves of plant species decomposing for one year on soils of four different sites.
Model calibration
The initial values of the model compartments representing the four C chemical classes were attributed by partitioning total organic matter mass according to a partitioning vector s i (Table 2) describing undecomposed litter quality, as follows:
where \( {s}_i={I}_i/{\displaystyle {\sum}_{i=1}^4{I}_i} \) is the initial relative abundance of the i-th molecular class, calculated as the integral value (I i ) of the corresponding 13C NMR spectral region, expressed as fraction of the total contributions of the 4 chemical shift intervals.
The maximum potential mineralisation rate (k i ) of each plant biomolecular class has been estimated assuming that the baseline decay of each molecular type follows a simple exponential model (Olson 1963), when it is not limited by environmental conditions and/or intermolecular protection (i.e. interactions among different biomolecules or metabolites producing inhibition of organic matter decay rate). Then, we considered the decay rates of organic matter decomposing in controlled, but not limiting conditions. In particular, k i values for the molecular classes were derived from decay rates determined for the most rapidly decomposing material (i.e. Coronilla emerus L. leaf litter) during the earliest decomposition stage (i.e. the first 30 days of litterbag incubation). This by assuming that the decomposition rates, for that specific material, were the largest due to concomitant optimal environmental conditions and almost negligible intermolecular protection. Then, for each i-th chemical class, the value of k i was calculated as follows:
where C i (t = 0) and C i (t = 30) are the mass values of the molecular content from C. emerus litter undecomposed and after 30 days of incubation, respectively, calculated as a function of mass loss and NMR spectral data.
The intermolecular protection coefficients (p i , Eq. 2 and Table 2) were estimated by fitting model simulations of mass loss dynamics for the molecular distribution in all organic materials decomposing in the controlled conditions of temperature and water availability. In particular, we used the relative abundance s i of the chemical classes (Eq. 6) derived from NMR spectral data for the plant organic matter recorded after 30, 90, and 180 days of decomposition, for a total of 228 datapoints (4 chemical classes × 3 decomposition dates × 19 litter materials, i.e. all species except C. emerus). This approach was based on the assumption that the reduction of decay rate observed in the 19 litter materials, in comparison to the fastest-decomposing C. emerus material, was due not only to differences of chemical composition among the plant materials (i.e. relative abundance of the chemical classes) but also to affinity interactions among the different biomolecules, as implemented by the protection coefficients.
The fitted equation was a simplified form of Eq. (1) with initial values of C i and k i assigned from Eqs. (6) and (7), respectively, while k T = k W = 0 was due to controlled, not limiting environmental conditions. Microbial turnover was evaluated by considering an average metabolic ratio, setting m e = 0.1 (i.e. assuming that 10 % of total plant carbon mineralised at every step contributed to microbial mass). Since the composition of microbial communities feeding on plant material can be highly variable in real conditions (e.g. Moller et al. 1999; Esperschütz et al. 2013), and the microbial inoculum applied with soil in our experiment in controlled conditions was not directly characterized, Eq. (5) was assessed in all simulations by assuming an average composition having an equal relative mass fractions of fungi and bacteria (i.e. B = F = 0.5). This approach, though apparently simplistic, may be reliable with the assumption that the variability of decay rates due to microbial community composition is negligible at common levels of microbial diversity (Dang et al. 2005).
An iterative procedure of linear programming was applied to the fitted equation based on the numerical optimization “simplex” algorithm (Himmelblau 1972; Nelder and Mead 1965) consisting in changing the parameter values (i.e. p i ) one at a time, running a model simulation with daily time step, recording the model fit of experimental data (i.e. C i at 30, 90, and 180 days for the 19 litter types), and repeating until reaching the maximum model fit, in terms of coefficient of determination (R2) between observed and simulated C i and minimization of the residuals.
Model simulations and validation
In order to validate the OMDY decomposition model, a set of simulation exercises was run, and the outputs statistically compared to the validation dataset. For model simulations, Eq. (1) was fitted with molecular class-specific values of baseline decay rates, intermolecular protection coefficients and microbial turnover parameters, as described above, and applied in model runs that simulated experimental conditions at the field sites. The organic matter moisture equation (Eq. 4) was fitted on the basis of plant leaves properties recorded at the field sites, using average values both for thickness (Table 1) and porosity (L po = 0.65 mm3 air mm−3 litter). Real climatic time series for rainfall and temperature recorded at each site during the litterbag decomposition experiment (from September 2012 to September 2013) were used as model inputs (Supplementary Fig. S1). Finally, temperature- and moisture-dependent limiting effects on organic matter decomposition were fitted following previous applications in our survey area (Incerti et al. 2011).
Results
Changes of litter quality in decomposition experiments
Our experiments in controlled conditions showed both similarities and differences in the dynamics of plant molecular content among the tested organic materials. Considering the selected 13C NMR spectral chemical shift intervals corresponding to the OMDY model C molecular classes, the spectral signals resonating in the O-alkyl C region (68–78 ppm) were the most abundant in all undecomposed organic matters (Supplementary Table S3), with relative abundance ranging between 20 % and 30 % (Quercus pubescens and Ampelodesmos mauritanicus plant materials, respectively). A consistent and statistically significant variation in content of the corresponding molecular carbon was observed in all litter species during decomposition processes (Supplementary Table S4), with a mean percent decrease of 32 % after 180 days, and species values ranging between −10 % in the case of Picea abies litter, and −61 % for Hedera helix (Supplementary Table S3). The carbohydrate signals resonating in the di-O-alkyl C region (95–105 ppm) showed mean relative abundance of 7.0 % in undecomposed materials (ranging from 4.5 % of Alnus cordata to 9.3 % of Ampelodesmos mauritanicus, Supplementary Table S3), which generally decreased during decomposition, showing mean of 5.2 % after 180 days. However, significant differences among plant species were observed (Supplementary Table S4) as in the case of Quercus pubscens litter, which showed an increase in the region for carbohydrates after the first 30 days of decomposition, followed by minimal reductions thereafter.
On the other hand, the content of biomolecules giving signals in both alkyl C (10–20 ppm) and methoxyl and N-alkyl C (50–60 ppm) chemical shift intervals, which generally increased during decomposition. In the case of alkyl C compounds, content in plant materials changed on average from 3.0 % (range 1.4–4.7 %) in undecomposed matter to 5.3 % (3.4–9.2 %) after 180 days, while for methoxyl and N-alkyl C the corresponding increase was to 6.1 % (2.9–8.2 %) and 8.8 % (6.6–10.3 %), respectively (Supplementary Table S3). For both these molecular components, there was a significant variation in magnitude and direction as a function of plant species (Supplementary Table S4). In particular, molecules rich in alkyl C increased more in Festuca, Coronilla, Populus and Salix species (relative increases of 68.5, 67.4, 67.7, and 72.7 %, respectively) than in Lepidium and Castanea (+2.2 and +14 %, respectively), while methoxyl and N-alkyl C containing molecules showed the most increase in Festuca drymeia (+69.8 %), and a relative decrease in Lepidium sativum (−13 %) (Supplementary Table S3).
Mass loss dynamics and model simulations
Model calibration led to an accurate prediction of plant molecular distribution (Figs. 3 and 4) and mass loss (Fig. 5a-c) dynamics during decomposition in optimal controlled conditions. Considering plant molecular distribution, observations and simulations consistently showed an increase of alkyl C and methoxyl and N alkyl C (Fig. 3) and a decrease of O-alkyl C and di-O-alkyl C (Fig. 4) during decomposition. Highly significant linear regression of simulated and observed mass were obtained for both single species and all pooled data (Fig. 5d). It is noteworthy that highly significant predictions were obtained for twenty species covering a very wide spectrum of plant molecular content, ranging from slow decomposing sclerophyll leaves of Quercus ilex and needles of Cupressus sempervirens, Picea abies and Pinus halepensis (Fig. 5a), to variably decaying deciduous broadleaves (Fig. 5b) and rapidly decomposing grasses and herbs (Fig. 5c), and up to extremely rapid decaying plant leaves of nitrogen fixing leguminous Coronilla emerus and Medicago sativa (Fig. 5a and c).
Simulations of alkyl C and methoxyl and N-alkyl C dynamics in controlled conditions. Data refer to observed (dots) and simulated (lines) percent abundance of the two chemical classes, corresponding to 13C NMR spectral regions resonating at 10–20 ppm and 50–60 ppm, respectively, in 20 litter materials decomposing in controlled optimal conditions
Simulations of O-alkyl C and di-O-alkyl C dynamics in controlled conditions. Data refer to observed (dots) and simulated (lines) percent abundance of the two chemical classes, corresponding to 13C NMR spectral regions resonating at 68–78 ppm and 95–105 ppm, respectively, in 20 litter materials decomposing in controlled optimal conditions
Calibration of intermolecular interactions during decomposition. (a-c) Observations (symbols) and simulations (lines) of mass loss dynamics for leaves of 20 plant species decomposing in controlled optimal conditions (25 ± 2 °C, watered to field capacity). Data refer to means of 8 replicates (coefficient of variation <5 % in all cases, deviation bars omitted to improve readability). (d) Scatterplot and correlation (Pearson’s r and associated p-value, n = 240) between simulated and observed mass (C i ) of different molecular classes (i.e. i = I, II, III, IV, Table 2) in the 20 plant materials after 30, 90, and 180 days of decomposition
Model simulations, when applied in real environmental conditions, provided year-long time profiles of organic matter mass loss dynamics, that highly fitted the corresponding values observed in the four experimental fields, although differing by vegetation types and meteo-climatic conditions (Fig. 6, Supplementary Fig. S2). The model was able to capture remarkable differences in decomposition dynamics for different types of organic matter within each experimental site (Fig. 6), that is under equal temperature and moisture regime. Inter-specific differences of decomposition dynamics among plant materials were consistent with differences in plant molecular content as described by 13C-CPMAS-NMR spectra and implemented in the model of molecular classes (Fig. 6).
Decomposition dynamics of different organic materials in different environmental conditions. Data refer to mass loss simulations (lines) and observations (symbols, mean of 8 replicates) under real field conditions at four different experimental sites, for the fastest and slowest decomposing material at each site (data for all the 32 species from field experiments are reported in Supplementary Fig. S2). On the right, the 13C NMR spectra of undecomposed organic materials are reported, with indication of molecular classes (i.e. i = I, II, III, IV) used as input data for simulations
The model performance, in terms of linear regression between of simulated and observed mass loss data (32 plant species × 4 sampling dates, for a total of 128 data points) was significantly accurate (Pearson’s r = 0.929, Fisher’s F = 1.652, P < 0.0001), with no evident residual trend for both all data pooled or separated at different experimental field (Fig. 7).
Model validation against field data. Scatterplot and correlation (Pearson’s r and associated p-value) between simulated and observed mass in 32 leaf materials after 30, 90, 180, and 360 days of decomposition (n = 128). Symbols indicate different experimental fields (see Table 1 for further details)
Discussion
The OMDY model presented in this study accurately predicted plant organic matter decomposition dynamics in controlled optimal conditions, correctly reproducing mass loss data and variations of selected molecular classes for 20 different plant species, encompassing a wide range of plant molecular composition. The model simulations of medium-term organic matter decomposition (up to 1 year) in Mediterranean ecosystems, under different environmental conditions (from coastal maquis to mixed broadleaves forests) and plant molecular composition (32 plant species) was also significantly predictive of the observed mass loss dynamics. These satisfactory model performances are directly dependent on an innovative assessment of organic matter molecular distribution, based on the semi-quantitative information on the molecular content in plant organic matter as revealed by 13C-CPMAS-NMR spectroscopy. A further added value of the OMDY model is that it allows to reproduce the result of complex intermolecular interactions among different organic matter fractions occurring in plant leaves during decomposition, although the implementation of such processes is not directly explicit in the model structure.
Model performance and molecular quality implementation by 13C CPMAS NMR
Model calibration of the intermolecular interactions during decomposition provided highly accurate predictions, as observed for twenty species covering a very wide spectrum of molecular composition. Although this result was consequent to an efficient parameter optimisation, it is relevant to note that it was possible to predict inter-specific differences of decomposition dynamics, at least in controlled and optimal conditions of temperature and water supply, based on a detailed description of molecular distribution in selected 13C-CPMAS-NMR chemical shift intervals. In addition, this approach allowed an estimation of the decomposition rate and its variation over time for each molecular class that was not based on pre-defined functions, but emerging from the species-specific partitioning vector of type of molecular carbon coupled to microbial turnover. In particular, observations and simulations consistently showed a decrease of hydroxylated lipid O-alkyl C (δ 68–78) and carbohydrate di-O-alkyl C (δ 95–105) fractions, and an increase of lipid alkyl C (δ 10–20) and lignin methoxyl and peptidic N-alkyl C (δ 50–60) fractions in decomposing materials during the process. Although consistent among different materials, such pattern showed species-specific differences, which were correctly reproduced by the model. These spectral signals likely correspond to specific molecular components of organic C pools (Kögel-Knabner 2002; Piccolo et al. 2005; Berg and McClaugherty 2013): the aliphatic chain of lignin and of acetate of some hemicelluloses (δ 10–20), the methoxyl C of lignin (δ 50–60), the C2, C3, and C5 of simple labile sugars (δ 68–78), and the C1 of cellulose and other carbohydrates (δ 95–105). Moreover, this pattern is consistent with previous experimental evidence assessed by 13C NMR about molecular changes in decomposing litter or maturing compost (e.g. Mathers et al. 2007; Spaccini and Piccolo 2007; Bonanomi et al. 2013), which showed selective preservation of some molecular classes in respect to other ones.
By accounting for intermolecular interactions, the OMDY model shows that their effect on decomposition rates is considerable, thereby well integrating the intermolecular affinity among different molecules contained in the decomposing organic matter. In fact, different plant biomolecules can show diverse decay rates not only due to their size, but also to their specific molecular structure that may be different from that of neighbouring particles (Baldock and Skjemstad 2000; Piccolo et al. 2005). In fact, the selected preservation of highly hydrophobic compounds during microbial decomposition may act as a protective cage and slow down the mineralization of entrapped labile compounds (Piccolo et al. 2004; Fultz et al. 2013).
We are aware that our datasets for model validation were based on medium-term experiments lacking an exhaustive number of litter types of Mediterranean conditions. Moreover, our dataset was limited to foliar litter, whose molecular assessment by 13C NMR has been more frequently reported, compared to woody and root litter. Consequently, further studies should be planned to investigate the consistency of the OMDY model performance under different conditions, extending the analysis to woody and root materials, to other ecosystems (e.g. agro-ecosystems, grasslands, boreal and tropical forests) and to long-term decomposition datasets. A practical and extensive application of our model may be limited by the availability of solid-state 13C NMR spectroscopy, which is not commonly accessible to laboratories worldwide. However, a possible solution could be to build up a database with 13C-CPMAS-NMR spectra of undecomposed organic matter of major crops and dominant plant species in different world ecosystems. In this regard, our research group has already 13C-CPMAS-NMR spectra of more than 50 crops and natural plant species. Pooling together our data with that of other research groups in other parts of the world would provide a robust database suitable for applications to different biomasses.
Comparison with existing models
The OMDY model is similar to other existing tools in its description of limiting effects of temperature (review in Burke et al. 2003) and moisture on decay rates (e.g. Incerti et al. 2011), but it is also completely different from previous models implementing organic matter decomposition (e.g. DAYCENT, Parton et al. 1998; CENTURY, Parton 1996; CLM4, Bonan et al. 2013; LIDEL, Campbell et al. 2016).
First, our model is based on a totally new approach to quantifying organic matter molecular quality, based on a detailed description of molecular distribution by solid-state 13C–NMR, that allows predictions of the changes in the organic matter molecular content with its microbial transformation in soil (Piccolo et al. 2005; Bonanomi et al. 2013). Conversely, previous models commonly described organic matter chemical quality using simple, static, not-molecularly explicit carbon pools (e.g. labile, stable, and lignin; structure, metabolic, active, and slow) that are poorly sensitive to intermolecular interactions during decomposition, which are inherently defined by fixed baseline decay rates (e.g. Palosuo et al. 2005; Adair et al. 2008; Tuomi et al. 2009).
Previous models implied that each single carbon pool decomposed by its own dynamic based on a pool-specific rate, independently from other pools, with total organic matter carbon simply given by the sum of all pools. Conversely, the OMDY model innovatively accounts for the intermolecular interactions occurring during organic matter progressive mineralization and affecting the decomposition rates of different molecular components. Previous modelling work attempted to implicitly capture this source of variability by implementing one-pool decomposition equations with decay rate changing with time, according to different curves (Yang and Janssen 2000; Rovira and Rovira 2010). In the OMDY model, intermolecular interactions are implemented as feedbacks between organic matter molecular composition and the decomposition rates of the different molecular classes. In particular, the specific molecular decomposition rates are multiplied by a intermolecular protection parameter that varies over time as a function of the relative abundance of molecular components in the organic matter. We acknowledge that in such implementation the occurrence and type of interactions among different molecular classes are only implicitly represented. Handling a finer implementation of such processes would be problematic, requiring to explicitly represent the occurrence of interactions in relation to the physical location of different molecular classes. However, even with an implicit representation, our model simulations showed that the overall effect of intermolecular interactions (i.e. the parameter P) do vary for different types of litter and during the process, decisively affecting litter decay rate and the accuracy of model predictions.
Some experimental data indicate that high initial decomposition rates can result in slower organic matter decay at the later stages (Hobbie et al. 2012). Such observations are perfectly reproduced by our model implementation, in which high initial decomposition rates lead to a change in molecular composition of the decomposing matrix that translates into a larger value of the intermolecular protection parameter, which, in turn, alters the decay process. Interestingly, our mathematical formulation of the equations describing the intermolecular protection and microbial turnover also shows that, at least after one year of decomposition, the mass of organic matter cannot reach the zero value, but asymptotically stabilizes around minimal limit values.
A further critical issue in current decomposition models is the different underlying philosophies with respect to the influence of soil mineral N and P on litter dynamics. Nitrogen content and N related indexes such as C/N and lignin/N ratios have been often used as chemical parameters in organic matter models (Shibu et al. 2006; Adair et al. 2008). However, the reliability of such indicators throughout the decomposition process has been often criticized based on empirical evidence (Berg and McClaugherty 2013; Hättenschwiler et al. 2011; Bonanomi et al. 2013). In fact, exercises on previous models indicated that the exclusion of N from decomposition model variables does not preclude the possibility to correctly predict short- and medium-term organic matter decomposition dynamics (Palosuo et al. 2005; Incerti et al. 2011). In a recent comparative analysis on the application of different models in order to predict long-term decomposition in different biomasses, Bonan et al. (2013) observed that reliable modelling predictions of C-N dynamics and N mobilization are largely context-dependent. The inconsistency between simulations and observations could be related to the different implementations of the effects of mineral nutrient by different models. In this respect, our simulations implicitly assume that conditions of N availability do not limit decomposition process. While our choice appears justifiable, further applications of the OMDY model to existing datasets on long-term decomposition (e.g. LIDET, Bonan et al. 2013) should help to clarify whether it is possible to fit observations using our improved evaluation of molecular composition of organic matter, irrespective of N and P dynamics.
References
Adair EC, Parton WJ, Del Grosso SJ, Silver WL, Harmon ME, Hall SA, Burke IC, Hart SC (2008) Simple three-pool model accurately describes patterns of long-term litter decomposition in diverse climates. Glob Change Biol 14:2636–2660
Aerts R (1997) Climate, leaf litter chemistry and leaf litter decomposition in terrestrial ecosystems: a triangular relationship. Oikos 79:439–449
Attiwill PM, Adams MA (1993) Nutrient cycling in forests. New Phytol 124:561–582
Austin AT, Vivanco L (2006) Plant litter decomposition in a semi-arid ecosystem controlled by photodegradation. Nature 442: 555–558
Baker NR, Allison SD (2015) Ultraviolet photodegradation facilitates microbial litter decomposition in a Mediterranean climate. Ecology 96:1994–2003
Baldock JA, Skjemstad JO (2000) Role of the soil matrix and minerals in protecting natural organic materials against biological attack. Org Geochem 31:697–710
Berg B, Matzner E (1997) The effect of N deposition on the mineralization of C from plant litter and humus. Environ Rev 5:1–25
Berg B, McClaugherty C (2013) Plant litter: decomposition, humus formation and carbon sequestration. Springer-Verlag, Berlin, Heidelberg, 3rd ed., 315 pp.
Bonan GB, Hartman MD, Parton WJ, Wieder WR (2013) Evaluating litter decomposition in earth system models with long-term litterbag experiments: an example using the community land model version 4 (CLM4). Glob Change Biol 19:957–974
Bonanomi G, Incerti G, Antignani V, Capodilupo M, Mazzoleni S (2010) Decomposition and nutrient dynamics in mixed litter of Mediterranean species. Plant Soil 331:481–496
Bonanomi G, Incerti G, Barile E, Capodilupo M, Antignani V, Mingo A, Lanzotti V, Scala F, Mazzoleni S (2011) Phytotoxicity, not nitrogen immobilization, explains plant litter inhibitory effects: evidence from solid-state 13C NMR spectroscopy. New Phytol 191:1018–1030
Bonanomi G, Incerti G, Giannino F, Mingo A, Lanzotti V, Mazzoleni S (2013) Litter quality assessed by solid state 13C NMR spectroscopy predicts decay rate better than C/N and lignin/N ratios. Soil Biol Biochem 56:40–48
Bonanomi G, Capodilupo M, Incerti G, Mazzoleni S (2014) Nitrogen transfer in litter mixture enhances decomposition rate, temperature sensitivity, and C quality changes. Plant Soil 381:307–321
Bradford MA, Berg B, Maynard DS, Wieder WR, Wood SA (2016) Understanding the dominant controls on litter decomposition. J Ecol 104:229–238
Burke IC, Kaye JP, Bird SP, Hall SA, McCulley RL, Sommerville GL (2003) Evaluating and testing models of terrestrial biogeochemistry: the role of temperature in controlling decomposition. In: Canham CD, Lauenroth WK (eds) Models in ecosystem science. Princeton University Press, Princeton, NJ, pp. 225–253
Campbell EE, Parton WJ, Soong JL, Paustian K, Hobbs NT, Cotrufo MF (2016) Using litter chemistry controls on microbial processes to partition litter carbon fluxes with the litter decomposition and leaching (LIDEL) model. Soil Biol Biochem 100:160–174
Coleman K, Jenkinson DS (1996) RothC-26.3 - a model for the turnover of carbon in soil. In: Powlson DS, Smith P, Smith JU (eds) Evaluation of soil organic matter models. Using Existing Long-Term Datasets. Springer-Verlag, Heidelberg, pp. 237–246
Conte P, Spaccini R, Piccolo A (2004) State of the art of CPMAS 13C-NMR spectroscopy applied to natural organic matter. Prog Nucl Mag Res Sp 44:215–223
Coq S, Souquet JM, Meudec E, Cheynier V, Hättenschwiler S (2010) Interspecific variation in leaf litter tannins drives decomposition in a tropical rain forest of French Guiana. Ecology 91:2080–2091
Cornelissen JHC, Thompson K (1997) Functional leaf attributes predict litter decomposition rate in herbaceous plants. New Phytol 135:109–114
Cornwell WK, Cornelissen JHC, Amatangelo K, Dorrepaal E, Eviner VT, Godoy O, Hobbie SE, Hoorens B, Kurokawa H, Pérez-Harguindeguy N, Quested HM, Santiago LS, Wardle DA, Wright IJ, Aerts R, Allison SD, van Bodegom P, Brovkin V, Chatain A, Callaghan TV, Díaz S, Garnier E, Gurvich DE, Kazakou E, Klein JA, Read J, Reich PB, Soudzilovskaia NA, Vaieretti MV, Westoby M (2008) Plant species traits are the predominant control on litter decomposition rates within biomes worldwide. Ecol Lett 11:1065–1071
Cotrufo MF, Wallenstein MD, Boot CM, Denef K, Paul E (2013) The microbial efficiency-matrix stabilization (MEMS) framework integrates plant litter decomposition with soil organic matter stabilization: do labile plant inputs form stable soil organic matter? Glob Change Biol 19:988–995
Dang CK, Chauvet E, Gessner MO (2005) Magnitude and variability of process rates in fungal diversity-litter decomposition relationships. Ecol Lett 8:1129–1137
Davidson EA, Janssens IA (2006) Temperature sensitivity of soil carbon decomposition and feedbacks to climate change. Nature 440:165–173
De Deyn GB, Cornelissen JH, Bardgett RD (2008) Plant functional traits and soil carbon sequestration in contrasting biomes. Ecol Lett 11:516–531
Del Grosso SJ, Parton WJ, Mosier AR, Hartman MD, Brenner J, Ojima DS, Schimel DS (2001) Simulated interaction of carbon dynamics and nitrogen trace gas fluxes using the DAYCENT model. In: Schaffer M, Ma L, Hansen S (eds) Modeling carbon and nitrogen dynamics for soil management. CRC Press, Boca Raton, FL, pp. 303–332
Del Grosso SJ, Parton WJ, Mosier AR, Holland EA, Pendall E, Schimel DS, Ojima DS (2005) Modeling soil CO2 emissions from ecosystems. Biogeochemistry 73:71–91
Esperschütz J, Zimmermann C, Dümig A, Welzl G, Buegger F, Elmer M, Munch JC, Schloter M (2013) Dynamics of microbial communities during decomposition of litter from pioneering plants in initial soil ecosystems. Biogeosciences 10:5115–5124
Franko U, Oelschlägel B, Schenk S (1995) Simulation of temperature, and nitrogen dynamics using the model CANDY. Ecol Model 81:213–222
Fultz LM, Moore-Kucera J, Zobeck TM, Acosta-Martínez V, Wester DB, Allen VG (2013) Organic carbon dynamics and soil stability in five semiarid agroecosystems. Agric Ecosyst Environ 181:231–240
Gholz HL, Wedin DA, Smitherman SM, Harmon ME, Parton WJ (2000) Longterm dynamics of pine and hardwood litter in contrasting environments: toward a global model of decomposition. Glob Change Biol 6:751–765
Gillon D, Joffre R, Ibrahima A (1994) Initial litter properties and decay rate: a microcosm experiment on Mediterranean species. Can J Botany 72:946–954
Gillon D, Joffre R, Ibrahima A (1999) Can litter decomposability be predicted by near infrared reflectance spectroscopy? Ecology 80:175–186
Hargreaves GH, Samani ZA (1985) Reference crop evapotranspiration from temperature. Appl Eng Agric 1:96–99
Hättenschwiler S, Coq S, Barantal S, Handa IT (2011) Leaf traits and decomposition in tropical rainforests: revisiting some commonly held views and towards a new hypothesis. New Phytol 189:950–965
Himmelblau DM (1972) Applied nonlinear programming. McGraw-Hill, New York, NY, 498 pp.
Hobbie SE, Eddy WC, Buyarski CR, Adair EC, Ogdahl ML, Weisenhorn P (2012) Response of decomposing litter and its microbial community to multiple forms of nitrogen enrichment. Ecol Monogr 82:389–405
Huang Y, Stankiewicz BA, Eglinton G, Snape CE, Evans B, Latter PM, Ineson P (1998) Monitoring biomacromolecular degradation of Calluna vulgaris in a 23-year field experiment using solid state 13C-NMR and pyrolysis-GC/MS. Soil Biol Biochem 30:1517–1528
Hunt ER Jr, Piper SC, Nemani R, Keeling CD, Otto RD, Running SW (1996) Global net carbon exchange and intra-annual atmospheric CO2 concentrations predicted by an ecosystem process model and three-dimensional atmospheric transport model. Global Biogeochem Cy 10:431–456
Incerti G, Bonanomi G, Giannino F, Piermatteo D, Castaldi S, Fioretto A, Papa S, De Marco A, Fierro A, Maggi O, Persiani AM, Rutigliano FA, Feoli E, Virzo De Santo A, Mazzoleni S (2011) Litter decomposition in Mediterranean ecosystems: modelling the controlling role of climatic conditions and litter quality. Appl Soil Ecol 49:148–157
Kirschbaum MUF (1995) The temperature dependence of soil organic matter decomposition and the effect of global warming on soil organic carbon storage. Soil Biol Biochem 27:753–760
Kögel-Knabner I (2002) The macromolecular organic composition of plant and microbial residues as inputs to soil organic matter. Soil Biol Biochem 34:139–162
Lecerf A, Risnoveanu G, Popescu C, Gessner MO, Chauvet E (2007) Decomposition of diverse litter mixtures in streams. Ecology 88:219–227
Li C, Zhuang Y, Frolking S, Galloway J, Harriss R, Moore B III, Schimel D, Wang X (2003) Modeling soil organic carbon change in croplands of China. Ecol Appl 13:327–336
Liski J, Nissinen A, Erhard M, Taskinen O (2003) Climatic effect on litter decomposition from arctic tundra to tropical rainforest. Glob Change Biol 9:575–584
Liski J, Palosuo T, Peltoniemi M, Sievänen R (2005) Carbon and decomposition model Yasso for forest soils. Ecol Model 189:168–182
Lloyd J, Taylor JA (1994) On the temperature dependence of soil respiration. Funct Ecol 8:315–323
Mathers NJ, Jalota RK, Dalal RC, Boyd SE (2007) 13C-NMR analysis of decomposing litter and fine roots in the semi-arid Mulga lands of southern Queensland. Soil Biol Biochem 39:993–1006
Mazzoleni S, Bonanomi G, Giannino F, Incerti G, Piermatteo D, Spaccini R, Piccolo A (2013) New modeling approach to describe and predict carbon sequestration dynamics in agricultural soils. In: Piccolo A (ed) Carbon sequestration in agricultural soils. Springer, Berlin, pp. 291–307
Mazzoleni S, Bonanomi G, Incerti G, Chiusano ML, Termolino P, Mingo A, Senatore M, Giannino F, Cartenì F, Rietkerk M, Lanzotti V (2015) Inhibitory and toxic effects of extracellular self-DNA in litter: a mechanism for negative plant–soil feedbacks? New Phytol 205:1195–1210
Meentemeyer V (1978) Macroclimate and lignin control of litter decomposition rates. Ecology 59:465–472
Melillo JM, Aber JD, Muratore JF (1982) Nitrogen and lignin control of hardwood leaf litter decomposition dynamics. Ecology 63:621–626
Moller J, Miller M, Kjoller A (1999) Fungal-bacterial interaction on beech leaves: influence on decomposition and dissolved organic carbon quality. Soil Biol Biochem 31:367–374
Muetzelfeldt R, Massheder J (2003) The Simile visual modelling environment. Eur J Agron 18:345–358
Nebbioso A, Piccolo A (2011) Basis of a humeomics science: chemical fractionation and molecular characterization of humic biosuprastructures. Biomacromolecules 12:1187–1199
Nebbioso A, Vinci G, Drosos M, Spaccini R, Piccolo A (2015) Unveiling the molecular composition of the unextractable soil organic fraction (humin) by humeomics. Biol Fert Soils 51:443–451
Nelder JA, Mead R (1965) A simplex method for function minimization. Comput J 7:308–313
Olson JS (1963) Energy storage and the balance of producers and decomposers in ecological systems. Ecology 44:322–331
Palosuo T, Liski J, Trofymow JA, Titus BD (2005) Litter decomposition affected by climate and litter quality - testing the Yasso model with litterbag data from the Canadian intersite decomposition experiment. Ecol Model 189:183–198
Pane C, Spaccini R, Piccolo A, Scala F, Bonanomi G (2011) Compost amendments enhance peat suppressiveness to Pythium ultimum, Rhizoctonia solani and Sclerotinia minor. Biol Control 56:115–124
Parton WJ (1996) The CENTURY model. In: Powlson DS, Smith P, Smith JU (eds) Evaluation of soil organic matter models. Springer, Berlin, pp. 283–291
Parton WJ, Ojima DS, Cole CV, Schimel DS (1994) A general model for soil organic matter dynamics: sensitivity to litter chemistry, texture, and management. In: Quantitative Modelling of Soil Forming Processes, SSSA Special Publication 39, pp 147–167
Parton WJ, Hartman M, Ojima D, Schimel D (1998) DAYCENT and its land surface submodel: description and testing. Glob Planet Chang 19:35–48
Piccolo A (1996) Humus and soil conservation. In: Piccolo A (ed) Humic substances in terrestrial ecosystems. Elsevier, Amsterdam, pp. 225–264
Piccolo A (2002) The supramolecular structure of humic substances. A novel understanding of humus chemistry and implications in soil science. Adv Agron 75:57–134
Piccolo A, Campanella L, Petronio BM (1990) 13C-NMR spectra of humic substances extracted with different mechanisms. Soil Science Society of America J 54:750–755
Piccolo A, Spaccini R, Nieder R, Richter J (2004) Sequestration of a biologically labile organic carbon in soils by humified organic matter. Clim Chang 67:329–343
Piccolo A, Conte P, Spaccini R, Mbagwu JSC (2005) Influence of land use on the humic substances of some tropical soils of Nigeria. Eur J Soil Sci 56:343–352
Potter CS, Randerson JT, Field CB, Matson PA, Vitousek PM, Mooney HA, Klooster SA (1993) Terrestrial ecosystem production: a process model based on global satellite and surface data. Glob Biogeochem Cy 7:811–841
Preston CM, Nault JR, Trofymow JA (2009) Chemical changes during 6 years of decomposition of 11 litters in some Canadian forest sites. Part 2. 13C abundance, solid-state 13C NMR spectroscopy and the meaning of “lignin”. Ecosystems 12:1078–1102
Rodrigo A, Recous S, Neel C, Mary B (1997) Modelling temperature and moisture effects on C-N transformations in soils: comparison of nine models. Ecol Model 102:325–339
Rovira P, Rovira R (2010) Fitting litter decomposition datasets to mathematical curves: towards a generalised exponential approach. Geoderma 155:329–343
Rovira P, Vallejo VR (2007) Labile, recalcitrant, and inert organic matter in Mediterranean forest soils. Soil Biol Biochem 39:202–213
Running SW, Gower ST (1991) Forest-BGC, a general model of forest ecosystem processes for regional applications. II. Dynamic carbon allocation and nitrogen budgets. Tree Physiol 9:147–160
Shibu ME, Leffelaar PA, Van Keulen H, Aggarwal PK (2006) Quantitative description of soil organic matter dynamics – a review of approaches with reference to rice-based cropping systems. Geoderma 137:1–18
Sitch S, Smith B, Prentice IC, Arneth A, Bondeau A, Cramer W, Kaplan JO, Levis S, Lucht W, Sykes MT, Thonicke K, Venevsky S (2003) Evaluation of ecosystem dynamics, plant geography and terrestrial carbon cycling in the LPJ dynamic global vegetation model. Glob Change Biol 9:161–185
Smejkalova D, Spaccini R, Piccolo A (2008) Multivariate analysis of CPMAS 13C-NMR spectra of soils and humic matter as a tool to evaluate organic carbon quality in natural systems. Eur J Soil Sci 59:496–504
Smith P, Smith JU, Powlson DS, McGill WB, Arah JRM, Chertov OG, Coleman K, Franko U, Frolking S, Jenkinson DS, Jensen LS, Kelly RH, Klein-Gunnewiek H, Komarov AS, Li C, Molina JAE, Mueller T, Parton WJ, Thornley JHM, Whitmore AP (1997) A comparison of the performance of nine soil organic matter models using datasets from seven long-term experiments. Geoderma 81:153–225
Spaccini R, Piccolo A (2007) Molecular characterization of compost at increasing stages of maturity. 2. Thermochemolysis-GC-MS and 13C-CPMAS-NMR spectroscopy. J Agric Food Chem 55:2303–2311
Swift MJ, Heal OW, Anderson JM (1979) Decomposition in Terrestrial Ecosystems. In: Studies in Ecology, 5. Blackwell Scientific Publications, Oxford.
Taylor BR, Parkinson D, Parsons WFJ (1989) Nitrogen and lignin content as predictors of litter decay rates: a microcosm test. Ecology 70:97–104
Tian G, Brussaard L, Kang BT (1995) An index for assessing the quality of plant residues and evaluating their effects on soil and crop in the (sub-) humid tropics. Appl Soil Ecol 2:25–32
Tuomi M, Thum T, Järvinen H, Fronzek S, Berg B, Harmon M, Trofymow JA, Sevanto S, Liski J (2009) Leaf litter decomposition - estimates of global variability based on Yasso07 model. Ecol Model 220:3362–3371
Yang HS, Janssen BH (2000) A mono-component model of carbon mineralization with a dynamic rate constant. Eur J Soil Sci 51:517–529
Acknowledgments
This work was funded by the Italian Ministry of Scientific Research and University (MIUR) within the FISR program (project MESCOSAGR, coordinator prof. A. Piccolo).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Alfonso Escudero .
Electronic supplementary material
ESM 1
(PDF 242 kb)
Rights and permissions
About this article
Cite this article
Incerti, G., Bonanomi, G., Giannino, F. et al. OMDY: a new model of organic matter decomposition based on biomolecular content as assessed by 13C-CPMAS-NMR. Plant Soil 411, 377–394 (2017). https://doi.org/10.1007/s11104-016-3039-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-016-3039-2