Abstract
Aims
It has been reported that root exudates of Sorghum bicolor can inhibit nitrification in a bioassay using Nitrosomonas, and methyl 3-(4-hydroxyphenyl) propionate (MHPP) was identified as one of the nitrification inhibiting compounds. Therefore, we have investigated the effects of this compound on nitrogen dynamic, potential nitrification activity and on soil microorganisms.
Methods
We conducted soil incubation experiments using synthetic MHPP to evaluate its effect on changes in inorganic soil nitrogen pools, on nitrification activity and on abundance of ammonia-oxidizing bacteria and archaea. Addition of MHPP at two concentrations equivalent to 70 and 350 μg C g−1 soil was compared to glucose as a carbon source and to the commercially available nitrification inhibitor dicyandiamide (DCD).
Results
Soil amended with the high dose of MHPP and with DCD showed reduced nitrate content and low nitrification activity after 3 and 7 days of incubation. This was mirrored by a 70 % reduction in potential nitrification activity compared to a nitrogen-only control. None of the incubation treatments affected non-target microbial counts as estimated by 16S rRNA gene copy numbers, however, the high dose of MHPP significantly reduced the abundance of ammonia-oxidizing bacteria and archaea.
Conclusions
These findings suggest that MHPP is capable of suppressing nitrification in soil, possibly by reducing the population size and activity of ammonia-oxidizing microorganisms.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The biological oxidation of ammonium (NH4 +) or ammonia (NH3) via nitrite (NO2 −) to nitrate (NO3 −) is called nitrification. The nitrification process regulates the flow between two nitrogen pools, NH4 + and NO3 − (Ceccherini et al. 2007) and if nitrification is intensive, as in many agricultural fields, NO3 − can be lost due to leaching or denitrification. Leaching of NO3 − represents the most important mechanism of N loss from agricultural soils with estimates being as high as 61.5 Tg N yr−1 (Schlesinger 2009). Under low O2 concentration, NO3 − may also be reduced via denitrification to dinitrogen gas (N2) or nitrous oxide (N2O) a potent greenhouse gas and a natural catalyst of stratospheric ozone degradation. In addition, N2O can be produced during nitrification and in a distinct process called nitrifier denitrification (Wrage et al. 2001). To minimize N losses in agricultural fields that typically receive large amounts of fertilizer N as ammonium or urea, it would be desirable to regulate the nitrification process in soil.
To avoid problems related to nitrification different synthetic nitrification inhibitors have been developed (Prasad and Power 1995) but their widespread use in agriculture is limited by their considerable costs and their variable performance across different agro-climatic and soil environments. It has been hypothesized that some plants can naturally inhibit nitrification in soil (Rice and Pancholy 1972) and studies reported in the last years (Lata et al. 2004) confirmed such nitrification inhibition occurs via a process termed ‘Biological Nitrification Inhibition (BNI)’, which refers to the capacity of certain plants to inhibit nitrification by the exudation of specific secondary organic compounds from roots (Subbarao et al. 2007a). Field crops and pastures have been evaluated for BNI capacity. Pariasca et al. (2010) have found inhibition of nitrification in soil amended with rice (Oryza sativa L.) root exudates. Subbarao et al. (2009) discovered a nitrification inhibitor in root exudates of the tropical grass Brachiaria humidicola. Root exudates of rye grass, wheat, lettuce, salad rape and onion also showed signs of inhibiting nitrification (More and Waid 1971).
Zakir et al. (2008) further showed BNI activity in root exudates of Sorghum bicolor using a nitrification inhibition assay (Subbarao et al. 2006) that measures the activity of a recombinant Nitrosomonas europaea strain. Zakir et al. (2008) concluded that BNI activity was caused by multiple components present in the sorghum exudate and isolated the phenolic substance methyl 3-(4-hydroxyphenyl) propionate (MHPP) as the main active compound. The release of MHPP was stimulated by the presence of NH4 + in the solution used to collect root exudates and exudation rates of up to 10.8 mg of MHPP g−1 root DW d−1 were detected (Zakir et al. 2008). MHPP concentrations in the rhizosphere of sorghum have so far not been reported.
It is well known that sorghum produces a range of metabolites having allelopathic or phytotoxic effects and in some cases the phytotoxic effect could be attributed to specific compounds like sorgoleone (Einhellig and Souza 1992). Sorgoleone has also been shown to inhibit nitrification activity and recently a 3rd compound, sakuranetin, was said to contribute to BNI activity in sorghum exudates (Subbarao et al. 2012a). However, there is no clear consensus whether plant-released compounds suppress nitrification in soil directly, by some specific toxic effect on ammonia oxidizers, or indirectly by causing net N immobilization as a result of carbon addition.
Despite the importance of sorghum as field crop and its potential to inhibit nitrification due to the release of exudates such as MHPP, no further studies have been undertaken to characterize the effect of this compound on soil nitrification, nitrification activity and on the response of ammonia-oxidizing microorganisms. The oxidation of NH3 to NO2 −, mediated by the enzyme ammonia monooxygenase (AMO), represents the first and rate-limiting step in nitrification. For decades it was thought that this step was mediated solely by ammonia-oxidizing bacteria (AOB). The discovery in a vast range of environments (marine, sediment and soil) of a gene encoding the subunit of the enzyme ammonia monooxygenase (amoA) in microorganisms belonging to the domain of archaea, its transcription, and the evidence for autotrophic growth of an ammonia-oxidizing archaea, suggests that archaeal ammonia oxidation might contribute considerably to nitrification (Beman and Francis 2006; Park et al. 2006; Leininger et al. 2006; Zhang et al. 2010).
The objective of this study was to evaluate the potential of a synthetic MHPP to inhibit nitrification in soil, using soil incubation and potential nitrification essays. The effect MHPP on abundance of ammonia-oxidizing bacteria (AOB) and archaea (AOA) was further analyzed using quantitative real-time polymerase chain reaction (qPCR) targeting the ammonia monooxygenase (amoA) genes.
Materials and methods
Soil
The soil used in this study, a Dystric Regosol Climate Humid Mediterranean (FAO 1974), was collected from a fallow field at Peccioli near the city of Pisa (Italy). The soil had the following composition: 75.5 % sand, 15.5 % silt, 9 % clay, 2.1 % total carbon, 0.21 % total nitrogen, pH 6.8. Soil was sieved at 2 mm and pre-incubated at 40 % of water-filled pore space (WFPS) at 25 °C for 1 week before starting the incubation study.
Experimental set-up and incubation effect of MHPP on nitrogen dynamics
Soil microcosms consisted of 100 ml plastic bottles containing 20 g of soil (oven dry equivalent). Methyl 3-(4-hydroxyphenyl) propionate (MHPP; C10H12O3; MW: 180.2) was obtained from Aldrich Chemical Company, Dorset, (UK). Treatments were: (1) Nitrogen 100 mg * Kg−1 soil, added as ammonium sulfate and considered as the control sample; (2) Nitrogen plus MHPP at a concentration of 525 μg MHPP g−1 soil (equivalent to 350 μg C * g−1 soil; MHPP-high dose); (3) Nitrogen plus MHPP at a concentration of 105 μg MHPP g−1 soil (equivalent of 70 μg C * g−1 soil; MHPP low dose); (4) Nitrogen plus glucose (70 μg C * g−1 soil); (5) Nitrogen plus dicyandiamide, a well known inhibitor of nitrification, at a concentration of 10 μg * g−1 soil (DCD, C2H4N4). Glucose, one of the most common compound exuded by the majority of plants (Jones and Darrah 1996), was used only at one concentration because previous studies showed that nitrification was depressed by high amount of glucose (data not shown), presumably because ammonium availability decreased due to rapid growth of soil microorganisms feeding on readily available carbon.
Stock solutions of 71 mM (NH4)2SO4, 39.1 mM MHPP, 13 mM glucose and 4.8 mM dicyandiamide were prepared by using sterilized water and stored at −20 °C until use. Working solutions were prepared by mixing 1 ml of 71.3 mM ammonium sulfate solution with 1.5 ml or 0.30 ml of 39.1 mM of MHPP, ensuring the two carbon concentrations of 350 and 70 μg C * g−1 soil respectively; 1.5 ml of 13 mM glucose solution that ensured a carbon concentration of 70 μg C * g−1 soil; 0.5 ml of 4.8 mM DCD to have a concentration of 10 μg * g−1 soil. Additionally, a set of soil microcosms received distilled water only. All amendments were done in a single pulse with C inputs being comparable to those reported in other studies (Watkins et al. 2009; Shi et al. 2011).
Nitrogen dynamics were expressed on net values of NH4 + -N and NO3 − -N, that is the amount of inorganic nitrogen in treated soils minus the amount of inorganic nitrogen in soil treated with distilled water only. After each addition, soil was mixed carefully and incubated at 25 °C in a IC600 chamber (YAMATO, Japan) for 1 week. During incubation soil was maintained at 60 % of WFPS considered to offer the best condition for nitrification because diffusion of both substrates and gases (O2) are not restricted (Linn and Doran 1984). Soil samples were taken at 0 day (immediately after amendments), 3 and 7 days. NH4 +-N, NO3 − -N and total inorganic nitrogen were measured for each incubation time. Five replicates for each treatment were used. At each incubation times, 3 g of soil were used to measure inorganic N (NH4 +-N and NO3 − -N) by shaking the soil with 2 M KCl (soil/solution ratio 1:10) for 1 h. Soil suspensions were then filtered through Whatman filter N 41. The extracts were measured for inorganic N content by auto analyzer (Bran + Lubbe, Hamburg, Germany). At 7 days, four of the five independent soil subsamples, were sampled and stored at −20 °C and subsequently used for soil DNA extraction. Because we were also interested in comparing potential nitrification activity to abundance of ammonia-oxidizing microorganisms, an additional set of three replicates of soil microcosms were used to measure potential nitrification activity (see below) after 1 week of incubation.
Potential nitrification activity
The shaken slurry method (Hart et al. 1994) was used to determine potential nitrification activity (PNA). The procedure is briefly described as follows. Stock solution of 0.2 M KH2PO4 (27.22 g L−1), 0.2 M of K2HPO4 (34.83 g L−1) and 50 mM of (NH4)2 SO4 (6.60 g L−1) were prepared by using sterilized distilled water. A phosphorous-nitrogen (PN) working solution having a final concentration of 1 mM PO4 3− and 1.5 mM NH4 + at pH 7.4 was obtained. At 7 days of soil incubation, a set of three soil microcosms for each treatment was collected and 15 g of soil were transferred into in Erlenmeyer flasks and supplemented with 100 ml of working solution. Slurries were incubated for 24 h under continuously shaking at 180 rpm on an orbital shaker (SI-300R, AsOne, Japan) at 25 °C. Aliquots of 10 ml were taken at 2, 4, 22 and 24 h intervals and centrifuged at 8000 g for 10 min at 5 °C. NO3 − -N was determined by Continuous Flow Analysis (Bran + Lubbe, Hamburg, Germany). Potential nitrification activity was calculated by regression analysis of NO3 − vs time.
Quantification of Bacterial and Archaeal 16S rRNA and amoA Genes by Real-Time PCR
Soil subsamples (2 g) were collected at day 0 for soil only and N treatments (T0) and again at 7 days (T7) for all treatments and stored at −20 0C. DNA was extracted from frozen soil (400 mg) using the Fast DNA Spin Soil Kit (MP Biomedicals) and further purified using Ultraclean15 DNA Purification Kit (MoBio Laboratories, CA, USA). Concentration of extracted DNA was determined by QubitTM using the QUANT-iT dsDNA HS Assay Kit (Invitrogen). Four independent replicates were used to determine the copy numbers of bacterial and archaeal 16S rRNA and amoA genes.
The PCR quantification was based on the fluorescent dye SYBR-Green I, using the MiniOpticonTM System CFB-3120 (Bio-Rad). Bacterial and archaeal amoA genes were quantified using the primers shown in Table 1, SYBRs Premix Ex TaqTM (TaKaRa, Japan), 1 ul (0.1 ng) of template DNA, in MiniOpticonTM System CFB-3120 (Bio-Rad). Amplifications were carried out as follows: 30 s at 95 °C, 40 cycles at 95 °C for 5 s, 10 s at 55 °C and 15 s at 72 °C, followed by a gradual increase in temperature from 55 °C to 96 °C during the dissociation stage (to monitor the presence of non specific amplification products). A negative control without template was included in each PCR run. Data analysis was carried out using the Opticon Monitor Software (BioRad).
Possible inhibition of the real-time PCR was assessed by running a series of 10-fold dilutions of the extracted DNA and determining the amplification efficiency of each diluted sample. Inhibition was observed without dilution, but high amplification efficiencies of 95–99 % were obtained for both the AOB and the AOA quantifications after a 10-fold dilution. A 10-fold dilution of each sample was thus used for the final analysis.
Amplified PCR products were gel-purified, ligated into the pGEM-T Easy Vector (Promega, Medison), and the ligation products were used to transform Escherichia coli JM109 competent cells following the manufacturer’s instructions. The plasmid DNA of positive clones were extracted using the PureYieldTM Plasmid Miniprep System (Promega, Medison), their sequence determined using the pUC/M13 forward primer and their identity confirmed using DDBJ Blast (http://blast.ddbj.nig.ac.jp/top-j.html). The plasmid DNA concentration was determined by Nanodrop® ND-1000 UV–vis Spectrometer (NanoDrop Technologies, Wilmington, DE). The copy number of target genes was calculated directly from the concentration of the extracted plasmid DNA. Ten-fold serial dilutions of a known copy number of the plasmid DNA were subjected to a real-time PCR assay in triplicate to generate an external standard curve. High efficiency of around 98–100 % were obtained for AOB amoA amplification, with the R value ranging between 0.997 and 0.999, and efficiencies of 96–100 % were obtained for AOA amoA amplification, with the R value ranging between 0.998 and 0.999.
Statistical analysis
All statistical tests were performed using the software Statistix 9 (Analytical Software, Tallahassee, FL). If necessary, data were log10 transformed to normalize the distribution. Differences between treatment means were tested by one-way analysis of variance (ANOVA), and if treatment effects were found to be significant at the level of P < 0.05, least significant difference (LSD) were used to separate means. Two-sided Dunnett’s Multiple Comparisons with a Control were used to determine whether gene copy numbers (16S and amoA) differed significantly between control soil samples taken prior to incubation (T0) and at day 7.
Results
Effect of MHPP on nitrogen dynamics
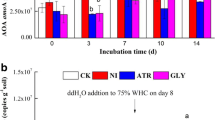
In soil incubation experiments 100 mg of ammonium had been added per Kg of soil and about 80 % of the NH4 + added was recovered by a 2 M KCl extraction at day 0 (Fig. 1a). N dynamics then differed between treatments during the 1-week incubation period. In the nitrogen treatment (soil incubated with ammonium sulfate only), ammonium recovered after 3 days of incubation decreased to 68.0 mg NH4 +-N Kg−1 with a further reduction to 2.7 mg NH4 +-N Kg−1 soil at day 7 (Fig. 1a). This was accompanied by an increase in NO3 − from 10 mg at day 0 to 40 mg (day 3) and 81 mg Kg−1 soil (day 7), indicating that nitrification was rapid in the control treatment (Fig. 2a).
a Net NH4 +-N, b NO3 −-N concentrations, c total mineral N in soil at 0, 3 and 7 days of incubation. Nitrogen (100 μg NH4 +-N g−1) was added in all treatments. N = nitrogen only, Glucose (70 μg C * g−1 soil), MHPP low dose (70 μg C * g−1soil), MHPP high dose (350 μg C * g−1), DCD (10 μg * g−1 soil). Mean value and standard error are shown (n = 5). The two error bars represent LSD (P < 0.05) at each of incubation time
Nitrification potential activity in soil sampled after 7 day incubation with the following treatments: Soil only (no addition of N or other compounds), Nitrogen (100 μg NH4 +-N g−1 the following treatments received the same dose of N), MHPP high dose (350 μg C * g−1), MHPP low dose (70 μg C * g−1soil), DCD (10 μg* g−1 soil). Mean value and standard error are shown (n = 3). The different letters above the bars indicate statistical differences (P < 0.05)
The glucose treatment showed a very different pattern with a significant (P < 0.05) decline in NH4 + within 3 days that was not accompanied by a corresponding increase in NO3 −-N (Fig. 1a, b), suggesting that some of the added N was temporarily immobilized by soil microflora. However, by day 7 ammonium and nitrate levels in the glucose treatment resembled those of the control, without significant differences. The addition of MHPP at a low-dose had small effects on ammonium concentrations at both 3 (55.4 mg NH4 +-N Kg−1, P < 0.004) and 7 days (11.1 mg NH4 +-N Kg−1, P < 0.01), however, nitrate concentrations were not affected by this treatment and closely resembled those seen for the control treatment (Fig. 1b).
In contrast N dynamics were very different in the high-dose MHPP and the DCD treatments. After 3 days more than 70 % of the ammonium was recovered in these two treatments (75.7 mg NH4 +-N Kg−1 for MHPP high dose; 70.1 mg NH4 +-N Kg−1 for DCD) and after 7 days around 30 % of the ammonium remained (29.3 mg NH4 +-N Kg−1 for MHPP high dose, P < 0.001; 34.0 mg NH4 +-N Kg−1 for DCD, P < 0.001), compared to less than 3 % in the control. These slower decreases in ammonium concentrations were mirrored by low nitrate concentrations that only reached 28.1 and 44.9 mg NO3 −-N Kg−1 after 7 days for the high MHPP and the DCD treatment, respectively, compared to more than 80 mg NO3 −-N Kg−1 after 7 days in the control, glucose and low MHPP treatments.
In addition, total inorganic nitrogen in the MHPP high dose treatment remained unchanged at day 3 (Fig. 1c) suggesting that NH4 + immobilization did not occur while it probably was a factor for the glucose treatment where little over 50 % of the total N present at day 0 was not accounted for at day 3. Between days 3 and 7, some NH4 + immobilization probably occurred in the high MHPP since total N was about 30 mg lower than in other treatments. However, NH4 + concentrations in the soil remained significantly higher compared to the N and glucose treatments indicating that the repression of nitrification was also responsible for the low nitrate level observed during the incubation.
Results thus showed that N dynamics in the glucose treatment were entirely different from any other treatment including the control (Fig. 1). The apparent disappearance of N in the glucose treatment at day 3 was indicative of NH4 + being immobilized in microbial biomass. The glucose treatment was thus not a suitable control for the amount of C added in other treatments, presumably because MHPP and DCD were more stable compounds than glucose and C far less available to spur microbial growth, particularly during the first 3 days. The glucose control treatment was therefore not used in subsequent experiments.
Determination of potential nitrification in soil samples taken after the 7-day incubation period revealed significant treatment effects (Fig. 2). Potential nitrification activity was high in the N amended soil (83 mg NO3 − -N Kg−1 d−1) but decreased by about 80 % in soil samples treated with both MHPP levels and DCD.
Bacterial or archaeal 16S rRNA and amoA gene abundance in soil
A quantitative PCR assay was used to estimate the population size in soil samples of two philogenetically distinct groups of soil microorganisms, bacteria and archaea (Fig. 3a). In soil extracted at time zero (T0), population size estimates based on 16S rRNA gene copy number were 5.6 × 108 per gram of soil for bacteria and 1.0 × 106 per gram of soil for archaea (Log10 transformed values are shown in Fig. 3a). T0 estimates for extracts of the raw and the N amended soil were not significantly different (data not shown); an average of both treatments is therefore shown in Fig. 3a and b. The 7-day incubation period had no significant effect on population size as estimated by the 16S rRNA gene copy (Fig. 3a), indicating that neither MHPP nor DCD affect non-target microorganisms.
Bacterial (grey bars) and archaeal (black bars) 16S rRNA (a) and amoA gene (b) copy numbers (mean ± standard error n = 4) in the different treatments after 1 week of soil incubation compared to copy numbers at the start of the experiment (T0). Two-sided Dunnett’s Multiple Comparisons with a control (T0) were used to determine statistical significance between means at P < 0.05 (*) or P < 0.01 (**)
To estimate whether treatments specifically affected ammonia-oxidizers within the bacterial or archaeal populations the copy number of bacterial and archaeal amoA genes were measured by quantitative PCR. At T0 the bacterial amoA copy number was 1.0 × 105 per gram of soil and this more than doubled to 2.2 × 105 copies per gram of soil after 7 days of incubation in the N-amended soil (Fig. 3b). Slightly less but still significant increases were detected for DCD and low dose of MHPP. The only treatment not showing a significant increase was the high dose of MHPP (1.2 × 105 copies per gram of soil). Comparisons within treatments at T7 further showed that the high MHPP treatment was significantly lower compared to the N, soil only and low MHPP treatments (LSD, p < 0.05; data not shown). Archaeal amoA gene abundance was one order of magnitude higher compared to bacterial amoA and none of the treatments led to a significant increase above the value of 1.1 × 106 per gram of soil detected at T0 (Fig. 3b). Seven days of incubation without addition of N (soil only) significantly reduced copy number to 0.6 × 106 per gram of soil, a reduction that was also observed for the high dose of MHPP.
Discussion
Reducing nitrification rates in soils represents a strategy to mitigate the negative impact of nitrate on the environment (Subbarao et al. 2012b) and many synthetic compounds, called nitrification inhibitors, are available. Nevertheless, high cost and limited effectiveness in some soils have limited their widespread adoption in farming. Biological Nitrification Inhibition (BNI) in which plants inhibit nitrification in soil via root exudates, could represent a valid alternative to synthetic inhibitors in regulating nitrification in soil. Here we tested whether the synthetic analog of methyl 3-(4-hydroxyphenyl) propionate (MHPP), a compound detected in sorghum root exudates and implicated in the nitrification inhibiting properties of these exudates (Zakir et al. 2008), is capable of reducing nitrification in soil and whether this was due to a direct inhibition effect as opposed to the indirect N immobilization effect. Results showed that MHPP lowered net nitrification, potential nitrification activity and amoA gene copy number, particularly of ammonia-oxidizing bacteria.
Low nitrification rates have at times been explained by a decline in NH4 + supply rather than through toxicity of specific compounds to nitrifiers (Schimel et al. 1996). It has therefore been argued that addition of organic compounds that may represent a carbon source for soil microorganisms may favor heterotrophs that are better competitor for NH4 + than autotrophs. As a result ammonium oxidizers would have limited NH4 + supply available (NH4 + immobilization) and soil nitrification would have been reduced indirectly by addition of a carbon source like MHPP. Furthermore, when NH4 + concentration is low and its spatial availability is limited, heterotrophic NO3 − immobilization (assimilation) could also occur (Rice and Tiedje 1989).
The possibility of NH4 + immobilization being of importance was investigated in more detail and several lines of evidence suggested that this was of minor importance and that MHPP had a more direct effect on ammonia-oxidizers. First, nitrogen dynamics of the MHPP (high) treatment mirrored those seen for the commercially available nitrification inhibitor DCD and was distinctly different from the N and glucose control treatments, both for nitrate and ammonium concentrations (Fig. 1). Second, the sum of both N forms at day 3 indicated that NH4 + immobilization did not occur in the MHPP treatment, while it was a factor for the glucose treatment where rapid disappearance of NH4 + was not followed by NO3 − production. Thus, lower nitrate concentrations at day 3 in MHPP (high) and DCD treatments can be attributed to reduced nitrification and not to any change in substrate (NH4 +) availability. This was further confirmed by the analysis of potential nitrification activity by the soil-slurry method (Hart et al. 1994). The ammonia-oxidizing activity was high in the nitrogen only treatment but inhibited (around 70 % inhibition compared to the N control) at both MHPP concentrations and in the DCD treatment. Since potential nitrification activity was measured in soil samples supplemented with an excess of NH4 + (the NH4 + concentration was monitored to assure NH4 + did not become limiting during incubation) and since the assay was continuously shaken to ensure aerobic condition, it can be ruled out that substrate unavailability or denitrification played a role in the reduced nitrification rates seen in the MHPP and DCD treatments.
The last line of evidence for a direct effect of MHPP was provided by the analysis of amoA gene copy number. Determination of amoA gene copy number showed that the 7 day incubation with a high dose of MHPP did not increase copy number relative to the number determined in the pre-incubation bulk soil, whereas all other treatments roughly doubled copy numbers of the bacterial amoA gene. Therefore, the observed inhibition of potential nitrification was probably due to a direct effect of compounds used on ammonia oxidizing bacteria.
Despite this overall agreement of the data with our hypothesis that MHPP acts as a biological nitrification inhibitor, some of our results need further discussions. In the 7 day soil incubation the low dose of MHPP did not reduce nitrification while a very strong effect was observed on potential nitrification activity. This apparent inconsistency between nitrification rate (net NO3 − pool) and potential nitrification activity of the low MHPP dose may be due to the different conditions existing for both methods: In the shaken soil slurry method, the MHPP reached the sites harboring ammonia-oxidizers due to an increased diffusion in the slurry. Although soil incubation for measuring net NO3 − pools was carried out under optimal soil moisture condition, some limitations in the diffusion could have affected the effectiveness of MHPP, especially in the low dose that may not have been as uniformly distributed.
In contrast to gene copy numbers for bacterial amoA that indicated an increase in bacterial populations over the 7 day incubation for all but the high MHPP treatment, archaeal amoA gene copy numbers remained mostly stable or decreased slightly (soil only and high MHPP). Little is known about the contribution of archaea to soil nitrification and about the interactions between archaea and bacteria during this process. We had decided to include the quantification of archaeal gene copy numbers in this study more out of interest than due to some specific hypothesis and results proved to be inconclusive.
Within the bacterial amoA gene copy number the increase in the DCD treatment after 7 days was unexpected since this treatment was comparable to the high MHPP dose in inhibiting nitrification.
It is well known that DCD is bacteriostatic rather than bactericidal (i.e., bacteria are only depressed or inhibited in their activities but not killed) (Amberger 1989; Trenkel 1997). In contrast it is possible that the effect of MHPP changes from bacteriostatic to bactericidal at higher concentrations. However, we do not have enough data to confirm this hypothesis.
Evidence that plants can inhibit nitrification in soils has been shown previously for crop plants other than sorghum. For example, a compound called brachialactone, isolated from root exudates of the grass Brachiaria humidicola, has been shown to repress potential nitrification rate in soil (Subbarao et al. 2009). Smits et al. (2010) found inhibition of nitrification in matgrass swards. Moreover, after addition of root exudates collected from Leymus racemosus (Lam.) Tzvelev, nitrification was repressed for more than 60 days (Subbarao et al. 2007b).
The BNI effect of MHPP appears to be more short-lived and dose dependent. NH4 + immobilization in the high MHPP treatment seen between days 3 and 7 (25 % of total N present at days 0 and 3 were not detected at day 7) suggests microbial decomposition of MHPP after 3 days. Under field conditions this loss of activity would be offset by a continuous or pulsed release of MHPP throughout the growth period. However, whether the release of MHPP and other potential nitrification inhibitors by sorghum is indeed continuous or peaks at certain growth stages remains to be determined. Zakir et al. (2008) reported exudation rates of 10.8 mg MHPP per g root dry weight and day, based on collections in NH4Cl solution. We added 0.525 mg MHPP per 20 g soil in our study (0.105 mg MHPP per 20 g soil in the low MHPP dose), equal to 1/20 to 1/100 of daily exudation (per g root). Whether these doses resulted in MHPP concentrations that would represent rhizosphere concentrations in a sorghum field cannot be estimated with certainty. It is likely that only a fraction of the entire root system would be releasing MHPP, which would increase exudation rates in these active root zones. It is further unknown how far MHPP would diffuse out in to the soil, which in turn would determine the soil volume affected by MHPP. Given that exudation rates of 10.8 mg MHPP per g root dry weight (based on the entire root) and day are likely higher in younger root tissue, and that MHPP is accumulating over a few days, one may speculate that our low dose of MHPP resulted in lower ‘rhizosphere’ concentrations than may occur in a sorghum field and that the higher dose may be more realistic.
We had based our choice of MHPP doses in part on C equivalents typically used in incubation studies (Watkins et al. 2009; Shi et al. 2011). The high dose of MHPP (350 μg C * g−1 soil) is at the high end of such C additions, however, studies highlighted how allelochemicals can be released into the soil in unexpectedly large quantities. The allelochemical 8-hydroxyquinoline exuded from Centaurea diffusa roots has been detected in soil at concentrations between 200 and 600 μg C * g−1 soil (Vivanco et al. 2004). Concentration of catechin, a phenolic compound exuded from the roots of Centaurea maculosa, have varied from 0–650 μg C * g−1 soil over time (Perry et al. 2007), suggesting that secondary metabolites can be released in pulses.
With regard to any potential benefits of MHPP release on reduced nitrification under field conditions it has be kept in mind that sorghum root exudates are a mixture of compounds and that MHPP could affect nitrification synergistically with other exudates having BNI activity. For example, two compounds exuded by sorghum roots identified as sakuranetin and sorgoleone, identified in the hydrophilic (sakuranetin) and hydrophobic (sorgoleone) phases of sorghum root exudates, have been shown to inhibit Nitrosomonas activity in a culture-bioassay to a higher degree than MHPP (Subbarao et al. 2012a). However, when sakuranetin was added into the soil, its BNI function was completely lost showing no effects in reducing soil nitrification.
It will require several further studies to unravel the potentially complex spatial and temporal interactions between multiple BNI compounds in sorghum root exudates.
In this study we showed for the first time that a compound exuded by roots of sorghum, MHPP, can act as biological inhibitor of nitrification in soil, and that this is likely due to MHPP reducing potential nitrification activity and abundance of ammonia-oxidizing microorganisms. From the confirmation of MHPP as a biological nitrification inhibitor to applications in practical agriculture is a long way, however, our results may encourage further efforts to characterize BNI in crop plants, particularly with the aim of identifying genotypes with superior BNI capacity that could eventually be used in crop improvement (Pariasca-Tanaka et al. 2010). The development of such genotypes would represent a very cost-effective way to enhance fertilizer efficiency in farming while reducing some of the negative environmental effects associated with N fertilizer application.
References
Amberger A (1989) Research on dicyandiamide as a nitrification inhibitor and future outlook. Commun Soil Sci Plant Anal 20:1933–1955
Beman JM, Francis CA (2006) Diversity of ammonia-oxidizing archaea and bacteria in the sediments of a hypernutrified subtropical estuary: Bahia del Tobari, Mexico. Appl Environ Microbiol 72:7767–7777
Ceccherini MT, Ascher J, Pietramellara G, Mocali S, Viti C, Nannipieri P (2007) The effect of pharmaceutical waste-fungal biomass, treated to degrade DNA, on the composition of eubacterial and ammonia oxidizing populations of soil. Biol Fertil Soils 44:299–306
Einhellig FA, Souza IF (1992) Phytotoxicity of sorgoleone found in grain sorghum root exudates. J Chem Ecol 18:1–11
FAO (1974) Legend of the Soil Map of the World. UNESCO, Paris
Felske A, Holger RI, Wokerink A, Stackebrandt E, Akkermans ADL (1997) Ribosome analysis reveals prominent activity of an uncultured member of the class Actinobacteria in grassland soils. Microbiology 143:2983–2989
Hart SC, Stark JM, Davidson EA, Firestone MK (1994) Nitrogen mineralization, immobilization, and nitrification. In: Weaver RW, Angle JS, Bottomley BS (eds) Methods of Soil Analysis. Part 2. Microbiological and Biochemical Properties. Soil Science Society of America, Madison, pp 985–1018
Jones DL, Darrah PR (1996) Re-sorption of organic compounds by roots of Zea mays L. and its consequences in the rhizosphere. 3. Characteristics of sugar influx and efflux. Plant Soil 178:153–160
Lata JC, Degrange V, Raynaud X, Maron PA, Lensi R, Abbadie L (2004) Grass populations control nitrification in savanna soils. Funct Ecol 18:605–611
Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C (2006) Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806–809
Linn DM, Doran JW (1984) Effect of water-filled pore space on carbon dioxide and nitrous oxide production in tilled and non tilled soils. Soil Sci Soc Am J 48:1264–1272
Lueders T, Friedrich MW (2003) Evaluation of PCR amplification bias by terminal restriction fragment length polymorphism analysis of small-subunit rRNA and mcrA genes by using defined template mixtures of methanogenic pure cultures and soil DNA extracts. Appl Environ Microbiol 69:320–326
More DRE, Waid JS (1971) The influence of washing of living roots on nitrification. Soil Biol Biochem 3:69–83
Nübel U, Engelen B, Felske A et al (1996) Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol 178:5636–5643
Pariasca TJ, Nardi P, Wissuwa M (2010) Nitrification inhibition activity, a novel trait in root exudates of rice. AoB Plants. doi: 10.1093/aobpla/plq014
Park HD, Wells GF, Bae H, Criddle CS, Francis CA (2006) Occurrence of ammonia-oxidizing archaea in wastewater treatment plant bioreactors. Appl Environ Microbiol 72:5643–5647
Perry LG, Thelen GC, Ridenour WM, Callaway RM, Paschke MW et al (2007) Concentrations of the allelochemical (±)-catechin in Centaurea maculosa soils. J Chem Ecol 33:2337–2344
Prasad R, Power PJ (1995) Nitrification inhibitors for the agriculture health and environment. Adv Agron 54:233–281
Rice E, Pancholy SK (1972) Inhibition of nitrification by climax ecosystems. Am J Bot 59:1033–1040
Rice CW, Tiedje JM (1989) Regulation of nitrate assimilation by ammonium in soils and in isolated soil microorganisms. Soil Biol Biochem 21:597–602
Rotthauwe JH, Witzel K-P, Liesack W (1997) The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63:4704–4712
Schimel JP, Van Cleve K, Cates RG, Clausen TP, Reichardt PB (1996) Effects of balsam poplar (Populus balsamifera) tannins and low molecular weight phenolics on microbial activity in taiga floodplain soil: Implications for changes in N cycling during succession. Can J Bot 74:84–90
Schlesinger WH (2009) On the fate of anthropogenic nitrogen. Proc Natl Acad Sci 106:203–208
Shi S, Richardson AE, O’Callaghan M, DeAngelis KM, Jones EE, Stewart A, Firestone MK, Condron LM (2011) Effects of selected root exudate components on soil bacterial communitiesFEMS. Microbiol Ecol 77:600–610
Smits NAC, Hefting MM, Kamst-van Agterveld MP, Laanbroek HJ, Paalman AJ, Bobbink R (2010) Nitrification along a grassland gradient: Inhibition found in matgrass swards. Soil Biol Biochem 42:635–641
Subbarao GV, Ishikawa T, Ito O, Nakahara K, Wang HY, Berry WL (2006) A bioluminescence assay to detect nitrification inhibitors released from plant roots: a case study with Brachiaria humidicola. Plant Soil 288:101–112
Subbarao GV, Rondon M, Ito O, Ishikawa T, Rao IM, Nakahara K, Lascano C, Berry WL (2007a) Biological nitrification inhibition (BNI)—Is it a widespread phenomenon? Plant Soil 294:5–18
Subbarao GV, Ban T, Masahiro K, Ito O, Samejima H, Wang HY, Pearse SJ, Gopalakrishnan S, Nakahara K, Hossain AKMZ, Tsujimoto H, Berry WL (2007b). Can biological nitrification inhibition (BNI) genes from perennial Leymus racemosus (Triticeae) combat nitrification in wheat farming?
Subbarao GV, Nakahara K, Hurtado MP, Ono H, Moreta DE, Salcedo AF, Yoshihashi AT, Ishikawa T, Ishitani M, Ohnishi-Kameyama M, Yoshida M, Rondon M, Rao IM, Lascano CE, Berry WL, Ito O (2009) Evidence for biological nitrification inhibition in Brachiaria pastures. Proc Nat Acad Sci U S A 106:17302–17307
Subbarao GV, Nakahara K, Ishikawa T, Ono H, Yoshida M, Yoshihashi T, Zhu Y, Zakir HAKM, Deshpande SP, Hash CT, Sahrawat KL (2012a) Biological nitrification inhibition (BNI) activity in sorghum and its characterization. Plant Soil. doi 10.1007/s11104-012-1419-9
Subbarao GV, Sahrawat KL, Nakahara K, Ishikawa T, Kudo N, Kishii M, Rao IM, Hash CT, George TS, Srinivasa RP, Nardi P, Bonnett D, Berry W, Suenaga K, Ito O, Lata JC (2012b) Biological Nitrification Inhibition (BNI):—A Novel Strategy to Regulate Nitrification in Agricultural Systems. Advances in Agronomy Vol 114
Trenkel, M.E. (1997). Controlled-Release and Stabilized Fertilizers in Agriculture. International Fertilizer Industry Association
Treusch AH, Leininger S, Kletzin A, Schuster SC, Klenk HP, Schlepler C (2005) Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ Microbiol 7:1985–1995
Vivanco JM, Bais HP, Stermitz FR, Thelen G, Callaway R (2004) Biogeographical variation in community response to root allelochemistry: novel weapons and exotic invasion. Ecol Lett 7:285–292
Watkins AJ, Nicol GW, Shaw LJ (2009) Use of an artificial root to examine the influence of 8-hydroxyquinoline on soil microbial activity and bacterial community structure. Soil Biol Biochem 41:580–585
Wrage N, Velthof GL, Beusichem MLV, Oenema O (2001) Role of nitrifier denitrification in the production of nitrous oxide. Soil Biol Biochem 33:1723–1732
Zakir HAKM, Subbarao GV, Pearse SJ, Gopalakrishnan S, Ito O, Ishikawa T, Kawano N, Nakahara K, Yoshihashi T, Ono H, Yoshida M (2008) Detection, isolation and characterization of a root-exuded compound, methyl 3-(4-hydroxyphenyl) propionate, responsible for biological nitrification inhibition by sorghum (Sorghum bicolor). New Phytol 180:442–451
Zhang LM, Offre PR, He LZ, Verhamme DT, Nicol GW, Prosser JI (2010) Autotrophic ammonia oxidation by soil thaumarchaea. Proc Natl Acad Sci U S A 107:17240–17245
Acknowledgments
P. Nardi was funded by the Japan Society for Promotion of Science (JSPS) postdoctoral fellowship. We are grateful to Dr. M.T. Ceccherini (University of Firenze) for valuable discussions and critical comments on the manuscript. We also thank to Dr. G. Renella (University of Firenze) for kindly providing the soil used in this research and Taro Matsuda (JIRCAS) for technical support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Liz Shaw.
Rights and permissions
About this article
Cite this article
Nardi, P., Akutsu, M., Pariasca-Tanaka, J. et al. Effect of methyl 3-4-hydroxyphenyl propionate, a Sorghum root exudate, on N dynamic, potential nitrification activity and abundance of ammonia-oxidizing bacteria and archaea. Plant Soil 367, 627–637 (2013). https://doi.org/10.1007/s11104-012-1494-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-012-1494-y