Abstract
Aims
Potatoes have an inadequate rooting system for efficient acquisition of water and minerals and use disproportionate amounts of irrigation and fertilizer. This research determines whether significant variation in rooting characteristics of potato exists, which characters correlate with final yield and whether a simple screen for rooting traits could be developed.
Methods
Twenty-eight genotypes of Solanum tuberosum groups Tuberosum and Phureja were grown in the field; eight replicate blocks to final harvest, while entire root systems were excavated from four blocks. Root classes were categorised and measured. The same measurements were made on these genotypes in the glasshouse, 2 weeks post emergence.
Results
In the field, total root length varied from 40 m to 112 m per plant. Final yield was correlated negatively with basal root specific root length and weakly but positively with total root weight. Solanum tuberosum group Phureja genotypes had more numerous roots and proportionally more basal than stolon roots compared with Solanum tuberosum, group Tuberosum genotypes. There were significant correlations between glasshouse and field measurements.
Conclusions
Our data demonstrate that variability in rooting traits amongst commercially available potato genotypes exists and a robust glasshouse screen has been developed. By measuring potato roots as described in this study, it is now possible to assess rooting traits of large populations of potato genotypes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A major challenge for agriculture in the future is to feed a predicted world population of 9 billion by 2050. This challenge is made greater by an anticipated change in weather patterns, brought about by climate change, with rainfall and temperature predicted to be increasingly variable and drought more frequent in many food-insecure regions (Lobell et al. 2008; Schiermeier 2008). In the past, many widely-grown crop cultivars were selected under ad libitum conditions of nutrients and water, and their yields and relative performances could differ considerably if environmental or financial costs required changes in agricultural practices such as reduced irrigation, tillage or fertilizer use (Tilman et al. 2002; Sachs et al. 2010). The potential for improving water use efficiency through improving crop transpiration efficiency (Condon et al. 2004), and improving water and fertiliser use efficiency through improving root systems, is now widely recognised (Lynch and Ho 2005; Lynch 2007; Henry et al. 2009; Zhu et al. 2009; Gewin 2010) and has been heralded as the second green revolution (Lynch 2007).
Potatoes are the fourth largest crop in terms of production, and the world’s most important non-grain food crop. Potato crops are particularly sensitive to drought (Yuan et al. 2003), and consequently require a disproportionate amount of irrigation to maintain yield and tuber quality (Porter et al. 1999; Fabeiro et al. 2001). In the UK, agriculture consumes 4 % of total water withdrawals, but whilst potato production uses only 2.5 % of arable land, it consumes almost 50 % of the irrigation water applied to field crops (White et al. 2005a). Potato crops, with their shallow root systems (Porter et al. 1999), and poor ability to penetrate compacted soil (Gregory and Simmonds 1992) in addition to being drought sensitive, have poor nitrogen (N) (Zebarth et al. 2004) and phosphorus (P) utilization efficiency (Dechassa et al. 2003; White et al. 2005a; Hammond et al. 2011). Their requirement for large fertilizer applications in combination with increased irrigation, contributes to environmental degradation by leaching of N and runoff of P to watercourses (Zebarth et al. 2004; White et al. 2005a). Potato roots are estimated to recover < 70 % of broadcast N and < 10 % of broadcast P fertiliser (White et al. 2005a), and it has been suggested (White et al. 2005a) that a larger, deeper root system would improve this. The “ideotype” for a potato root system can be hypothesised to be soil water deficit responsive, transporting ABA to the shoot, and enabling efficient stomatal closure (Wilkinson and Hartung 2009), as well as being economical with carbon, by producing aerenchyma (Fan et al. 2003; Zhu et al. 2009) or increased specific root length (SRL = root length per unit root mass) (White et al. 2005a). For example, potato is known to respond to P deficit by increasing surface area using adventitious roots, root hairs (Dechassa et al. 2003), mycorrhizal associations (Niemira et al. 1995), and maximising root length density (longer root length per unit carbon invested) (Opena and Porter 1999). There is also evidence for exudation of organic acids, RNases, phosphatases and upregulation of Pi transporters, to aid in nutrient mobilisation and uptake (Dechassa and Schenk 2004; Zimmermann et al. 2004; White et al. 2005a, b). In addition, to these known traits of potato roots, they should be architecturally plastic to enable efficient foraging of patchy and locally depleted resources such as phosphorus (Ho et al. 2003) and nitrate (Dunbabin et al. 2003), as well as being long (to increase the area of soil explored (Lynch and Brown 2001)), deep (to access water (Watt et al. 2008)), and thin (high SA / vol. to facilitate uptake (Steudle 2000; Rewald et al. 2011)). The root system must also serve the function of protecting itself against pathogens and parasites (Bradshaw et al. 2009).
In the case of potato, plant breeding is a long-term process; it may take 50 years of multi-trait selection to combine, in a single cultivar, disease and pest resistance along with acceptable yields and processing qualities (Bradshaw et al. 2009), and it would therefore be beneficial to identify useful root traits in genotypes already valued for other qualities. Although root system architecture is complex and driven by a suite of quantitative trait loci (QTL) (Dorlodot et al. 2007), there is evidence for stable differences between cultivars, breeding lines and wild relatives (Iwama 2008) although little of the available biodiversity has been exploited (Bradshaw et al. 2006). Including group Andigena genotypes increases the genetic diversity in breeding tetraploids with tetrasomic inheritance such as S. tuberosum group Tuberosum (Bradshaw et al. 2006), while the complexity associated with breeding tetraploids is bypassed by including diploid groups such as group Phureja, and producing diploid crosses. In the search for improved performance in the reduced input systems of the future, it is important to assess genetically divergent but cultivated groups for desirable traits which could be combined with modern cultivars to improve yield and quality and exploit hybrid vigour (Bradshaw et al. 2006). With increased interest in using root traits as a selection criterion for improving yields (White et al. 2005a; Lynch 2007), little information about the variation of these traits in available germplasm and the difficulty of fast and accurate phenotyping (Richards et al. 2010; Zhu et al. 2011), our aim was to measure genotypic variation in root traits of a range of potato genotypes. Including measurements previously identified as informative for root system study such as the ratio of root weight to total plant weight (root weight ratio (RWR)) and the ratio of root length to root mass (specific root length (SRL)) as discussed by Hodge (2009), we aimed to measure the field grown root system in detail to observe whether root classes such as basal and stolon roots (Fig. 1g) differed between genotypes and whether any of these characters could predict final yield in the field. We included group Tuberosum genotypes and named cultivars as well as group Phureja and group Andigena genotypes, to assess the variation present in these groups. The same traits were measured after 2 weeks growth in sand in the glasshouse to identify consistent characters and to develop a simple glasshouse screen for root traits. This information will enable genotypes to be chosen on the basis of their root characteristics and better inform growers and breeding programs.
Materials and methods
Plant material
Following a pilot trial (Wishart et al. 2009), 28 potato genotypes were selected for a larger scale investigation. A range of genotypes, comprising cultivars and advanced breeding lines, was chosen (Table 1) and grown in the field for 10 weeks until tuber initiation. A further eight replicate blocks were grown until maturity for yield. The same materials were grown in the glasshouse in pots until 2 weeks post emergence. Genotypes with a similar time to tuber initiation were chosen to minimise differences resulting from the effects of tuber filling and all named cultivars are classified as “maincrop” by the British Potato Variety Database (http://varieties.potato.org.uk).
Field experiment
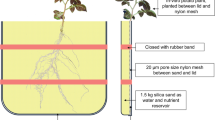
The field trial was carried out at Gourdie Farm, Liff, Dundee (56°28′N 03°03′W) on a free-draining, sandy silt loam, brown forest soil, association Balrownie, series Buchanyhill (parent material, colluvium; rock type, 590 undifferentiated sandstone). Twenty eight genotypes were selected; seven S. tuberosum L. group Tuberosum named cultivars, four group Tuberosum genotypes chosen for extreme carbon isotope ratios, two S. tuberosum group Tuberosum × S. tuberosum group Phureja diploid cross parents of a mapping population and an unnamed genotype (12601ab1) parent of an established mapping population, four genotypes derived from the S. tuberosum group Andigena, and ten S. tuberosum group Phureja genotypes selected from the Commonwealth Potato Collection (CPC) (Table 1). Three sets of parental lines from S. tuberosum group Tuberosum mapping populations were included (Table 1). They were grown in 4 randomized blocks, with individual plants separated from one another by double spacing (0.75 m between plants) in all directions relative to conventional planting practice and surrounded by a guard row (Fig. 1a and b). Eight further replicate blocks were grown to maturity and harvested for final yield. The plants were managed using standard practices with a total NPK fertiliser input of 1,050 kg / hectare (ha) delivering 147 kg / ha N, 147 kg / ha P2O5 and 220 kg / ha K2O. K supplement was added at 80 kg / ha of K2O to give a total K2O of 300 kg / ha. Herbicide treatments were applied throughout May and insecticide and fungicide (to control blight) through July and August. The four blocks used for trait measurements were grown for 10 weeks with destructive sampling at the tuber initiation stage. Individual plants, one plant per genotype per block, were excavated by a team of three or four people, using a large fork to loosen the plant roots and small hand forks to extract as much root as possible in-situ (Fig. 1b and c). Broken roots were also collected and the shoots and associated roots taken to the laboratory for counting and measuring. Above ground material was divided into plantlets by detaching stems and their associated roots from the mother tuber. It is accepted that water use efficiency can be estimated indirectly by measuring the carbon isotope composition of plant material and therefore a single leaflet from the first fully expanded leaf was freeze dried, ground in a ball mill and 1 mg samples were analysed for δ13C using continuous-flow isotope ratio mass spectrometry (CF-IRMS) (Europa Scientific Instruments, Crewe). Carbon isotopic natural abundances are reported as δsample = (Rsample − Rstandard) / Rstandard × 1000 0/00 where Rsample and Rstandard are the heavy / light isotope ratios of sample and standard. The leaves were counted and all shoot material dried at 70 °C for 7 days to obtain dry weight. Stolons were measured, and stolons and stolon nodes were counted. Roots from the stolon/stem junction and from nodes on stolons were collected and classified as “stolon roots” and “stolon node roots”, respectively (Fig. 1g). They were stored in 70 % alcohol before measuring lengths on a flatbed scanner using WinRHIZO software (Arsenault et al. 1995) after which plant material was dried at 70 °C and dry weight recorded. Similarly, roots that were not associated with a stolon, but present at the junction of the stem and mother tuber, were counted and classified as “basal roots” (Fig. 1g). These root lengths were also measured using WinRHIZO before drying and weighing. The remaining “stems” now devoid of stolons and tuber, any remaining mother tuber, and “new tubers” forming, were also dried at 70 °C and weighed separately. Root material unintentionally separated from the plant, either in the field or during processing, was classified as “extra root” and added to all “total root” measurements. The ratio of root weight to shoot weight and root weight ratio (RWR: the ratio of total root weight to total plant weight), were also calculated (see Table 2). Stem and stolon were considered “below ground” traits, as distinct from “shoot” which was the above ground biomass (plantlet and leaves) (Fig. 1g). Specific root length (SRL) (Eissenstat 1991) was calculated as root length per unit root weight (m g−1) for each root class (basal root or stolon root) and the number of roots per unit root weight (no g−1). Each block was harvested as quickly as possible, although the scale of the experiment resulted in each block taking 2–3 days to be sampled and processed. The fourth block, therefore, grew for 2 weeks longer than the first block.
Glasshouse experiment
The same 28 genotypes were grown in pots in a randomized block under glass, with five replicates per cultivar (Fig. 1d). Tubers were weighed and planted at 10 cm depth into dry, coarse, river sand watered and allowed to drain. Plants were grown for two weeks, post emergence, at a day temperature of 20 °C and a minimum night temperature of 15 °C in 18 L pots which were 30 cm diameter and 26 cm deep. Plants were fed modified Hoagland’s nutrient solution at 2 ml kg−1 media day−1 (Hoagland and Arnon 1950). After 2 weeks the plants were carefully removed from the pots (Fig. 1e). The roots were washed free of sand, and subjected to exactly the same measurements as were carried out on the field material (Fig. 1f; Table 2). Genotype LT1 failed to grow and could not therefore be included in comparisons with the field results.
Statistical analyses
Analyses were conducted in GENSTAT (VSN International, Heml Hempstead, UK) and data were subject to standard transformations (log10 and √x) when required. Summary statistics are included in Table 2.
To check for any bias in the measurements, ANOVAs were first run with “Recorder” as a factor. ANOVA with blocking, to account for each experimental replicate, was used to assess variations by “genotype” or “group” (only group Tuberosum and group Phureja were included as having sufficient genotypes to measure within group variation), (Table 1). Post hoc pairwise comparisons were run, with Bonferroni corrections, using the Tukey, or, when there were unequal sample sizes and variances, the Games-Howell procedure. Relationships within the data were assessed by correlation.
Correlations with final yield were calculated on mean data (n = 8 replicates) using the final yield measurements from the 8 replicate blocks. Partial correlations, controlling for mother tuber weight (glasshouse data) or block (field data), were performed on within-plant comparisons of traits; for example, basal root length with basal root weight and basal root number. Correlations were also performed on root measurements with above ground measurements.
Ratios were calculated to standardise root measurements. Length of a single root in each class (basal root or stolon root) was also calculated as the ratio of length per number of roots in that class. Relative proportions of root class as basal root/stolon root were also calculated. Harvest index was calculated from mean final yield for each genotype as a proportion of the total plant dry weight (Table 1). All general linear models were run in SPSS (PASW statistics 18).
For the statistical analyses of the glasshouse experiment genotypic differences were analysed by ANOVA including “tuber weight” as a covariate to adjust for variation in the size of the mother tuber.
Principal coordinate analysis was carried out on both above- and below-ground traits from both field and glasshouse data sets (Table 2 denoted). Principal components analysis using a correlation matrix was carried out on basal root, stolon root and total root counts and measurements (Table 2 denoted $) and general ANOVA was used to identify any effect of genotype or group relationships between the PC scores or latent vectors (coordinates), and plant traits were identified by correlation.
Results
Field experiment
Significant differences between genotypes were found for all the traits examined (20 of the 22 traits at p < 0.001; Table 2). The total number of roots ranged from 92 ± 43 (mean ± sd ) in Estima (group Tuberosum) to 500 ± 130 in Mayan Twilight (group Phureja) (Fig. 2a). The number of basal roots and stolon roots also differed significantly (Fig. 2a). Total root length varied significantly from Estima (group Tuberosum) (40.31 ± 23.15 m) to HB171/13 (progeny of a group Tuberosum × group Phureja diploid cross, T × P) which had the longest total root length (112.4 ± 31.40 m; Fig. 2b). The total basal and stolon root lengths also varied significantly between genotypes (Fig. 2b).
Variation in the total number of roots (F27,107 = 5.10, p < 0.001), number of basal roots (F27,107 = 5.99, p < 0.001) and stolon roots (F27,107 = 2.01, p = 0.002) for each genotype (mean and lsd. for total root No). Total number of roots = number of basal roots + number of stolon roots + extra roots (unclassified) (a). Variation in mean total root length (F27,107 = 4.32, p < 0.001), basal root length (F27,107 = 5.95, p < 0.001) and stolon root length (F27,107 = 1.98, p = 0.011) for each genotype (mean and least significant difference (lsd.) for total root length) Total root length = length of basal roots + length of stolon roots + length of extra roots (unclassified) (b)
Mean specific root length (SRL) (stolon root and basal root length per gram of dry weight) also varied significantly. Basal root SRL varied from 12 ± 5 m g−1 in Cara (group Tuberosum) to 35 ± 8 m g−1 in NTB 16 (group Andigena) (Fig. 3). DB257/28 (group Phureja) had the shortest average basal root length (0.1 ± 0.06 m) while NTB 16 (group Andigena) had the longest (0.74 ± 0.25 m). Individual stolon root lengths also varied from 0.13 ± 0.02 m in Mayan Twilight (group Phureja) to 0.5 ± 0.24 m in NTB 81 (group Andigena). There were significant differences among the genotypes in basal root diameter (F27,98 = 2.74, p < 0.001), with LT1and HT1 roots significantly thinner than genotypes 271/39 and 842P75 (group Phureja) and NTB145 (group Andigena) (p = 0.018, 0.049, 0.034 respectively). Across all genotypes, basal root diameter (0.60 ± 0.06 mm, n = 98) was highly significantly different (F(2,302) = 21.55; p < 0.001) from stolon root diameter (0.65 ± 0.05, n = 105) mm. Basal root and extra root did not differ in diameter (p = 0.998). As expected, many traits were found to correlate; for example, stolon root length with stolon root number and weight and similarly basal root length correlated with basal root number and weight.
Comparison of groups
Statistical comparisons were carried out on group Phureja and group Tuberosum which had sufficient representative genotypes (Table 2). While there was no significant difference in total root weight or total root length, there was a significant difference in total number of roots and in lengths and proportions of the different root classes (basal root vs stolon root). The group Phureja genotypes had more basal roots, and more stolon roots than the group Tuberosum genotypes (Fig. 4a). Group Phureja genotypes had greater stolon root SRL (F1,84 = 5.01, p = 0.02) and significantly shorter individual basal (F1,84 = 5.35, p = 0.028) and stolon roots (F1,84 = 5.42, p = 0.029) compared with the group Tuberosum genotypes (Fig. 4b & c). The group Phureja genotypes also had a greater proportion of basal root to stolon root than group Tuberosum genotypes (Fig. 4d). There was no significant difference among the taxonomic groups in total shoot weight (p = 0.932),, stem weight (p = 0.384), number of stolons (p = 0.067) or total plant weight (p = 0.915), although there was a significant difference in the number of leaves and plantlets as group Phureja genotypes had a significantly greater number of leaves (p = 0.001) and plantlets (p = 0.003) compared with the group Tuberosum genotypes and while the number of stolons did not differ, the group Phureja genotypes had significantly shorter stolons than the group Tuberosum genotypes (Table 2). The four group Andigena genotypes in this study had longer mean individual basal roots (72.64 ± 50.1 cm) compared with group Phureja (18.03 ± 10.96 cm), group Tuberosum (26.37 ± 17.34 cm) and T × P (26.61 ± 11.86 cm) and also the longest individual stolon roots (42.85 ± 20.04 cm). The two T × P crosses in the study had a greater number of roots compared with group Tuberosum (Fig. 4a), similar to the group Phureja parent.
(a) Variation in the total number of roots (F1,84 = 27.18, p < 0.001), number of basal roots (F1,84 = 58.07, p < 0.001) and stolon roots (F1,84 = 11.68, p = 0.001) for each group. Statistical test results shown for group Phureja and group Tuberosum only. (b) The average length of each basal root differed significantly between T and P genotypes (F (1,84) = 4.98, p = 0.028); and (c) the average length of each stolon root differed significantly between T and P genotypes (F (1,84) = 4.93, p = 0.029)1). (d) The proportion of basal roots to stolon roots was significantly different between T and P genotypes (F1,84 = 20.27, p < 0.001). (All graphs show means, error bars are standard deviations and letters denote significant differences (post hoc), GLM weighted by block) (Table 1 gives details of genotypes, groups and abbreviations)
Multivariate analysis
Principal coordinate analysis of above and below ground traits (traits included are marked by x in Table 2) separated the genotypes (p = 0.002), but not the groups, by “size” in the first dimension, which explained 46.1 % of the variation. However, in the second dimension, which explained 19.3 % of the variation, there was significant separation by genotype and also by group (both p < 0.001), and significant correlations between the latent vectors and many root traits (results not shown). Principal components analysis of basal root, stolon root and total root traits (traits included are marked $ in Table 2), identified the rooting characteristics separating the groups. There was clear separation in the second dimension, which accounted for 19.9 % of the variation, with total stolon root length (−0.413) and weight (−0.395) negatively associated with total basal root length (0.321), weight (0.236) and number (0.558) (Fig. 5a). The third dimension loadings separated the number of roots from their length and weight (data not shown). ANOVA of the principal component scores identified significant mean differences amongst genotypes and groups in the first three dimensions. The group Phureja genotype mean scores were significantly separated from the group Tuberosum genotypes in the first dimension by the total size of root and in the second dimension by their higher ratio of basal to stolon roots (Fig. 5b).
The Principal component (PC) latent vectors (loadings) from dimension 1 (explaining 54.67 % of the variation) and dimension 2 (explaining 19.4 % of the variation) of root traits across all genotypes labelled by trait from the PC analysis of root characters from the field. Basal root length (basal rt L) and weight (basal rt wt) were negatively associated with stolon root length (stolon rt L) and weight (stolon rt wt) (a). Mean PC scores from the first two dimension for each group (lsds for each dimension included). Dimension 1 (significant by genotype (F27,94 = 2.63, p < 0.00), and group (F3,94 = 4.44, p = 0.006) separated the genotypes on the basis of size (54.67 % of the variation explained). Dimension 2, explaining 19.94 % of the variation, separated the Group Tuberosum genotypes from Group Phureja genotypes due to the relative proportion of stolon roots vs many basal roots present in the Group Phureja genotypes (b)
Stable isotope data
Genotype explained 76 % of the variation in δ13C isotope discrimination (F27,104 = 8.82, p < 0.001). The genotype HB171/13 (T × P ) had the most negative measure of δ13C (−29.52 ± 0.88) and the group Tuberosum genotype LT1 had the least negative δ13C (−26.9 ± 0.31). Total number of roots (basal root and stolon root number) and many other traits were significantly correlated with δ13C (Table 2). Total root weight (Spearman’s r = 0.31, p = 0.002; Fig. 6a) and total number of roots (Spearman’s r = 0.35, p < 0.001) were more strongly correlated with - δ13C than total shoot weight (Spearman’s r = 0.25, p = 0.011). There were no significant correlations of - δ13C with numbers of plantlets or leaves or with mean final yield (Spearman’s r = 0.17, p = 0.4). The isotope measurements differed significantly amongst the groups (F3, 104 = 7.44, p < 0.001; Fig. 6b) with NTB group Andigena and group Tuberosum genotypes displaying the lower discrimination associated with increased water-use efficiency and significantly different from the group Phureja genotypes which were more negative δ13C.
A scatter plot of mean total root weight and mean - δ13C. There was a significant correlation between root weight and carbon isotope discrimination (− δ13C), Spearman’s r = 0.310 p = 0.001. The larger the root weight the greater the discrimination against the heavy isotope (a). Mean - δ13C differed significantly between group Phureja and group Tuberosum genotypes (F1,81 = 8.56, p = 0.004) (mean and standard deviations). Group Phureja genotypes discriminated against the heavy isotope and had a significantly more negative signature than group Tuberosum genotypes (Letters denote significant differences (post hoc) (Table 1 gives details of genotypes, groups and abbreviations) (b)
Yield
The group Tuberosum genotypes had significantly greater yields (3.5 ± 1.6 kg) than the group Phureja genotypes (2.1 ± 0.9 kg) (F1, 22 = 6.08, p = 0.023) (Fig. 7a & b). Correlations, between the traits (mean values) and the mean yield in the field for each genotype, identified those which were significantly correlated (Table 2); further correlations which were only found in group Tuberosum cultivars were identified “T” in column 1, (Table 2). The strongest correlation was a negative correlation with basal specific root length (SRL m g−1) (Spearman’s r = −0.65, p < 0.001; basal SRLs for each genotype are shown in Fig. 3). The stolon SRLs (m g−1) and the relative proportion of basal roots to stolon roots and the root weight to total plant weight (RWR) were also negatively correlated with final yield (Spearman’s r = − 0.43, p = 0.024; r = − 0.47, p = 0.013, Pearson’s r = − 0.57, p = 0.002, respectively). Total shoot weight, stolon weight, length and number, stolon root weight and total root weight were significantly positively correlated with final yield (Table 2). The best general linear model for yield (yield in the field = 0.062*total shoot weight – 0.001* basal root length - 0.002* basal root no g−1 + 2.07) explained 65 % of the variation in yield. In the model, shoot weight explained 33 %, basal root length 25 %, and the number of basal roots per gram weight (no g−1) 8 %, of the variation in final yield. If only group Tuberosum genotypes were included in the model, total shoot weight explained 67 % of the variation in yield while 61 % of the variation in final yield of the group Tuberosum genotypes was, in another model, associated negatively with the SRL of basal roots. The harvest index (HI) ranged from 99.1 % for Pentland Dell (group Tuberosum ) to 95.4 % for the group Phureja genotype 84.2P75 and the HI for the entire group Phureja genotypes, except DB257/28, were lower than for ten of the twelve group Tuberosum genotypes. Golden Wonder and HT2 had the lowest HI of the group Tuberosum genotypes (Table 1).
Final yield differed significantly among the genotypes (F27,222 = 24.16, p < 0.00, n = 8) (means and standard deviations with pairwise post hoc comparison shown for T × P genotype 99FT/5) (a). Final yield differed significantly amongst the groups (F3,222 = 19.24, p < 0.001). T genotypes (and T × P genotypes) produced higher yields than P genotypes (b)
Glasshouse experiment
Significant differences were found in all the traits (all p < 0.001; Table 2: Glasshouse). The total number of roots varied from 69 ± 24 in Sarpo Mira (group Tuberosum) to 535 ± 221 in DB271/39 (group Phureja). Inca Dawn (group Phureja) had the shortest total root length (11 ± 7 m) while genotype 71P10 (group Phureja) had the longest (57.89 ± 6.64 m). Inca Dawn (group Phureja) had the shortest total basal root length (0.5 ± 0.4 m) and the shortest total stolon root length (10.5 ± 7.2 m) while genotype 71P10 (group Phureja) had the longest total basal root length (23 ± 5.9 m) and the longest total stolon root length (33 ± 6.2 m). Inca Dawn (group Phureja) had the smallest total root weight (0.17 ± 0.08 g) and 71P10 (group Phureja) had the heaviest total root weight (1.6 ± 0.3 g). The general linear model (total root length = 3253.7* total root weight + 134.3* total shoot weight + 0.28*SRL stolon root + 0.22*SRL basal root - 1,780), explained 93 % of the variation in total root length. In this model 62 % of the variation was explained by total root weight, 11 % by basal root SRL, 10 % stolon root SRL, and 1 % shoot weight. The same parameters explained 85 % of the variation in root length in the field grown plants.
Partial correlations (controlling for mother tuber weight as these plants were only grown for 2 weeks post emergence), again identified the expected relationships between stolon root length and stolon root weight and number of stolon roots as well as basal root length with basal root weight and number of basal roots.
Comparison of glasshouse and field results
There was a significant correlation between glasshouse and field measurements for fifteen traits; for example, the mean total root length, total root weight and total number of roots were significantly correlated between glasshouse and field data (Pearson’s r = 0.4, p = 0.04; r = 0.45, p = 0.02; Spearman’s r = 0.64, p < 0.001 respectively; Table 2, Fig. 8). Eight of these significant correlations are presented in Fig. 8.
There was a significant correlation between plants grown in the field and the same genotypes grown in pots for two weeks for a the length of each stolon root (m) (rs = 0.54, p = 0.003), b the length of each basal root (m) (rs = 0.66, p < 0.001), c the number of basal roots (rs = 0.51, p = 0.008), d the number of stolon roots (rs = 0.53, p = 0.005), e the weight of stolons (g) (rs = 0.53, 0.005), f total root weight (g) (r = 0.45, p = 0.02), g total number of roots (rs = 0.64, p < 0.001), h total root length (m) (r = 0.4, p = 0.04)
There was no significant difference in the mean number of roots between the glasshouse and field data (t = 0.85, p = 0.402) but there was a significant difference in root length (t = 6.98, p < 0.001) and root weight (t = 10.95, p < 0.001). There was also no significant difference between the glasshouse and field data in plantlet number (median = 5 in both data sets; Table 2). In the glasshouse, after 2 weeks of growth the group Tuberosum genotypes had proportionally more basal roots to stolon roots compared with the group Phureja genotypes (0.66 ± 0.34 compared with 0.39 ± 0.23; p < 0.001) and significantly heavier basal root weight (0.37 ± 0.17 g compared with 0.26 ± 0.19 g; p = 0.02), in spite of the group Phureja genotypes having a greater number of basal roots (77.92 ± 54.05 compared with 50.38 ± 25.04; p = 0.009). The field data showed the opposite with the group Phureja genotypes having a greater proportion of basal root to stolon root (Fig. 4d).
A general linear model of the traits measured (mean values) in the glasshouse with mean yield from the field as the dependent variable, produced a highly significant positive relationship (yield in the field = 6.06*RMA (glasshouse) + 4.18*stolon weight (glasshouse) + 0.01*number of nodes (glasshouse)) explaining 60 % of the variation in yield. The genotypes with the greatest root to shoot ratio, (alone explaining 44 % of the variation), heaviest stolons (8 % of the variation) and most nodes (10 % of the variation) after 2 weeks growth in the glasshouse were the genotypes that produced the heaviest tuber yields in the field.
Discussion
We measured a large range of traits, including those thought to be important for resource capture (Watt et al. 2008; Richards et al. 2010). The data from both glasshouse and field experiments clearly demonstrated significant variation, in many of the traits, between genotypes and the differences found were consistent across different years (data not shown) and experiments. Variation in commercial varieties has been previously reported (Steckel and Gray 1979 (four cultivars); Stalham and Allen 2001 (sixteen cultivars)) and in spite of the narrow genetic base of cultivated potatoes (discussed in White et al. 2005a), significant variation was also found in the group Tuberosum genotypes in this study for many traits such as total root length (Estima and Pentland Dell the shortest, Cara the longest of the named cultivars), root number (Estima significantly less than Cara and Golden Wonder) and root weight (Pentland Dell the lightest, Cara the heaviest). Cara is known to have an unusually long period of growth (Stalham and Allen 2001) and had the greatest root length in our study (113 m). While previously reported root diameters for a range of potato cultivars and wild relatives ranged from 0.25 mm to 0.41 mm (Iwama 2008), in this study the minimum root diameter was 0.5 mm (measured by WinRHIZO ) and the finest roots may, therefore, have been poorly represented with the total root length being under-estimated. Although Iwama (2008) comments on the unreliability of root system comparisons due to large variation in values reported in the literature, his comparisons of root dry weight (g hill−1) of ten cultivars in the plough layer (1–5 g) agree with those reported here (median 3.2 g range 2.2–4.2 g).
We identified significant variation between the groups in this study; group Tuberosum (twelve cultivars) and group Phureja (ten cultivars), in particular. Only these groups had sufficient genotypes to confidently evaluate inter-group variation. Group Phureja genotypes had significantly more leaves and more roots of both classes than the group Tuberosum genotypes. A larger root system might be advantageous in many environments (Gregory et al. 1978; Kirkegaard et al. 2007; Watt et al. 2008) and during all stages of development (White et al. 2005a). However, the group Tuberosum genotypes had less total root length with fewer, longer roots and proportionally more stolon roots than the group Phureja genotypes. The group Phureja genotypes were characterised by many, short roots and a predominance of basal roots over stolon roots. Therefore, similarities in total root weight but highly significant differences in total root length (ANOVA group Table 2) for example, identified group differences in thickness, individual root lengths and overall structure of the root. Potato roots are poor at penetrating compacted soil (Gregory and Simmonds 1992) and in water stressed conditions where a high density of roots near the surface might result in root drying and reduced yields (White et al. 2005a) the numerous shorter roots and predominance of the deeper (below the mother tuber) basal roots of the group Phureja genotypes could be a useful trait in breeding for sustainable agriculture.
Basal roots (Fig. 1) might be important for water uptake and anchorage while stolon roots spreading from the stolons horizontally at higher levels might be more important in nutrient acquisition and tuber formation. In this field experiment, the two group Phureja × group Tuberosum hybrid genotypes were characterised by numerous roots (430.9 ± 67) as found in the group Phureja parent (317 ± 23) (Fig. 4a) but also long stolons, long individual basal roots (0.27 ± 0.04 m) (Fig. 4b) and stolon roots (0.31 ± 0.07 m) (Fig. 4c) and the high yields observed in the group Tuberosum parent (Fig. 7b). These characteristics were also found in the glasshouse experiment. Similar evidence for the incorporation of root characters from parental genotypes into offspring (despite the tetraploid complications (Bradshaw 1994)) was reported by Iwama (2008). The four group Andigena cultivars were characterised by extremely long but few roots. Individual basal roots were twice as long (0.64 ± 0.13 m) as in the group Tuberosum genotypes (Fig. 4b). The significant differences found between the groups emphasise the importance of mining groups other than group Tuberosum for complementary traits useful to breeders. The traits observed in the T × P crosses included in this study illustrate that trait inheritance is complex but through hybrid vigour and the chance inheritance of useful traits, as for example more numerous roots in our two group Phureja × group Tuberosum hybrid genotypes, desirable improvements may be achieved as suggested by Bradshaw et al. (2006). The relative influence of different root classes and rooting characteristics under differing water and nutrient regimes could be tested experimentally and, by using the range of traits found here, the potato would be a good model to explore the idea of a “designed root, tailored to the environment” discussed by Lynch (2007) and others (Gewin 2010).
δ13C discrimination is generally accepted as a useful indirect measure of water–use efficiency as it closely reflects the variation in CO2 concentration across the leaf. One route for improving leaf-level water-use efficiency is to improve transpirational efficiency (Condon et al. 2004) and the lower the discrimination against the heavy isotope (less negative δ13C ) the lower the stomatal conductance and potentially the better the water-use efficiency. The most negative δ13C was found in the genotype with the greatest total length of roots HB171/13 (T × P). The least discrimination (least negative δ13C) was found in one of the genotypes (group Tuberosum LT1) chosen for extreme δ13C in a previous survey and it was one of the lowest three genotypes for root number, weight, length and final yield. The greater incorporation of heavy isotope found in LT1 could be as a result of good water-use efficiency or it could be expected in small plants with smaller root systems, having lower photosynthetic rates per unit leaf area and lower stomatal conductance (Meinzer and Grantz 1990; Condon et al. 2004). The potatoes in this study, although rainfed, would not have been subject to water deficit and the lack of a relationship between δ13C discrimination and yield and the positive correlation with biomass also discussed by Condon et al. (2004) and Khazaei et al. (2009) in relation to grain crops, is likely to have resulted from high photosynthetic capacity in association with substantial and regular rainfall (Condon et al. 2004). The correlations with root traits were stronger than correlations with above ground traits supporting differences in stomatal conductance (Condon et al. 2004) driven by root signalling (Wilkinson and Hartung 2009). The isotope signature under these conditions reflected size of plant, however the three genotypes named in Fig. 6a (NTB 145, NTB 81 and HT1) display less discrimination than expected from their root size and one (HT1) was selected from a previous experiment as a genotype displaying an extreme outlying isotope signature.
Group Tuberosum cultivars have been selected in European environments for yield so many correlates between simple traits and final yield in the field might be anticipated. Group Phureja genotypes did not produce as large yields (Fig. 7a & b) or as many traits which correlated with final yield under these field conditions. The four largest yields were from group Tuberosum cultivars (Cara, Maris Piper, Sarpo Mira and Stirling). The group Phureja × group Tuberosum genotype 99FT/5 ranked fifth for yield and the top three group Tuberosum genotypes had significantly greater yields than the top yielding group Phureja (Mayan Gold) (Fig. 7a).
The importance of photosynthate in tuber bulking is well documented (White et al. 2005a; Levy and Veilleux 2007) and in this study we found a negative correlation between root weight to total plant weight ratio and final tuber yield across all groups (Table 2). Enhanced root respiratory demand for carbon has been suggested (Balemi and Schenk 2009) to account for inefficiencies in nitrogen and phosphorus utilisation in potato genotypes with an increased proportion of root dry matter. Shoot weight alone accounted for 37 % of the variation in yield (see GLM for yield above), and under these field conditions, 73.7 ± 11.3 % of the total plant dry weight was shoot weight. Root dry weight correlated weakly, but significantly and positively, with final yield (Table 2) as found in previous studies (Sattelmacher et al. 1990; Iwama 2008). The GLM also included a significant negative effect of basal root length and basal SRL associated with the higher yielding genotypes (Cara for example). Previous studies have reported greater water uptake capacity and higher photosynthetic and transpiration rates in plants with greater root to shoot ratio (Meinzer and Grantz 1990) while thinner roots; longer root length per unit root weight (increased SRL) are associated with increased nutrient interception (White et al. 2005a) indicating an advantage to plants possessing these traits. In this study the opposite was found as genotypes with the lowest root weight per unit shoot weight, and root weight as a proportion of total plant weight, (RWR), were the high yielding group Tuberosum genotypes Maris Piper and Sarpo Mira. Pentland Dell with the highest RWR, root weight per unit shoot weight and SRL relatively high compared with Cara, Fig. 3, had a final yield 40 % less than the cultivar Cara in spite of having the highest harvest index overall (Table 1). The RWR was highly significantly, but negatively correlated with final yield but not as highly correlated as, for example, the SRL of basal roots which was highly negatively correlated with final yield (r = − 0.65; p < 0.001; Table 2 correlations vs final yield). Genotypes LT1 and NTB16 had the largest basal root SRLs and Inca Dawn and LT1 had the largest stolon root SRLs and the lowest density root tissue. These genotypes had low yields under these conditions. The genotypes with low basal root SRLs, and the most dense basal root tissue, produced greater tuber yields under these replete (nutrients and water) conditions.
The significant differences in genotype SRLs (both basal and stolon roots) contrast with the WinRHIZO data for root diameter (few significant differences amongst genotypes for basal roots and none pairwise for stolon roots). Previous suggestions to explain such inconsistencies have been lignification of the hypodermis (Eissenstat 1991) or presence of aerenchyma (Zhu et al. 2009). It would be of interest to determine if there could be aerenchyma in genotype NTB145, for example, which had a significantly larger basal root diameter than LT1 but no difference in SRL (LT1 was one of the thinnest roots with the second highest SRL, Fig. 3). Zhu et al. (2009) reported enhanced drought tolerance in maize cultivars with more aerenchyma as, under water stress, more carbon can be allocated to root exploration. As Group Tuberosum genotypes have been selected for resistance to pathogens (Bradshaw et al. 2009), it could be that lignification is more extensive in genotypes with significantly lower SRLs but little change in root diameter.
Plants were grown for two weeks post emergence in the glasshouse in order to determine whether, after a relatively short growing time, useful measurements could be taken which correlated with any of those taken from field grown plants. It is well understood that measurements from pot grown plants (Mokany and Ash 2008) and from non-soil techniques (Zhu et al. 2011) may not provide a true representation of root characteristics in agriculturally relevant environments (Zhu et al. 2011). It has also been discussed by Hodge (2004) that for predicting performance in heterogeneous habitats, no single trait of root architecture has been found. We developed a systematic measuring system (Fig. 1g), over several smaller trials, to provide what we considered a useful range of root trait and plant measurements. We appreciated that intraspecific variation due to the inevitable lack of replication possible at this scale, suggested as necessary by Hulshof and Swenson (2010), along with growth plasticity, linked to the different experimental systems (glasshouse versus field), might result in too much variation to identify patterns in the data. However, the glasshouse results were consistent with those in the field for many of the traits we measured, and the best general linear models (GLMs) for root length included the same traits; root weight, shoot weight, basal SRL and stolon SRL, from both data sets with more variation in total root length being explained in the glasshouse data compared with the field experiment (95 % vs 85 %). In the first two weeks of growth the group Tuberosum genotypes put a greater investment into roots, basal in particular, with significantly greater RWR compared with the group Phureja genotypes. This resource allocation changed after tuberization (10 weeks) in the group Tuberosum genotypes with increased allocation to above ground parts relative to roots, stolons and stolon roots. The negative relationship shown clearly by the PCA (Fig. 5a) between resources to stolon roots and to basal roots was found after 10 weeks growth as well as in the glasshouse. Clearly the importance of carbon allocation to different root classes and to root or shoot will depend on the relative importance of different stresses whether nutrient or water depletion, soil penetration (Iwama 2008) or pest attack, or all of these.
Mean numbers of plantlets, basal roots and stolon roots were not significantly different between glasshouse and field experiments. Many other traits were correlated between glasshouse and field experiments such as total root length, total root weight and total root number (Table 2; correlations field vs pot and Fig. 8). Field measures of four of these traits (stolon length and weight, number of nodes and the total root weight) also correlated significantly with final yield. From a measure of, for example, dry root weight and number of leaves after two weeks growth in the glasshouse under the conditions used here (large pots) it would be possible to identify genotypes with many leaves and large roots, two of the main characteristics associated with large final yield in the field. Iwama’s study (2008) demonstrated that lines selected for greater dry root weight, for example, performed better under drought conditions. By applying the technique used in this study to available mapping populations, it should be possible to identify markers which segregate with specific rooting characteristics and overcome the problems of screens conducted in non-soil systems (discussed by Zhu et al. 2011). Future work testing known genotypes against particular stresses would be necessary to evaluate which traits would be most beneficial for improved resource and water-use efficiency.
Conclusions
In this paper we have developed a system for measuring below ground traits. We have identified root trait variation amongst closely related but distinct Solanum groups. We have also designed a relatively high throughput screen for these traits that could be used to identify QTL and markers for the traits thus speeding up the deployment of these traits into agriculturally relevant genotypes to improve resource capture efficiency in the field. Taken together this information has great potential for enabling identification of particular root traits, such as root number and individual root length and SRL, that could be manipulated by breeding to improve the sustainability of potato production.
Abbreviations
- ABA:
-
Abscisic acid
- DW:
-
Dry weight
- Glm:
-
General linear model
- HI:
-
Harvest index
- L:
-
Length
- No:
-
Number
- Rt(s):
-
Root(s)
- SRL:
-
Specific root length
- SA:
-
Surface area
- Vol:
-
Volume
- Wt:
-
Weight
References
Arsenault JL, Pouleur S, Messier C, Guay R (1995) WinRHIZO, a root-measuring system with a unique overlap correction method. Hortic Sci 30:906
Balemi T, Schenk MK (2009) Genotypic difference of potato in carbon budgeting as a mechanism of phosphorus utilization efficiency. Plant Soil 322:91–99
Bradshaw JE (1994) Quantitative genetics theory for tetrasomic inheritance. In: Bradshaw JE, Mackay GR (eds) Potato genetics. CAB International, Wallingford, pp 71–99
Bradshaw JE, Bryan EJ, Ramsay G (2006) Genetic resources (including wild and cultivated Solanum species) and progress in their utilisation in potato breeding. Potato Res 49:49–65
Bradshaw JE, Dale MFB, Mackay GR (2009) Improving the yield, processing quality and disease and pest resistance of potatoes by genotypic recurrent selection. Euphytica 170:215–227
Condon AG, Richards RA, Rebetzke GJ, Farquhar GD (2004) Breeding for high water-use efficiency. J Exp Bot 55:2447–2460
Dechassa N, Schenk MK (2004) Exudation of organic anions by roots of cabbage, carrot, and potato as influenced by environmental factors and plant age. J Plant Nutr Soil Sci 167:623–629
Dechassa N, Schenk MK, Claassen N, Steingrobe B (2003) Phosphorus efficiency of cabbage (Brassica oleraceae L. var. capitata), carrot (Daucus carota L.), and potato (Solanum tuberosum L.). Plant Soil 250:215–224
Dorlodot S, Forster B, Pages L, Price A, Tuberosa R, Draye X (2007) Root system architecture: opportunities and constraints for genetic improvement of crops. Trends Plant Sci 12:474–481
Dunbabin V, Diggle A, Rengel Z (2003) Is there an optimal root architecture for nitrate capture in leaching environments? Plant Cell Environ 26:835–844
Eissenstat D (1991) On the relationship between specific root length and the rate of root proliferation:a field study using citrus rootstocks. New Phytol 118:63–68
Fabeiro C, de Santa M, Olalla F, de Juan JA (2001) Yield and size of deficit irrigated potatoes. Agric Water Manag 48:255–266
Fan MS, Zhu JM, Richards C, Brown KM, Lynch JP (2003) Physiological roles for aerenchyma in phosphorus-stressed roots. Funct Plant Biol 30:493–506
Gewin V (2010) An underground revolution. Nature 466:552–553
Gregory PJ, Simmonds LP (1992) Water relations and growth in potatoes. In: Harris PM (ed) The potato crop: the scientific basis for improvement. Chapman and Hall, London, pp 214–246
Gregory PJ, McGowan M, Biscoe PV, Hunter B (1978) Water relations of winter wheat. I. Growth of the root system. J Agric Sci 91:91–102
Hammond JP, Broadley MR, Bowen HC, Spracklen WP, Hayden RM, White PJ (2011) Gene expression changes in phosphorus deficient potato (Solanum tuberosum L.) leaves and the potential for diagnostic expression markers. PLoS One 6(9):e24606
Henry A, Kleinman PJA, Lynch JP (2009) Phosphorus runoff from a phosphorus deficient soil under common bean (Phaseolus vulgaris L.) and soybean (Glycine max L.) genotypes with contrasting root architecture. Plant Soil 317:1–16
Ho MD, McCannon BC, Lynch JP (2003) Optimization modeling of plant root architecture for water and phosphorus acquisition. J Theor Biol 226:331–340
Hoagland DR, Arnon DI (1950) The water-culture method for growing plants without soil. Calif Agric Exp Station Circ 347:1–32
Hodge A (2004) The plastic plant: root responses to hetrogeneous supplies of nutrients. New Phytol 162:9–24
Hodge A (2009) Root decisions. Plant Cell Environ 32:628–640
Hulshof CM, Swenson NG (2010) Variation in leaf functional trait values within and across individuals and species: an example from a Costa Rican dry forest. Funct Ecol 24:217–223
Iwama K (2008) Physiology of the potato: New insights into root system and repercussions for crop management. Potato Res 51:333–353
Khazaei H, Mohammady S, Zaharieva M, Monneveux P (2009) Carbon isotope discrimination and water use efficiency in Iranian diploid, tetraploid and hexaploid wheats grown under well-watered conditions. Genet Resour Crop Evol 56:105–114
Kirkegaard JA, Lilley JM, Howe GN, Graham JM (2007) Impact of subsoil water use on wheat yield. Aust J Agric Res 58:303–315
Levy D, Veilleux RE (2007) Adaptation of potato to high temperatures and salinity—a review. Am J Potato Res 84:487–506
Lobell DB, Burke MB, Tebaldi C, Mastrandrea MD, Falcon WP, Naylor RL (2008) Prioritizing climate change adaptation needs for food security in 2030. Science 319:607–610
Lynch JP (2007) Roots of the second green revolution. Aust J Bot 55:493–512
Lynch JP, Brown KM (2001) Topsoil foraging—an architectural adaptation of plants to low phosphorus availability. Plant Soil 237:225–237
Lynch JP, Ho MD (2005) Rhizoeconomics: carbon costs of phosphorus acquisition. Plant Soil 269:45–56
Meinzer FC, Grantz DA (1990) Stomatal and hydraulic conductance in growing sugarcane; stomatal adjustment to water transport capacity. Plant Cell Environ 13:383–388
Mokany K, Ash J (2008) Are traits measured on pot grown plants representative of those in natural communities? J Veg Sci 19:119–126
Niemira BA, Safir GR, Hammerschmidt R, Bird GW (1995) Production of prenuclear minitubers of potato with peat-based arbuscular mycorrhizal fungal inoculum. Agron J 87:942–946
Opena GB, Porter GA (1999) Soil management and supplemental irrigation effects on potato: II. Root growth. Agron J 91:426–431
Porter GA, Opena GB, Bradbury WB, McBurnie JC, Sisson JA (1999) Soil management and supplemental irrigation effects on potato: I. Soil properties, tuber yield, and quality. Agron J 91:416–425
Rewald B, Ephrath JE, Rachmilevitch S (2011) A root is a root is a root? Water uptake rates of Citrus root orders. Plant Cell Environ 34:33–42
Richards RA, Rebetzke GJ, Watt M, Condon AG, Spielmeyer W, Dolferus R (2010) Breeding for improved water productivity in temperate cereals: phenotyping, quantitative trait loci, markers and the selection environment. Funct Plant Biol 37:85–97
Sachs J, Remans R, Smukler S, Winowiecki L, Andelman SJ, Cassman KG, Castle D, Defries R, Denning G, Fanzo J, Jackson LE, Leemans R, Lehmann J, Milder JC, Naeem S, Nziguheba G, Palm CA, Pingali PL, Reganold JP, Richter DD, Scherr SJ, Sircely J, Sullivan C, Tomich TP, Sanchez PA (2010) Monitoring the world’s agriculture. Nature 466:558–560
Sattelmacher B, Kuene R, Malagamba P, Moreno U (1990) Evaluation of tuber bearing Solanum species belonging to different ploidy levels for its yielding potential at low soil fertility. Plant Soil 129:227–233
Schiermeier Q (2008) A long dry summer. Nature 452:270–273
Stalham MA, Allen EJ (2001) Effect of variety, irrigation regime and planting date on depth, rate, duration and density of root growth in the potato (Solanum tuberosum) crop. J Agric Sci 137:251–270
Steckel JRA, Gray D (1979) Drought tolerance in potatoes. J Agric Sci 92:375–381
Steudle E (2000) Water uptake by plant roots: an integration of views. Plant Soil 226:45–56
Tilman D, Cassman KG, Matson PA, Naylor R, Polasky S (2002) Agricultural sustainability and intensive production practices. Nature 418:671–677
Watt M, Magee LJ, McCully ME (2008) Types, structure and potential for axial water flow in the deepest roots of field-grown cereals. New Phytol 178:135–146
White PJ, Broadley MR, Hammond JP, Thompson AJ (2005a) Optimising the potato root system for phosphorus and water acquisition in low-input growing systems. Asp Appl Biol 73:111–118
White PJ, Broadley MR, Greenwood DJ, Hammond JP (2005b) Proceedings of the International Fertiliser Society 568. Genetic modifications to improve phosphorus acquisition by roots. IFS, York, UK. ISBN 0853102058
Wilkinson S, Hartung W (2009) Food production: reducing water consumption by manipulating long-distance chemical signaling in plants. J Exp Bot 60:1885–1891
Wishart J, George TS, Brown LK, Thompson JA, Ramsay G, Bradshaw JE, White PJ, Gregory PJ (2009) Variation in rooting habit of potatoes: potential for improving resource capture. International Symposium “Root Research and Applications”, RootRAP 2–4 September, BOKU-Vienna Austria http://rootrap.boku.ac.at/fileadmin/files/RRcd/session02/oral/069.pdf
Yuan B, Nishiyama S, Kang Y (2003) Effects of different irrigation regimes on the growth and yield of drip-irrigated potato. Agric Water Manag 63:153–167
Zebarth BJ, Tai G, Tarn R, de Jong H, Milburn PH (2004) Nitrogen use efficiency characteristics of commercial potato cultivars. Can J Plant Sci 84:589–598
Zhu J, Brown KM, Lynch JP (2009) Root cortical aerenchyma improves drought tolerance of maize (Zea mays L.). Plant Cell Environ 33:740–749
Zhu J, Ingram PA, Benfey PN, Elich T (2011) From lab to field. New approaches to phenotyping root system architecture. Curr Opin Plant Biol 14:310–317
Zimmermann P, Regierer B, Kossmann J, Frossard E, Amrhein N, Bucher M (2004) Differential expression of three purple acid phosphatases from potato. Plant Biol 6:519–528
Acknowledgments
We thank all who helped in the field with digging up the potato root systems including Catherine Rose, Jackie Thompson, Lionel Dupuy, Gladys Wright, Ankush Prashar and Gaynor Mackenzie. We also thank the JHI field staff for setting-up and maintenance of the field trials. The research was also funded by the Scottish Government Work Package 1.7 “Profitable and sustainable agriculture” (2005–2011) and Work Package 3.3 “The soil, water and air interface and its response to climate and land use change” (2011–2016).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Ellis Hoffland.
Rights and permissions
About this article
Cite this article
Wishart, J., George, T.S., Brown, L.K. et al. Measuring variation in potato roots in both field and glasshouse: the search for useful yield predictors and a simple screen for root traits. Plant Soil 368, 231–249 (2013). https://doi.org/10.1007/s11104-012-1483-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-012-1483-1